Antibiotic Susceptibility and Resistance Genes in Oral Clinical Isolates of Prevotella intermedia, Prevotella nigrescens, and Prevotella melaninogenica
Abstract
1. Introduction
2. Results
2.1. Frequency of P. intermedia, P. nigrescens, and P. melaninogenica
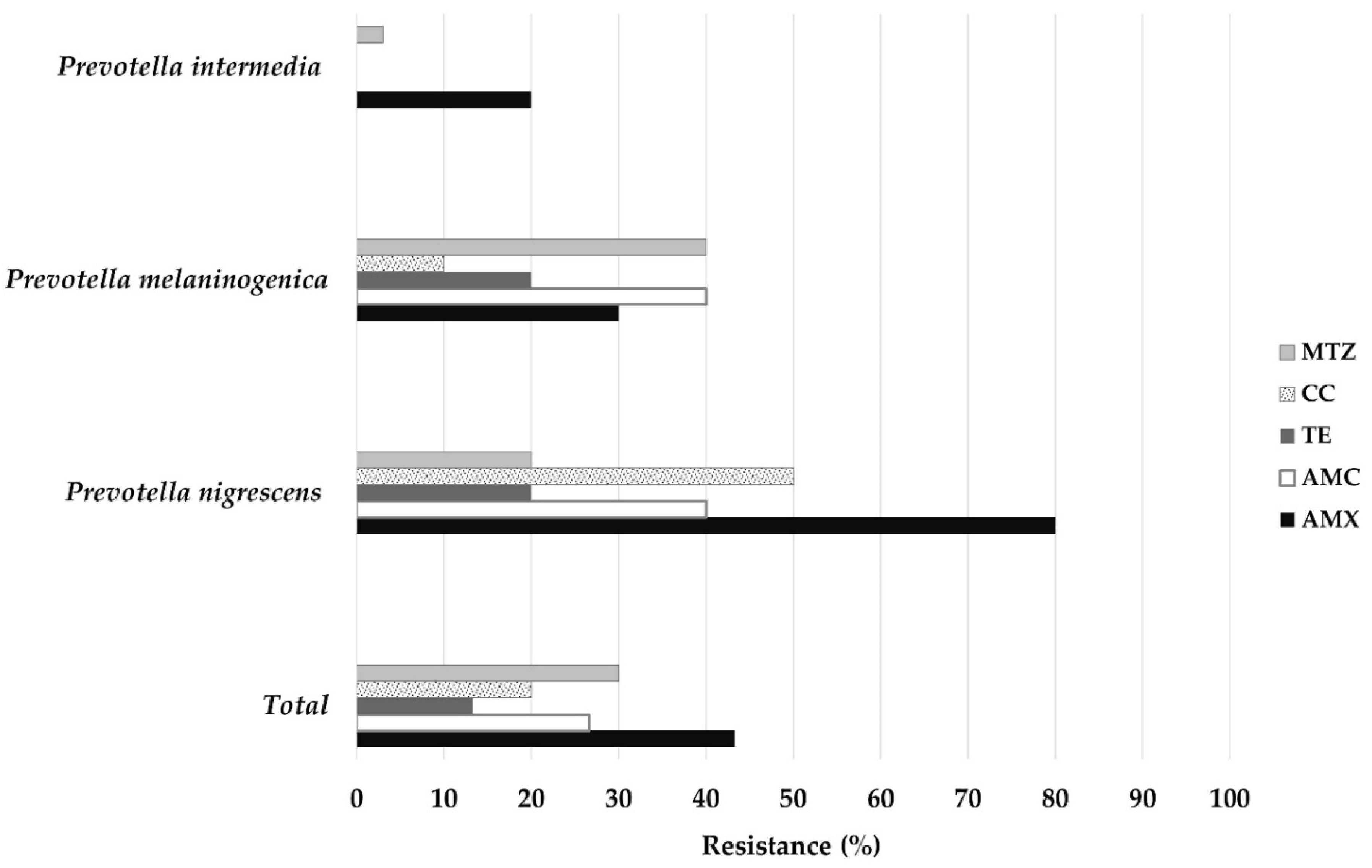
2.2. Evaluation of Antibiotic Susceptibility in Isolates of P. intermedia, P. nigrescens, and P. melaninogenica
2.3. Prevalence of Resistance Genes in P. intermedia, P. nigrescens, and P. melaninogenica
2.4. Genotype–Phenotype Relationship of P. intermedia, P. nigrescens, and P. melaninogenica Isolates
3. Discussion
4. Materials and Methods
4.1. Population and Samples
4.2. Microbiological Cultures and Species Identification
4.3. Species Identification and Detection of Resistance Genes
4.4. In Vitro Antibiotic Susceptibility Testing
4.5. Gene Detection
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krom, B.P.; Kidwai, S.; Ten Cate, J.M. Candida and other fungal species: Forgotten players of healthy oral microbiota. J. Dent. Res. 2014, 93, 445–551. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Chen, J.; He, J.; Miao, X.; Xu, M.; Wu, X.; Xu, B.; Yu, L.; Zhang, W. Antimicrobial Resistance and Prevalence of Resistance Genes of Obligate Anaerobes Isolated From Periodontal Abscesses. J. Periodontol. 2014, 85, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Lafaurie, G.I.; Contreras, A.; Barón, A.; Botero, J.; Mayorga-Fayad, I.; Jaramillo, A.; Giraldo, A.; González, F.; Mantilla, S.; Botero, A.; et al. Demographic, clinical, and microbial aspects of chronic and aggressive periodontitis in Colombia: A multicenter study. J. Periodontol. 2007, 78, 629–639. [Google Scholar] [CrossRef]
- Arredondo, A.; Blanc, V.; Mor, C.; Nart, J.; León, R. Azithromycin and erythromycin susceptibility and macrolide resistance genes in Prevotella from patients with periodontal disease. Oral Dis. 2019, 25, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Bancescu, G.; Didilescu, A.; Bancescu, A.; Bari, M. Antibiotic susceptibility of 33 Prevotella strains isolated from Romanian patients with abscesses in head and neck spaces. Anaerobe 2015, 35, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Dubreuil, L.; Veloo, A.C.; Sóki, J.; ESCMID Study Group for Anaerobic Infections (ESGAI). Correlation between antibiotic resistance and clinical outcome of anaerobic infections; mini-review. Anaerobe 2021, 72, 102463. [Google Scholar] [CrossRef] [PubMed]
- Veloo, A.C.; van Winkelhoff, A.J. Antibiotic susceptibility profiles of anaerobic pathogens in The Netherlands. Anaerobe 2015, 31, 19–24. [Google Scholar] [CrossRef]
- Arzese, A.R.; Tomasetig, L.; Botta, G.A. Detection of tetQ and ermF antibiotic resistance genes in Prevotella and Porphyromonas isolates from clinical specimens and resident microbiota of humans. J. Antimicrob. Chemother. 2000, 45, 577–582. [Google Scholar] [CrossRef]
- Alauzet, C.; Mory, F.; Teyssier, C.; Hallage, H.; Carlier, J.P.; Grollier, G.; Lozniewski, A. Metronidazole resistance in Prevotella spp. and description of a new nim gene in Prevotella baroniae. Antimicrob. Agents Chemother. 2010, 54, 60–64. [Google Scholar] [CrossRef][Green Version]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 26th ed.; CLSI Supplement M100S; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016; ISBN 1–56238–923–8. [Google Scholar]
- De Lima, B.R.; Nicoloso, G.F.; Fatturi-Parolo, C.C.; Ferreira, M.B.C.; Montagner, F.; Casagrande, L. Prevotella strains and lactamic resistance gene distribution in different oral environments of children with pulp necrosis. Int. Endod. J. 2018, 51, 1196–1204. [Google Scholar] [CrossRef]
- Veloo, A.C.M.; Baas, W.H.; Haan, F.J.; Coco, J.; Rossen, J.W. Prevalence of antimicrobial resistance genes in Bacteroides spp. and Prevotella spp. Dutch clinical isolates. Clin. Microbiol. Infect. 2019, 25, 1156.e9–1156.e13. [Google Scholar] [CrossRef] [PubMed]
- Lie, M.A.; van der Weijden, G.A.; Timmerman, M.F.; Loos, B.G.; van Steenbergen, T.J.; van der Velden, U. Occurrence of Prevotella intermedia and Prevotella nigrescens in relation to gingivitis and gingival health. J. Clin. Periodontol. 2001, 28, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Pianeta, R.; Iniesta, M.; Castillo, D.M.; Lafaurie, G.I.; Sanz, M.; Herrera, D. Characterization of the Subgingival Cultivable Microbiota in Patients with Different Stages of Periodontitis in Spain and Colombia. A Cross-Sectional Study. Microorganisms 2021, 9, 1940. [Google Scholar] [CrossRef] [PubMed]
- Toprak, N.U.; Akgul, O.; Sóki, J.; Soyletir, G.; Nagy, E. ESCMID Study Group for Anaerobic Infections (ESGAI). Detection of beta-lactamase production in clinical Prevotella species by MALDI-TOF MS method. Anaerobede 2020, 65, 102240. [Google Scholar] [CrossRef]
- Arredondo, A.; Blanc, V.; Mor, C.; Nart, J.; León, R. Resistance to β-lactams and distribution of β-lactam resistance genes in subgingival microbiota from Spanish patients with periodontitis. Clin. Oral Investig. 2020, 24, 4639–4648. [Google Scholar] [CrossRef]
- Binta, B.; Patel, M. Detection of cfxA2, cfxA3, and cfxA6 genes in beta-lactamase producing oral anaerobes. J. Appl. Oral Sci. 2016, 24, 142–147. [Google Scholar] [CrossRef]
- Webb, K.A.; Olagoke, O.; Baird, T.; Neill, J.; Pham, A.; Wells, T.J.; Ramsay, K.A.; Bell, S.C.; Sarovich, D.S.; Price, E.P. Genomic diversity and antimicrobial resistance of Prevotella species isolated from chronic lung disease airways. Microb. Genom. 2022, 8, 000754. [Google Scholar] [CrossRef]
- Kulik, E.M.; Lenkeit, K.; Chenaux, S.; Meyer, J. Antimicrobial susceptibility of periodontopathogenic bacteria. J. Antimicrob. Chemother. 2008, 61, 1087–1091. [Google Scholar] [CrossRef]
- Sanai, Y.; Persson, G.R.; Starr, J.R.; Luis, H.S.; Bernardo, M.; Leitao, J.; Roberts, M.C. Presence and antibiotic resistance of Porphyromonas gingivalis, Prevotella intermedia, and Prevotella nigrescens in children. J. Clin. Periodontol. 2002, 29, 929–934. [Google Scholar] [CrossRef]
- Ardila, C.M.; Granada, M.I.; Guzmán, I.C. Antibiotic resistance of subgingival species in chronic periodontitis patients. J. Perodont. Res. 2010, 45, 557–563. [Google Scholar] [CrossRef]
- Scornec, H.; Bellanger, X.; Guilloteau, H.; Groshenry, G.; Merlin, C. Inducibility of Tn916 conjugative transfer in Enterococcus faecalis by subinhibitory concentrations of ribosome-targeting antibiotics. J. Antimicrob. Chemother. 2017, 72, 2722–2728. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Canigia, L.; Cejas, D.; Gutkind, G.; Radice, M. Detection and genetic characterization of β-lactamases in Prevotella intermedia and Prevotella nigrescens isolated from oral cavity infections and peritonsillar abscesses. Anaerobe 2015, 33, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.; Sartelli, M.; Haque, S.Z. Dental Infection and Resistance—Global Health Consequences. Dent. J. 2019, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Mayorga-Fayad, I.; Lafaurie, G.; Contreras, A.; Castillo, D.M.; Barón, A.; del Rosario, A.M. Microflora subgingival en periodontitis crónica y agresiva en Bogotá, Colombia: Un acercamiento epidemiológico. Biomédica 2007, 27, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Ashimoto, A.; Chen, C.; Bakker, I.; Slots, J. Polymerase chain reaction detection of 8 putative periodontal pathogens in subgingival plaque of gingivitis and advanced periodontitis lesions. Oral Microbiol. Immunol. 1996, 11, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Fosse, T.; Madinier, I.; Hannoun, L.; Giraud-Morin, C.; Hitzig, C.; Charbit, Y.; Ourang, S. High prevalence of cfxA β-lactamase in aminopenicillin-resistant Prevotella strains isolated from periodontal pockets. Oral Microbiol. Immunol. 2002, 17, 85–88. [Google Scholar] [CrossRef]
- Ioannidis, I.; Sakellari, D.; Spala, A.; Arsenakis, M.; Konstantinidis, A. Prevalence of tetM, tetQ, nim and bla TEM genes in the oral cavities of Greek subjects: A pilot study. J. Clin. Periodontol. 2009, 36, 569–574. [Google Scholar] [CrossRef]
- Lacroix, J.M.; Walker, C.B. Detection and incidence of the tetracycline resistance determinant tet(M) in the microflora associated with adult periodontitis. J. Periodontol. 1995, 66, 102–108. [Google Scholar] [CrossRef]
- Lacroix, J.M.; Walker, C.B. Detection and prevalence of the tetracycline resistance determinant tetQ in the microbiota associated with adult periodontitis. Oral Microbiol. Immunol. 1996, 11, 282–288. [Google Scholar] [CrossRef]
- Reig, M.; Galan, J.C.; Fernando Baquero, F.; Perez, J.C. Macrolide Resistance in Peptostreptococcus spp. Mediated by ermTR: Possible Source of Macrolide-Lincosamide-Streptogramin, B. Resistance in Streptococcus pyogenes. Antimicrob. Agents Chemother. 2001, 45, 630–632. [Google Scholar] [CrossRef][Green Version]

| Healthy (%) | Gingivitis (%) | Periodontitis (%) | |
|---|---|---|---|
| P. melaninogenica | 40 | 30 | 30 |
| P. nigrescens | 10 | 40 | 50 |
| P. intermedia | 0 | 10 | 90 |
| Global Resistance | |||||
|---|---|---|---|---|---|
| Antibiotics | n | Range 1 (µg/mL) | MIC 50 2 (µg/mL) | MIC 90 3 (µg/mL) | R |
| n (%) | |||||
| AMX | 30 | <0.25–>64 | 0.5 | 32 | 13 (43.3) |
| AMC | 30 | <0.25–>64 | 1 | 16 | 8 (26.6) |
| TE | 30 | <0.25–>64 | 2 | 16 | 4 (13.3) |
| CC | 30 | <0.25–>64 | 0.25 | >64 | 6 (20) |
| MTZ | 30 | <0.25–>64 | 1 | >64 | 9 (30) |
| Prevotella intermedia | |||||
| Antibiotics | n | Range 1 (µg/mL) | MIC 50 2 (µg/mL) | MIC 90 3 (µg/mL) | R |
| n (%) | |||||
| AMX | 10 | <0.25–8 | <0.25 | 4 | 2 (20) |
| AMC | 10 | <0.25–2 | <0.25 | 1 | 0 |
| TE | 10 | <0.25–4 | <0.25 | 4 | 0 |
| CC | 10 | <0.25–4 | <0.25 | 1 | 0 |
| MTZ | 10 | <0.25–>64 | 1 | >64 | 3 (30) |
| Prevotella nigrescens | |||||
| Antibiotics | n | Range 1 (µg/mL) | MIC 50 2 (µg/mL) | MIC 90 3 (µg/mL) | R |
| n (%) | |||||
| AMX | 10 | <0.25–>64 | 16 | 64 | 8 (80) |
| AMC | 10 | 0.5–>64 | 4 | 64 | 4 (40) |
| TE | 10 | <0.25–16 | 2 | 16 | 2 (20) |
| CC | 10 | <0.25–>64 | 1 | >64 | 5 (50) |
| MTZ | 10 | <0.25–64 | 1 | 32 | 2 (20) |
| Prevotella melaninogenica | |||||
| Antibiotics | n | Range 1 (µg/mL) | MIC 50 2 (µg/mL) | MIC 90 3 (µg/mL) | R |
| n (%) | |||||
| AMX | 10 | <0.25–32 | 0.25 | 16 | 3 (30) |
| AMC | 10 | <0.25–64 | 2 | 32 | 4 (40) |
| TE | 10 | <0.5–>64 | 2 | 64 | 2 (20) |
| CC | 10 | <0.25-64 | 0.25 | 1 | 1 (10) |
| MTZ | 10 | <0.25–>64 | 2 | >64 | 4 (40) |
| Bacteria | n | cfxA F (%) | cfxA2 F (%) | blaTEM F (%) | tetM F (%) | tetQ F (%) | nimAB F (%) | nimAEFI F (%) | ermF (%) |
|---|---|---|---|---|---|---|---|---|---|
| P. intermedia | 10 | 0 | 0 | 2 (20) | 4 (40) | 4 (40) | 1 (10) | 0 | 0 |
| P. nigrescens | 10 | 4 (40) | 1 (10) | 4 (40) | 3 (30) | 7 (70) | 3 (30) | 1 (10) | 4 (40) |
| P. melaninogenica | 10 | 1 (10) | 4 (40) | 2 (20) | 4 (40) | 2 (20) | 1 (10) | 0 | 2 (20) |
| Total | 30 | 5 (16, 6) | 5 (16, 6) | 8 (26, 6) | 11 (36, 6) | 13 (43, 3) | 5 (16, 6) | 1 (3, 3) | 6 (20) |
| Gen | Sequence (5′→3′) | Amplicon Size (bp) | Reference |
|---|---|---|---|
| 16S—P. intermedia * | TTTGTTGGGGAGTAAAGCGGG TCAACATCTCTGTATCCTGCGT | 575 | [26] |
| 16S—P. nigrescens * | ATGAAACAAAGGTTTTCCGGTAAG CCCACGTCTCTGTGGGCTGCGA | 804 | [26] |
| 16S—P. melaninogenica * | TACAATGGAGAGTTTGATCC CGATCCTTGCGGTCACGGAC | 1453 | This study |
| cfxA ** | GCAAGTGCAGTTTAAGATT GCTTTAGTTTGCATTTTCATC | 934 | [27] |
| cfxA2 ** | CAAAGYGACAAYAATGCCTGCG TSACGAAGRCGGCWAT | 426 | [27] |
| BlaTEM † | ATGAGTATTCAACATTTCCG CCAATGCTTAATCAGTGAGG | 858 | [28] |
| tetM†† | GACACGCCAGGACATATGG TGCTTTCCTCTTGTTCGAG | 397 | [29] |
| tetQ ** | GGCTTCTACGACATCTATTA CATCAACATTTATCTCTCTG | 755 | [30] |
| ermF | TTTCGGGTCAGCACTTTACTA ACTTTCAGGACCTACCTCATA | 476 | [31] |
| nimAB†† | GGCTACAAGCAGCATGTC TGCATACTTTGCTCTTC | 377 | This study |
| nimAEFI†† | TGCATACTTTGCTCTTC ATGTTCAGAGAAATGCGGCG | 455 | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castillo, Y.; Delgadillo, N.A.; Neuta, Y.; Hernández, A.; Acevedo, T.; Cárdenas, E.; Montaño, A.; Lafaurie, G.I.; Castillo, D.M. Antibiotic Susceptibility and Resistance Genes in Oral Clinical Isolates of Prevotella intermedia, Prevotella nigrescens, and Prevotella melaninogenica. Antibiotics 2022, 11, 888. https://doi.org/10.3390/antibiotics11070888
Castillo Y, Delgadillo NA, Neuta Y, Hernández A, Acevedo T, Cárdenas E, Montaño A, Lafaurie GI, Castillo DM. Antibiotic Susceptibility and Resistance Genes in Oral Clinical Isolates of Prevotella intermedia, Prevotella nigrescens, and Prevotella melaninogenica. Antibiotics. 2022; 11(7):888. https://doi.org/10.3390/antibiotics11070888
Chicago/Turabian StyleCastillo, Yormaris, Nathaly Andrea Delgadillo, Yineth Neuta, Andrés Hernández, Tania Acevedo, Edwin Cárdenas, Andrea Montaño, Gloria Inés Lafaurie, and Diana Marcela Castillo. 2022. "Antibiotic Susceptibility and Resistance Genes in Oral Clinical Isolates of Prevotella intermedia, Prevotella nigrescens, and Prevotella melaninogenica" Antibiotics 11, no. 7: 888. https://doi.org/10.3390/antibiotics11070888
APA StyleCastillo, Y., Delgadillo, N. A., Neuta, Y., Hernández, A., Acevedo, T., Cárdenas, E., Montaño, A., Lafaurie, G. I., & Castillo, D. M. (2022). Antibiotic Susceptibility and Resistance Genes in Oral Clinical Isolates of Prevotella intermedia, Prevotella nigrescens, and Prevotella melaninogenica. Antibiotics, 11(7), 888. https://doi.org/10.3390/antibiotics11070888








