Abstract
The excessive use of antibiotics has triggered the appearance of new resistant strains, which is why great interest has been taken in the search for new bioactive compounds capable of overcoming this emergency in recent years. Massive sequencing tools have enabled the detection of new microorganisms that cannot be cultured in a laboratory, thus opening the door to the search for new biosynthetic genes. The great variety in oceanic environments in terms of pressure, salinity, temperature, and nutrients enables marine microorganisms to develop unique biochemical and physiological properties for their survival, enhancing the production of secondary metabolites that can vary from those produced by terrestrial microorganisms. We performed a search for type I PKS genes in metagenomes obtained from the marine sediments of the deep waters of the Gulf of Mexico using Hidden Markov Models. More than 2000 candidate genes were detected in the metagenomes that code for type I PKS domains, while biosynthetic pathways that may code for other secondary metabolites were also detected. Our research demonstrates the great potential use of the marine sediments of the Gulf of Mexico for identifying genes that code for new secondary metabolites.
1. Introduction
New antibiotics are urgently required to combat the emerging multi-drug resistant pathogens and new infectious agents [1]. In their natural environments, microorganisms produce a wide range of secondary metabolites with a variety of chemical structures [2]. As most of these microorganisms have never been cultured, identified, or classified, their great chemical richness remains unexplored [3]. However, this is changing as a result of the development of genomic techniques that do not depend on conventional cultivation, enabling the rapid progress of phylogenetic studies based on rDNA [4,5,6].
Secondary metabolites are produced by biosynthetic gene clusters, which are organized groups of two or more genes that encode a biosynthetic pathway to produce specialized metabolites [7]. Nonribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs) are two of the largest classes of biosynthetic gene clusters, encompassing most of the known antibiotics and antifungals [8]. With three types, I, II, and III, PKSs contain ketosynthase (KS) domains and a variety of other accessory domains, such as β-ketoacyl reductase (KR), dehydratase (DH), enoylreductase (ER), and methyltransferase (MT) [9,10]. Type I PKSs (PKS I) are multifunctional enzymes structurally organized into modules. An individual PKS enzyme can harbor one or multiple functional modules, each of which consist of several distinct active sites (domains) for each enzymatic step [10]. Type I PKSs can be further classified into modular or iterative classes, with the latter using the same domain many times, iteratively, to synthesize the polyketide. Modular PKSs are large multidomain enzymes in which each domain is used only once in the synthesis process [11,12].
Covering approximately 70% of the surface of the earth, the oceans are an invaluable source of new natural products [13]. With marine chemodiversity, one of the targets for the search for natural products as a source of new therapeutic drugs, multiple studies have been undertaken in this field to meet the growing demand for more effective antibiotics for combatting multiple diseases [14]. Analyzing the biodiversity of PKS I is an important tool for identifying new bioactive molecules capable of meeting the growing demand for more effective antibiotics. The study of these domains in marine sediments enables new PKS gene clusters to be identified, while ascertaining their abundance can facilitate the unlocking of their biosynthetic potential. The present study analyzes the main and accessory domains of PKS I in order to evaluate the potential of microbiomes from marine sediments of the Gulf of Mexico (GoM) to produce secondary metabolites.
2. Results
2.1. Metagenomic Reads Assembly and Coverage Analysis
Among the main problems encountered by the present study during the assembly of environmental samples were the low coverage and the formation of chimeras [15,16,17]. Assembling environmental samples is a complex task, despite the existence of multiple algorithms that can be used to minimize these problems. The search for biosynthetic genes can be an even more difficult process, as these genes generally contain repetitive domains that tend to be cut into multiple contigs [18]. We used the SPAdes genome assembler version 3.14.1 (metaSPAdes mode) [19,20] to assemble five metagenomes from marine sediments taken from the GoM, obtaining an average of 95,040 coding sequences from assemblies varying in size from 13.8 Mbp to 99.8 Mbp, and a relatively low N50. However, in all cases, N50 > 500 bp (~3.3× read length) (Table 1).

Table 1.
Results for the assembly of metagenomes from marine sediments of the GoM.
While the initial size of the assemblies was greater than 500 Mbp, after filtering with a contig cutoff >500 bp, the size decreased significantly, probably due to their high levels of fragmentation and low contiguity in them. The longest contigs corresponded to sample C13 and those with a size > 40 Kbp. However, these curated metagenomes still harbored more than 475,202 coding sequences with potential as novel bioactive compound pathways or functional elements. Raw data and a full quality assessment corresponding to the metagenome assemblies are shown in Supplementary Material S1.
Due to the drastic decrease in the contigs’ size, the coverage and diversity was evaluated based on the short reads, using the Nonpareil software [21,22], with the objective of assessing the fraction of the biomes represented in our data set. While the average coverage obtained from the short reads (which could represent the null model of diversity) was approximately 0.2 for all the metagenomes analyzed, the fraction of the diversity and richness captured in the assemblies is in line with the diversity indices estimated from the short reads (Table 2). This finding suggests that, although fragmented, the curated assemblies still capture a genomic space that effectively replicates the diversity contained in the short reads. The coverage, raw taxonomy profiles, and full diversity analysis are presented in Supplementary Material S2.

Table 2.
Diversity indicators estimated from the metagenomic reads (null diversity model) and from the metagenomic assemblies.
2.2. Screening PKS I Genes and Phylogenetic Analysis
The PKS I domains were identified by HMMer search using a predefined set of models (Supplementary Material S3). The search with Hidden Markov Models allowed to select 2066 candidate sequences coding for possible domains of PKS I in the sequences predicted by Prodigal (Supplementary Material S4). Of these candidate sequences, the most represented domain was ER, with 833 sequences, followed by the MT, KS, and KR domains, while the lowest values were obtained for the DH and TE domains. Due to the high similarity between PKS I sequences and fatty acid synthases (FAS I), we decided to complement the Hidden Markov Models with a phylogenetic study to confirm the right selection of the PKS I domains (Figure 1), using the sequences of the FAS I domain as the external group. FAS I sequences are available in Supplementary Material S5. Clustering in different clades for the domains that encode for PKS I and FAS I reaffirms the reliability of the method applied for selecting sequences that encode for biosynthetic genes.
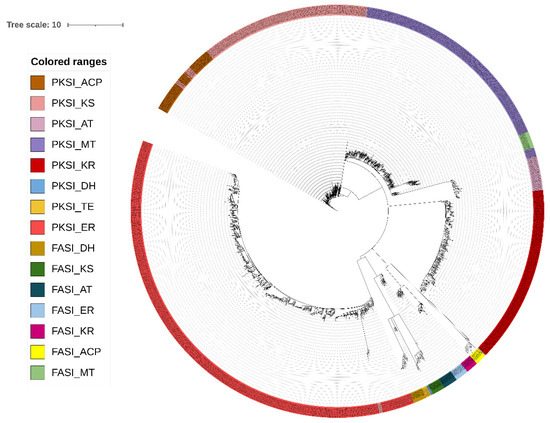
Figure 1.
Maximum likelihood tree for the PKS type I domains. PKS I: type I PKS; FASI: fatty acid synthases; AT: acyltransferase; ACP: acyl carrier proteins; ER: enoyl reductase: KR: ketoreductase; KS: ketosynthase; MT: methyltransferase; DH: dehydratase; TE: thioesterase. The raw Phylip format tree file is presented in Supplementary Material S6.
The normalization of the data and correspondence analysis indicate that the KS, KR, MT, ER, ACP, and AT domains are related (clustering close each other), while the TE and DH domains presented a minor relative abundance in our data (the TE domain was not found in the B7 and C13 metagenomes) (Figure 2). The distribution of these domains by taxonomic group (Figure 3) showed that the greatest diversity of PKS I domains belongs to the phyla Proteobacteria and Firmicutes, while, in the kingdom of Archaea the phylum Euryarchaeota is the most common carrier of these domains. The FOCUS taxonomic profiling results for contigs containing PKS I domains are shown in Supplementary Material S7.
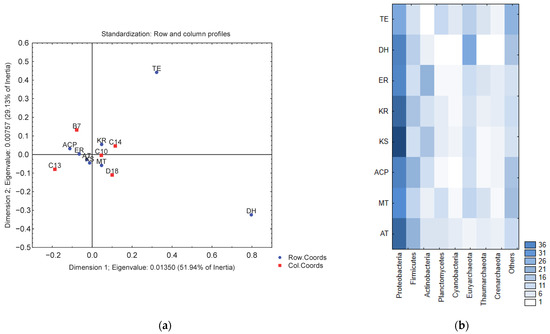
Figure 2.
(a) Correspondence analysis between PKS I domains and metagenomes (red squares: metagenomes; blue dots: PKS I domains). (b) Representation of PKS I domains by relevant taxonomic groups.
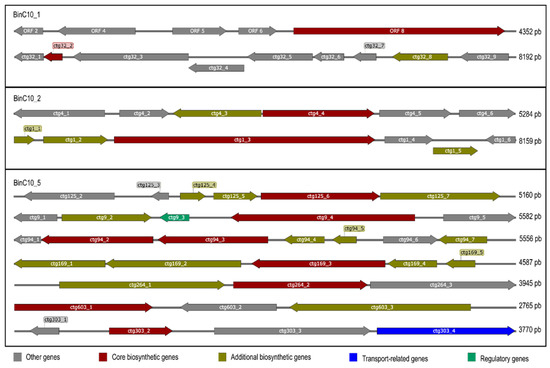
Figure 3.
Biosynthetic clusters detected by antiSMASH bacterial version 6.0 in the BinC10_1, C10_2, and C10_5. The arrows presented within each open reading frame indicate the direction of translation for each gene. The type of product detected by AntiSMASH is presented above each core biosynthetic gene. Contig number: ctg; RiPP: ribosomally synthesized and post-translationally modified peptides; RRE: RiPP recognition element; NRPS: nonribosomal peptide synthetases; T3PKS: type III PKS.
2.3. Marine Sediments of the GoM as a Source of Bioactive Compounds
The synthesis of secondary metabolites by bacteria helps to defend against predators and enables cell communication, among other functions, making secondary metabolites an excellent source of bioactive compounds for use in human therapies [23]. Deep-water marine sediments are a source of bioactive compounds that remain unexplored, due to the technical challenges of collecting the samples and the large area they occupy. The domains found in the five metagenomes obtained from sediments taken from the GoM were compared against the KEGG database to identify orthologues that may be involved in the synthesis of secondary metabolites. The results enabled the identification of 203 KO ortholog sequences (Table 3) involved in at least 14 metabolic pathways of bioactive synthesis (Supplementary Material S8).

Table 3.
Orthologues with possible biosynthetic function found when evaluating the PKS I domains present in the metagenomes of marine sediments of the GoM.
The most represented metabolic pathway was the biosynthesis of prodigiosin, an antimicrobial agent with little toxicity. Another pathway detected in the metagenomes obtained was the production of monoterpenoid, which consists of a ten-carbon backbone (two isoprene units) structure and can be divided into three subgroups: acyclic, monocyclic, and bicyclic [24]. Some monoterpenes have been described as presenting antimicrobial properties and painkilling effects [25]. Another compound detected was acarbose, an alpha-glucosidase inhibitor, which is described as a group of poorly absorbed antidiabetic agents [26]. Finally, domains related to the production of antibiotics, such as ansamycins, enediyne, vancomycin, streptomycin, and validamycin, were also present.
2.4. Exploring Biosynthetic Genes from Genome Bins
After binning was performed, the metagenome-assembled genomes (MAGs) were analyzed using the antiSMASH software, bacterial version 6.0, to find biosynthetic clusters. While most of the clusters obtained were incomplete, due to the short length of the contigs obtained, we were able to detect domains involved in the synthesis of the bioactive compounds from our samples.
The assembly of the C10 metagenome presented the highest quality (Table 1), in that it enabled the detection of several possible biosynthetic clusters. We were able to deconvolve three MAGs from the C10 metagenome: BinC10_1 (2 Mb in size and 73.74% completeness); BinC10_2 (700 Kb in size and 25% completeness); and BinC10_5 (2.9 Mb in size and 78.67% completeness) (Supplementary Material S9). Sequences related to ribosomally synthesized and post-translationally modified peptides (RiPPs) (Figure 3) were found in the BinC10_1, which was taxonomically consistent with the Desulfobacteraceae family (genomic Mash distance 0.07). The BinC10_2 contained domains that may code for NRPS and terpene, while BinC10_5 presented both the highest abundance of biosynthetic genes, which code for the RiPP recognition element (RRE), and the synthesis of terpene, ladderane, and type III PKS.
3. Discussion
Type I PKS produces a large family of medicinally important natural products. As PKS I multidomain proteins can be long and present a high degree of complexity, in most metagenomic sequences, the fragments of these proteins contain a simple domain [27]. The low number of multidomains found in our data could be due to the low level of metagenome coverage presented by the samples. However, by means of the use of Hidden Markov Models and phylogenetic relationships to facilitate the search for PKS, the present study was able to show the potential of these microbiomes to produce bioactive compounds from marine sediments. Each of the domains identified in the metagenomes found by the present study are likely to represent an entire PKS protein. Foerstner et al. (2008) built Hidden Markov Models to find PKS I domains, identifying PKS I domains from different metagenomes and annotating multiple proteins of unknown function in the UniRef database [27].
An average coverage (close to 0.2) consistent with complex and highly diverse communities (such as those found in marine sediments) was observed in our data set. The statistics obtained by the present study coincide with metagenomic observations obtained by other studies in complex environmental samples, such as soil, tropical forest, or seawater. Said studies obtained coverage levels that were always < 40%, which coincides with the greater level of diversity found in their samples than in other biomes, such as animal host microbiomes or enriched communities whose coverage has been found to be >60% by similar sequencing efforts [22,28]. Therefore, although coverage in metagenomics is still an important feature to consider, this metric depends more on the nature of the biome sampled than the data size [28]. We argue that the complexity of the communities represented in metagenomes B7 to D18 may influence the coverage values obtained by the present study. Moreover, we evaluate the diversity and richness indices as estimators of the number of species present in the samples, their distribution, and its representativeness. In all cases, the diversity and richness indices captured for the assemblies were not significantly different from those estimated from the short reads (Table 2). This indicates that the information contained in the assemblies captures the information from the null model at the taxonomic level, although the assembly is not expected to always express the entire diversity space of the entire sample (see gray columns in Table 2, wherein the closer the ratio is to 1, the more representative the metagenomic assembly). Finally, the Nonpareil diversity index concurs with those obtained by other studies for marine and other sandy soil communities, with expected values ranging from 21 to 25 (http://enve-omics.ce.gatech.edu/nonpareil/faq (accessed on 25 May 2022): B7 = 23.17; C10 = 23.05; C13 = 23.03; C14 = 22.92; and D18 = 22.96). This finding also supports the conclusion that the genomic space captured in our data is representative of the type of biomes analyzed.
Degenerate primers are usually used to identify KS and ACP domains in biodiversity studies or for the identification of new biosynthetic clusters [29], which limits the information available on the rest of the domains, especially the DH and TE domains. The correspondence analysis conducted by the present study indicates a limited presence of said domains (DH and TE) in the sediments and a lower ratio of the remaining domains of interest (KS, ACP, AT, ER, KR, and MT). Foerstner et al. (2008) identified PKS I domains in six metagenomes and the UniRef database, finding that, of the total domains identified (22,106), only 6.7% and 2.4% corresponded to DH and TE, respectively. This finding reflects either the low abundance of these domains in bacterial biosynthetic clusters or the scarcity of information about said domains, which presents a challenge to being able to identify them more reliably [27].
Our results are consistent with the biodiversity studies carried out in the GoM, which have found that its sediments largely contain the phyla Proteobacteria, Firmicutes, Actinobacteria, Plantomycetes, and Cyanobacteria [30,31], which have a proven potential as producers of bioactive compounds [32,33,34]. However, a large number of natural products have been isolated in Actinobacteria [23,35], and more than half of the KS genes in the Uniprot database belong to Actinobacteria [36,37]. The genus Streptomyces continues to be the predominant source of new chemistries, with 167 new metabolites reported during 2018, representing >69% of the marine-sourced bacterial natural products [38], although in recent years marine bioactives have been reported in other species, such as Roseovarius tolerans [39], Rhodovulum sulfidophilum [40], and Aequorivita sp. [41]. It cannot be ruled out that the reported predominance of Streptomyces is due to the fact that they are culturable microorganisms and they are highly represented in annotation databases. The phylum found by the present study to present the highest number of PKS I domains is Proteobacteria, which is the most abundant phylum found in marine environments (with an abundance between 50% and 80%); however, very few bioactive compounds have been described in these microorganisms [23].
The taxonomic profiling carried out by our study found that Actinomycetales, Clostridiales, Planctomycetales, Rhizobiales, and Spirochaetales are the orders that present more than two PKS domains in their sequences. While Clostridiales presents limited natural products, the genomic analysis conducted on these strict anaerobes shows the presence of natural product biosynthetic gene clusters that can code for entirely new products [42]. Graça et al. (2016) evaluated the genome of 40 taxa of Planctomycetales isolated from macroalgae obtained off the Portuguese coast, finding that 95% contained one or both of the bioactive genes PKS and NRPS; in addition, they also found antifungal and antibacterial activity in the bioactivity tests conducted on the samples [43]. The order Rhizobiales includes species of the genera Agrobacterium, Blastobacter, Mesorhizobium, and Ochrobactrum, which have been associated with bioactive compounds of marine origin, in contrast with Spirochaetales, for which no marine bioactive metabolites have been reported in the Comprehensive Marine Natural Products Database [44].
Prodigiosin is one of the secondary metabolites that are encoded in the metagenomes analyzed in this study. This red tripyrrole pigment, belonging to the prodiginines family, is produced by several bacterial genera, such as Serratia, Hahella, Streptomyces, Zooshikella, Vibrio, and Pseudomonas [45], and is known to have immunosuppressive, antifungal, antiviral, antimicrobial, anti-malarial, and anti-proliferative properties [46,47]. Genes encoding monoterpenoids were also detected in our data. The relatively low molecular weights of monoterpenoids and their intrinsic lipophilicity make these molecules suitable for administration via skin permeation and potential candidates for use in topical treatments, especially those used to relieve chronic pain [25]. Moreover, they have also been associated with various antimicrobial, hypotensive, anti-inflammatory, and antipruritic functions, among others [48]. Of the antidiabetic drugs, acarbose is associated with lesser gastrointestinal side effects than other alpha-glucosidase inhibitors and has been used as a single drug or in combination with other antidiabetic medications to control blood glucose levels in type 2 diabetic patients [26].
Among the antibiotic biosynthesis pathways found in the metagenomes analyzed are ansamycins, enediyne, vancomycin, streptomycin, and validamycin. Ansamycins are a group of antibiotics produced by several Actinomycetes strains and are molecules that have been proven to have very potent anticancer, antibacterial, and antiviral effects [49]. Enediyne natural products are among the most cytotoxic natural products ever discovered and are a promising source of next-generation antibody–drug conjugate payloads [50]. Vancomycin is enlisted as a drug of last resort for the treatment of resistant Gram-positive bacterial infections and is the first-choice antibiotic for the treatment of methicillin-resistant S. aureus infection [49].
The first of the class of aminoglycoside antibiotics to be discovered was the first antibiotic remedy for tuberculosis [49] and was derived from the Gram-positive bacteria of the genus Streptomyces [51]. Streptomycin is a broad-spectrum drug effective against both Gram-negative aerobic bacteria and staphylococci. Another aminoglycoside compound is validamycin [52], which has been used to control the rice sheath blight caused by Rhizoctonia solani for over 50 years in China, with no validamycin-resistant isolates reported in the field [52,53].
The search for biosynthetic clusters in the metagenome bins found the presence of RiPPs. Research conducted in the 20th century identified many classes of natural products, with four groups being particularly prevalent (terpenoids, alkaloids, polyketides, and non-ribosomal peptides), while, more recently, RiPPs have also been described [54]. RiPP biosynthesis starts with the ribosomal synthesis of a linear precursor peptide, and many RiPP biosynthetic proteins recognize and bind their cognate precursor peptide via a domain known as the RiPP recognition element (RRE) [55,56]. The various RiPPs that present antibiotic activity are widely addressed by Hudson and Mitchell (2018) [57]. Sequences encoding for RiPPs and RRE were found in BinC10_1, which belongs to the Desulfobateraceae family and represents a new species (MASH distance 0.07) with an unexplored genomic context.
Several biosynthetic clusters have also been detected in BinC10_5: RRE, terpene, ladderane, and type III PKS. While the terpene biosynthesis pathway is well known to be present in many plant and fungi genomes, it was recently proposed that it is also widely distributed in bacterial genomes [58]. Largely found as constituents of essential oils, terpenes are mostly hydrocarbons [59], and have been associated with medicinal and therapeutic properties such as those harnessed for anti-inflammatory therapies and the treatment of malaria, bacterial infections, and cardiovascular diseases [60]. Ladderane is exclusively present in the membranes of anaerobic ammonia-oxidizing (anammox) bacteria [61], which are able to oxidize ammonia via nitrite reduction into nitrogen gas. This process takes place in a separate intracytoplasmic organelle called the anammoxosome, which presents a high concentration of ladderane lipids that makes the membrane less permeable and, thus, provide a tight barrier against diffusion. This is assumed to be an important feature for retaining toxic intermediates, such as hydrazine (N2H4) and hydroxylamine (NH2OH), within the anammoxosome [62]. The identification of BinC10_5 using the Mash software has shown that this genome probably pertains to the Candidatus Scalindua genus (Mash distance 0.2), a genus described from natural habitats, especially from marine sediments and oxygen minimum zones [61,63,64]. Candidatus is a taxonomic status for uncultured prokaryotic cells [63], therefore it is likely that many of the genes detected in the present study code for entirely new antimicrobials.
Previous studies show that geographic location, latitude, and pH are determining factors in the diversity of biosynthetic genes and environmental microbiomes [64,65,66]. The GoM is one of the marine ecosystems most affected by the uncontrolled extraction of hydrocarbons and countless oil spills [67], which is why its bacterial biodiversity is distinctive and largely influenced by environmental factors specific to the region, especially to the presence of hydrocarbons [68]. The natural conditions of the ocean (low temperatures, high salinity, and high pressure), together with the hydrocarbon contamination, favor the natural selection of polyextremophilic microorganisms, which have gained the attention of biotechnologists due to their metabolic diversity and their ability to produce secondary metabolites. The marine sediments of the GoM are undoubtedly an important source of secondary metabolites to consider if we want to obtain new antimicrobials to combat the emerging resistant strains.
4. Materials and Methods
4.1. Sampling Sites and DNA Sequencing
The sediments were collected onboard the R/V Justo Sierra (UNAM) during the MMF-01 oceanographic campaign from 25 February to 18 March 2016. Eighteen sampling sites were selected from the Perdido and Coatzacoalcos regions and equally distributed between the two. Sampling was undertaken at a seafloor depth ranging from 550 to 3400 m using a box core, with subsampling then directly obtained from the box core using sterile syringes inserted at a depth of up to 10 cm. Each subsample was frozen and stored immediately in liquid nitrogen onboard and kept at −80 °C when they arrived to the laboratory, until nucleic acid extraction could be performed. For this study the results of the 16S-rDNA sequencing (see [69]) correspond to one sample from the Perdido region (B7, 1200 m) and the rest from the Coatzacoalcos region (C10, 550 m; C13, 2500 m; C14, 3200 m; and D18, 1500 m).
DNA was extracted from three independent syringes from each sampling site using the DNeasy PowerSoil Kit (Qiagen®, Hilden, Germany) following the protocol provided by manufacturer, with some modifications. During the lysis step, 275 µL of phenol-chloroform-isoamyl alcohol (25:24:1) solution was added, followed by 5 min incubation at room temperature to increase cell lysis. The elution step was performed twice using 50 µL of elution buffer with the column incubated for 10 min at room temperature. All centrifuge steps were performed at 14,000× g. Eluted DNA was quantified via UV absorption analysis (NanoDrop 2000 Spectrometer, Thermo Fisher Scientific, Waltham, US) and the quality of the extracted DNA was verified on an agarose gel electrophoresis. DNA was stored at −20 °C until further analysis.
Paired-end metagenome sequencing was performed in a NovaSeq instrument (2 × 150 bp) at MR DNA (Molecular Research LP, Shallowater, TX, USA).
4.2. Quality Control and Assembly
The quality of the short reads (151 pb) was assessed using the FastQC software version v0.11.5 and low-quality bases (Q < 30) and adapters were removed using AfterQC version 0.9.6, which was configured with the paired-end mode and default options [70]. The reads were assembled via SPAdes genome assembler v3.14.1 (metaSPAdes mode)—only-assembler option [20]. After contigs assembly, only contigs with a length of more than 500 bp were retained (3.3× read length). The quality of the assembly was evaluated with Quast Version: 5.0.2 option Meta [71]. The coding sequences were obtained with PRODIGAL v2.6.3 [72]. The taxonomic profiling of the contigs was performed on FOCUS using the default reference database [73].
4.3. Taxonomy Profiles, Coverage, and Diversity Estimations
The five metagenomes reported in the present study were profiled from both the short reads (null model for diversity) and the curated assemblies using the FOCUS software [73] with a enriched custom database comprising 14,551 genomes retrieved from the Assembly Database on the National Center for Biotechnology Information (NCBI). The genomes had to pertain to the type material in the NCBI database, in order to satisfy the current nomenclature standards [74]. This new representative database is available at https://github.com/ayixon/RaPDTool (accessed on 25 May 2022). The relative abundance profiles using FOCUS were directly loaded in the web biodiversity calculator (https://alyoung.com/labs/biodiversity_calculator.html, accessed on 25 May 2022), to obtain the Shannon index, the Equitability index, and the Margalef Richness index. The metagenome coverage was estimated using the Nonpareil software version 3.401 [21,22], applying the kmer algorithm on one of read files (R1 sister). The coverage curves were generated with the R functions Nonpareil.curve and Nonpareil.set.
4.4. Hidden Markov Model Search and Phylogenetic Tree Reconstruction
The Hidden Markov Models for each of the PKS I domains (acyltransferase (AT), acyl carrier proteins (ACP), enoyl reductase (ER), ketoreductase (KR), ketosynthase (KS), methyltransferase (MT), dehydratase (DH), and thioesterase (TE)) were downloaded from the Pfam database, version 33.1 [75]. The models were used to find protein sequences containing these domains with HMMER 3.3.2 (hmmsearch with --cut_tc option “trusted cutoff (TC)”) [76]. Sequences with a score greater than 40 and an E-value below 10−4 were selected and aligned using MAFFT v7.453 [77]. The alignments were manually curated, and the phylogenetic tree was built with IQ-TREE, multicore version 1.6.12, with the best-fit model WAG + F + R5, which was chosen according to the Bayesian Information Criterion [78,79]. The tree visualization was conducted using the iTOL tool [80].
4.5. Bioactive Potential in Marine Sediments and Environmental Draft Genome Reconstruction
The Kofam_scan tool, version to 1.3.0 (https://github.com/takaram/kofam_scan, accessed on 25 May 2022) [81], was applied on the five marine metagenomes to search for orthologues that may be involved in the synthesis of secondary metabolites. Putative biosynthesis pathways were explored using Kegg Mapper Server [82]. Draft environmental genomes (bins) were also constructed from the five metagenomes independently using Metabat2 [83] and then surveyed using AntiSMASH, bacterial version 6.0 [84], in order to detect the gene clusters that code for bioactive compounds. The bins were cured with RefineM [85], while their quality was analyzed using CheckM workflow: checkm lineage_wf [86]. The bins were identified via genomic comparison against the Genome Taxonomy Database, release [87].
Supplementary Materials
Supplementary files can be found through the link: https://doi.org/10.6084/m9.figshare.19769023 (accessed on 16 May 2022). File S1: Assembly of the different marine sediment samples obtained from the GoM; File S2: Taxonomy profiles, coverage, and diversity estimations; File S3: Hidden Markov Models (HMM) used to identify PKS I domains in metagenomes; File S4: Domain sequences identified by HMM in metagenomes; File S5: Sequences of fatty acid synthase (FAS) domains used as control or outer group in phylogeny; File S6: Phylogenetic tree file: File S7: Taxonomic profiling results for contigs containing PKS I domains, File S8: Orthologous search results with Kofamscan, File S9: Metagenome-assembled genomes BinC10_1, BinC10_2, BinC10_5.
Author Contributions
Conceptualization, M.F.-L., A.S.-R. and A.L.-L.; Formal analysis, M.F.-L. and A.S.-R.; Funding acquisition, A.L.-L.; Investigation, M.F.-L., A.S.-R., C.B. and K.S.-C.; Methodology, M.F.-L., A.S.-R. and A.L.-L.; Project administration, A.L.-L.; Resources, R.B.L. and A.L.-L.; Supervision, A.L.-L.; Visualization, M.F.-L. and A.S.-R.; Writing–original draft, M.F.-L. and A.S.-R.; Writing—review and editing, R.B.L. and A.L.-L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Council of Science and Technology of Mexico—Mexican Ministry of Energy—Hydrocarbon Trust, project 201441. This is a contribution of the Gulf of Mexico Research Consortium (CIGoM).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in both the article and in the Supplementary Materials.
Acknowledgments
We would like to thank all the members of crew of the R/V Justo Sierra for their contribution to a successful MMF-01-MET-02 oceanographic campaign.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kadri, S.S. Key Takeaways from the U.S. CDC’s 2019 Antibiotic Resistance Threats Report for Frontline Providers. Crit. Care Med. 2020, 48, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Katz, L.; Baltz, R.H. Natural product discovery: Past, present, and future. J. Ind. Microbiol. Biotechnol. 2016, 43, 155–176. [Google Scholar] [CrossRef] [PubMed]
- DeLong, E.F. Marine microbial diversity: The tip of the iceberg. Trends Biotechnol. 1997, 15, 203–207. [Google Scholar] [CrossRef]
- Pace, N.R. A molecular view of microbial diversity and the biosphere. Science 1997, 276, 734–740. [Google Scholar] [CrossRef]
- Hugenholtz, P. Exploring prokaryotic diversity in the genomic era. Genome Biol. 2002, 3, reviews0003.1. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hugenholtz, P.; Goebel, B.M.; Pace, N.R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 1998, 180, 4765–4774. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; De Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef]
- Cragg, G.M.; Newman, D.J. Natural products: A continuing source of novel drug leads. Biochim. Biophys. Acta Gen. Subj. 2013, 1830, 3670–3695. [Google Scholar] [CrossRef]
- Wong, F.T.; Khosla, C. Combinatorial biosynthesis of polyketides-a perspective. Curr. Opin. Chem. Biol. 2012, 16, 117–123. [Google Scholar] [CrossRef]
- Du, L.; Lou, L. PKS and NRPS release mechanisms. Nat. Prod. Rep. 2009, 27, 255–278. [Google Scholar] [CrossRef]
- Cane, D.E.; Walsh, C.T.; Khosla, C. Harnessing the biosynthetic code: Combinations, permutations, and mutations. Science 1998, 282, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Lal, R.; Kumari, R.; Kaur, H.; Khanna, R.; Dhingra, N.; Tuteja, D. Regulation and manipulation of the gene clusters encoding type-I PKSs. Trends Biotechnol. 2000, 18, 264–274. [Google Scholar] [CrossRef]
- Molinski, T.F.; Dalisay, D.S.; Lievens, S.L.; Saludes, J.P. Drug development from marine natural products. Nat. Rev. Drug Discov. 2009, 8, 69–85. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Qin, L. Metagenomics-based drug discovery and marine microbial diversity. Trends Biotechnol. 2005, 23, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Mavromatis, K.; Ivanova, N.; Barry, K.; Shapiro, H.; Goltsman, E.; McHardy, A.C.; Rigoutsos, I.; Salamov, A.; Korzeniewski, F.; Land, M.; et al. Use of simulated data sets to evaluate the fidelity of metagenomic processing methods. Nat. Methods 2007, 4, 495–500. [Google Scholar] [CrossRef]
- Pignatelli, M.; Moya, A. Evaluating the fidelity of De Novo short read metagenomic assembly using simulated data. PLoS ONE 2011, 6, e19984. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Y.; Fuhrman, J.A.; Sun, F.; Zhu, S. Assessment of metagenomic assemblers based on hybrid reads of real and simulated metagenomic sequences. Brief. Bioinform. 2020, 21, 777–790. [Google Scholar] [CrossRef]
- Meleshko, D.; Mohimani, H.; Tracanna, V.; Hajirasouliha, I.; Medema, M.H.; Korobeynikov, A.; Pevzner, P.A. BiosyntheticSPAdes: Reconstructing biosynthetic gene clusters from assembly graphs. Genome Res. 2019, 29, 1352–1362. [Google Scholar] [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. MetaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Rodriguez-R, L.M.; Konstantinidis, K.T. Nonpareil: A redundancy-based approach to assess the level of coverage in metagenomic datasets. Bioinformatics 2014, 30, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-R, L.M.; Gunturu, S.; Tiedje, J.M.; Cole, J.R.; Konstantinidisa, K.T. Nonpareil 3: Fast Estimation of Metagenomic Coverage and Sequence Diversity. mSystems 2018, 3, e00039-18. [Google Scholar] [CrossRef]
- Bhatnagar, I.; Kim, S.K. Immense essence of excellence: Marine microbial bioactive compounds. Mar. Drugs 2010, 8, 2673–2701. [Google Scholar] [CrossRef] [PubMed]
- Ludwiczuk, A.; Skalicka-Woźniak, K.; Georgiev, M.I. Terpenoids. In Pharmacognosy: Fundamentals, Applications and Strategy; Badal, R.D.S., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 233–266. ISBN 9780128021040. [Google Scholar]
- Perri, F.; Coricello, A.; Adams, J.D. Monoterpenoids: The Next Frontier in the Treatment of Chronic Pain? J. Multidiscip. Sci. J. 2020, 3, 195–214. [Google Scholar] [CrossRef]
- Khalili, N.; Safavipour, A. Evaluation of the Effects of Acarbose on Weight and Metabolic, Inflammatory, and Cardiovascular Markers in Patients with Obesity and Overweight. Int. J. Prev. Med. 2020, 8, 140. [Google Scholar] [CrossRef] [PubMed]
- Foerstner, K.U.; Doerks, T.; Creevey, C.J.; Doerks, A.; Bork, P. A computational screen for type I polyketide synthases in metagenomics shotgun data. PLoS ONE 2008, 3, 16–18. [Google Scholar] [CrossRef]
- Rodriguez-R, L.M.; Konstantinidis, K.T. Estimating coverage in metagenomic data sets and why it matters. ISME J. 2014, 8, 2349–2351. [Google Scholar] [CrossRef]
- Geers, A.U.; Buijs, Y.; Strube, M.L.; Gram, L.; Bentzon-Tilia, M. The natural product biosynthesis potential of the microbiomes of Earth—Bioprospecting for novel anti-microbial agents in the meta-omics era. Comput. Struct. Biotechnol. J. 2022, 20, 343–352. [Google Scholar] [CrossRef]
- Raggi, L.; García-Guevara, F.; Godoy-Lozano, E.E.; Martínez-Santana, A.; Escobar-Zepeda, A.; Gutierrez-Rios, R.M.; Loza, A.; Merino, E.; Sanchez-Flores, A.; Licea-Navarro, A.; et al. Metagenomic Profiling and Microbial Metabolic Potential of Perdido Fold Belt (NW) and Campeche Knolls (SE) in the Gulf of Mexico. Front. Microbiol. 2020, 11, 1825. [Google Scholar] [CrossRef]
- Ramírez, D.; Vega-Alvarado, L.; Taboada, B.; Estradas-Romero, A.; Soto, L.; Juárez, K. Bacterial diversity in surface sediments from the continental shelf and slope of the North West gulf of Mexico and the presence of hydrocarbon degrading bacteria. Mar. Pollut. Bull. 2020, 150, 110590. [Google Scholar] [CrossRef]
- Belova, S.E.; Saltykova, V.A.; Dedysh, S.N. Antimicrobial Activity of a Novel Freshwater Planctomycete Lacipirellula parvula PX69T. Microbiology 2020, 89, 503–509. [Google Scholar] [CrossRef]
- Letzel, A.C.; Pidot, S.J.; Hertweck, C. Genome mining for ribosomally synthesized and post-translationally modified peptides (RiPPs) in anaerobic bacteria. BMC Genom. 2014, 15, 983. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Obaidi, I.; Nagar, S.; Scalabrino, G.; Sheridan, H. The antiviral potential of algal-derived macromolecules. Curr. Res. Biotechnol. 2021, 3, 120–134. [Google Scholar] [CrossRef]
- Desriac, F.; Jégou, C.; Balnois, E.; Brillet, B.; Le Chevalier, P.; Fleury, Y. Antimicrobial peptides from marine proteobacteria. Mar. Drugs 2013, 11, 3632–3660. [Google Scholar] [CrossRef]
- Minowa, Y.; Araki, M.; Kanehisa, M. Comprehensive Analysis of Distinctive Polyketide and Nonribosomal Peptide Structural Motifs Encoded in Microbial Genomes. J. Mol. Biol. 2007, 368, 1500–1517. [Google Scholar] [CrossRef]
- Reddy, B.V.B.; Kallifidas, D.; Kim, J.H.; Charlop-Powers, Z.; Feng, Z.; Brady, S.F. Natural product biosynthetic gene diversity in geographically distinct soil microbiomes. Appl. Environ. Microbiol. 2012, 78, 3744–3752. [Google Scholar] [CrossRef]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2020, 37, 175–223. [Google Scholar] [CrossRef]
- Bruns, H.; Herrmann, J.; Müller, R.; Wang, H.; Wagner Döbler, I.; Schulz, S. Oxygenated N-Acyl Alanine Methyl Esters (NAMEs) from the Marine Bacterium Roseovarius tolerans EL-164. J. Nat. Prod. 2018, 81, 131–139. [Google Scholar] [CrossRef]
- Arashida, N.; Shimbo, K.; Terada, T.; Okimi, T.; Kikuchi, Y.; Hashiro, S.; Umekage, S.; Yasueda, H. Identification of novel long chain N-acylhomoserine lactones of chain length C20 from the marine phototrophic bacterium Rhodovulum sulfidophilum. Biosci. Biotechnol. Biochem. 2018, 82, 1683–1693. [Google Scholar] [CrossRef]
- Chianese, G.; Esposito, F.P.; Parrot, D.; Ingham, C.; de Pascale, D.; Tasdemir, D. Linear aminolipids with moderate antimicrobial activity from the antarctic gram-negative bacterium Aequorivita sp. Mar. Drugs 2018, 16, 187. [Google Scholar] [CrossRef]
- Li, J.S.; Barber, C.C.; Zhang, W. Natural products from anaerobes. J. Ind. Microbiol. Biotechnol. 2019, 46, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Graça, A.P.; Calisto, R.; Lage, O.M. Planctomycetes as novel source of bioactive molecules. Front. Microbiol. 2016, 7, 1241. [Google Scholar] [CrossRef] [PubMed]
- Lyu, C.; Chen, T.; Qiang, B.; Liu, N.; Wang, H.; Zhang, L.; Liu, Z. CMNPD: A comprehensive marine natural products database towards facilitating drug discovery from the ocean. Nucleic Acids Res. 2021, 49, D509–D515. [Google Scholar] [CrossRef] [PubMed]
- Darshan, N.; Manonmani, H.K. Prodigiosin and its potential applications. J. Food Sci. Technol. 2015, 52, 5393–5407. [Google Scholar] [CrossRef]
- Clements, T.; Rautenbach, M.; Ndlovu, T.; Khan, S.; Khan, W. A Metabolomics and Molecular Networking Approach to Elucidate the Structures of Secondary Metabolites Produced by Serratia marcescens Strains. Front. Chem. 2021, 9, 633870. [Google Scholar] [CrossRef]
- Bhagwat, A.; Padalia, U. Optimization of prodigiosin biosynthesis by Serratia marcescens using unconventional bioresources. J. Genet. Eng. Biotechnol. 2020, 18, 26. [Google Scholar] [CrossRef]
- Guimarães, A.G.; Quintans, J.S.S.; Quintans-Júnior, L.J. Monoterpenes with analgesic activity—A systematic review. Phyther. Res. 2012, 27, 1–15. [Google Scholar] [CrossRef]
- Vardanyan, R.; Hruby, V. Chapter 30—Antibiotics. In Synthesis of Best-Seller Drugs; Academic Press: Cambridge, MA, USA, 2016; pp. 573–643. ISBN 978-0-12-411492-0. [Google Scholar]
- Adhikari, A.; Shen, B.; Rader, C. Challenges and opportunities to develop enediyne natural products as payloads for antibody-drug conjugates. Antib. Ther. 2021, 4, 1–15. [Google Scholar] [CrossRef]
- Germovsek, E.; Barker, C.I.; Sharland, M. What do i need to know about aminoglycoside antibiotics? Arch. Dis. Child. Educ. Pract. Ed. 2017, 102, 89–93. [Google Scholar] [CrossRef]
- Bian, C.; Duan, Y.; Wang, J.; Xiu, Q.; Wang, J.; Hou, Y.; Song, X.; Zhou, M. Validamycin a induces broad-spectrum resistance involving salicylic acid and jasmonic acid/ethylene signaling pathways. Mol. Plant-Microbe Interact. 2020, 33, 1424–1437. [Google Scholar] [CrossRef]
- Bian, C.; Duan, Y.; Xiu, Q.; Wang, J.; Tao, X.; Zhou, M. Mechanism of validamycin A inhibiting DON biosynthesis and synergizing with DMI fungicides against Fusarium graminearum. Mol. Plant Pathol. 2021, 22, 769–785. [Google Scholar] [CrossRef] [PubMed]
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J.; et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013, 30, 108–160. [Google Scholar] [CrossRef] [PubMed]
- Burkhart, B.J.; Hudson, G.A.; Dunbar, K.L.; Mitchell, D.A. A prevalent peptide-binding domain guides ribosomal natural product biosynthesis. Nat. Chem. Biol. 2015, 11, 564–570. [Google Scholar] [CrossRef] [PubMed]
- Kloosterman, A.M.; Shelton, K.E.; van Wezel, G.P.; Medema, M.H.; Mitchell, D.A. RRE-Finder: A Genome-Mining Tool for Class-Independent RiPP Discovery. mSystems 2020, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Hudson, G.A.; Mitchell, D.A. RiPP antibiotics: Biosynthesis and engineering potential. Curr. Opin. Microbiol. 2018, 45, 61–69. [Google Scholar] [CrossRef]
- Cuadrat, R.R.C.; Ionescu, D.; Dávila, A.M.R.; Grossart, H.P. Recovering genomics clusters of secondary metabolites from lakes using genome-resolved metagenomics. Front. Microbiol. 2018, 9, 251. [Google Scholar] [CrossRef]
- Aldred, E.M.; Buck, C.; Vall, K. Terpenes. In Pharmacology; Churchill Livingstone: London, UK, 2009; pp. 167–174. ISBN 9780443068980. [Google Scholar]
- Cox-Georgian, D.; Ramadoss, N.; Dona, C.; Basu, C. Therapeutic and medicinal uses of terpenes. In Medicinal Plants; Joshee, N., Dhekney, S.P.P., Eds.; Springer: Berlin/Heidelberg, Germany, 2019; pp. 333–359. ISBN 9783030312695. [Google Scholar]
- van Niftrik, L.; Jetten, M.S.M. Anaerobic Ammonium-Oxidizing Bacteria: Unique Microorganisms with Exceptional Properties. Microbiol. Mol. Biol. Rev. 2012, 76, 585–596. [Google Scholar] [CrossRef]
- Antonsen, S.; Østby, R.B.; Stenstrøm, Y. Naturally Occurring Cyclobutanes: Their Biological Significance and Synthesis; Atta-ur-Rahman, Ed.; Elsevier: Amsterdam, The Netherlands, 2018; Volume 57, ISBN 9780444640574. [Google Scholar]
- Murray, R.G.; Stackebrandt, E. Taxonomic Note: Implementation of the Provisional Status Candidatus for Incompletely Described Procaryotes. Int. J. Syst. Bacteriol. 1995, 45, 186–187. [Google Scholar] [CrossRef]
- Rego, A.; Sousa, A.G.G.; Santos, J.P.; Pascoal, F.; Canário, J.; Leão, P.N.; Magalhães, C. Diversity of bacterial biosynthetic genes in maritime antarctica. Microorganisms 2020, 8, 279. [Google Scholar] [CrossRef]
- Charlop-Powers, Z.; Owen, J.G.; Reddy, B.V.B.; Ternei, M.; Guimaraes, D.O.; De Frias, U.A.; Pupo, M.T.; Seepe, P.; Feng, Z.; Brady, S.F. Global biogeographic sampling of bacterial secondary metabolism. eLife 2015, 2015, e05048. [Google Scholar] [CrossRef]
- Lemetre, C.; Maniko, J.; Charlop-Powers, Z.; Sparrow, B.; Lowe, A.J.; Brady, S.F. Bacterial natural product biosynthetic domain composition in soil correlates with changes in latitude on a continent-wide scale. Proc. Natl. Acad. Sci. USA 2017, 114, 11615–11620. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Salazar, J.; Loza, A.; Ornelas-Ocampo, K.; Gutierrez-Rios, R.M.; Pardo-López, L. Bacteria From the Southern Gulf of Mexico: Baseline, Diversity, Hydrocarbon-Degrading Potential and Future Applications. Front. Mar. Sci. 2021, 8, 625477. [Google Scholar] [CrossRef]
- Godoy-Lozano, E.E.; Escobar-Zepeda, A.; Raggi, L.; Merino, E.; Gutierrez-Rios, R.M.; Juarez, K.; Segovia, L.; Licea-Navarro, A.F.; Gracia, A.; Sanchez-Flores, A.; et al. Bacterial diversity and the geochemical landscape in the southwestern Gulf of Mexico. Front. Microbiol. 2018, 9, 2528. [Google Scholar] [CrossRef] [PubMed]
- Barcelos Santiago, C. Caracterización de las Comunidades Microbianas Presentes en los Sedimentos de Perdido y Coatzacoalcos del Golfo de México Mediante Análisis Metagenómicos; Centro de Investigación Científica y de Educación Superior de Ensenada: Ensenada, Mexico, 2018; 77p. [Google Scholar]
- Chen, S.; Huang, T.; Zhou, Y.; Han, Y.; Xu, M.; Gu, J. AfterQC: Automatic filtering, trimming, error removing and quality control for fastq data. BMC Bioinform. 2017, 18, 91–100. [Google Scholar] [CrossRef]
- Mikheenko, A.; Saveliev, V.; Gurevich, A. MetaQUAST: Evaluation of metagenome assemblies. Bioinformatics 2016, 32, 1088–1090. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Silva, G.G.Z.; Cuevas, D.A.; Dutilh, B.E.; Edwards, R.A. FOCUS: An alignment-free model to identify organisms in metagenomes using non-negative least squares. PeerJ 2014, 2, e425. [Google Scholar] [CrossRef]
- Federhen, S. Type material in the NCBI Taxonomy Database. Nucleic Acids Res. 2015, 43, D1086–D1098. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, 412–419. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, 29–37. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R.; Teeling, E. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Whelan, S.; Goldman, N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001, 18, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, 256–259. [Google Scholar] [CrossRef]
- Aramaki, T.; Blanc-Mathieu, R.; Endo, H.; Ohkubo, K.; Kanehisa, M.; Goto, S.; Ogata, H. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 2020, 36, 2251–2252. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef]
- Kang, D.D.; Li, F.; Kirton, E.; Thomas, A.; Egan, R.; An, H.; Wang, Z. MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 2019, 7, e7359. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. AntiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Parks, D.H.; Rinke, C.; Chuvochina, M.; Chaumeil, P.A.; Woodcroft, B.J.; Evans, P.N.; Hugenholtz, P.; Tyson, G.W. Recovery of nearly 8000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2017, 2, 1533–1542. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the genome taxonomy database. Bioinformatics 2020, 36, 1925–1927. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).