Rectal and Tracheal Carriage of Carbapenemase Genes and Class 1 and 2 Integrons in Patients in Neurosurgery Intensive Care Unit
Abstract
1. Introduction
2. Results
2.1. Patient’s Information
2.2. Antimicrobial Resistance Genes Detected in Clinical Samples
2.3. Bacterial Isolates and Asymptomatic Carriage
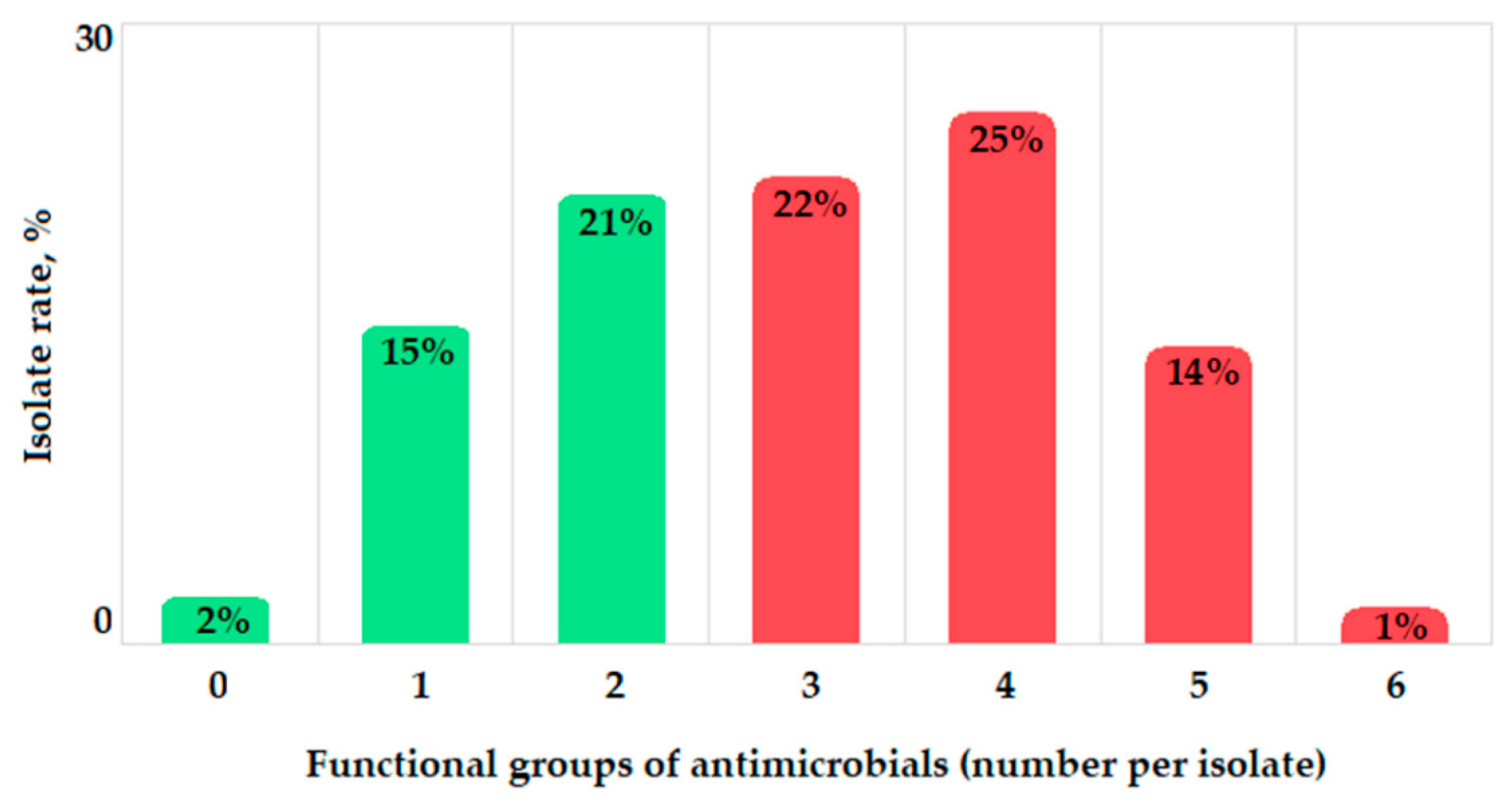
2.4. Antibacterial Resistance Phenotypes and Genotypes
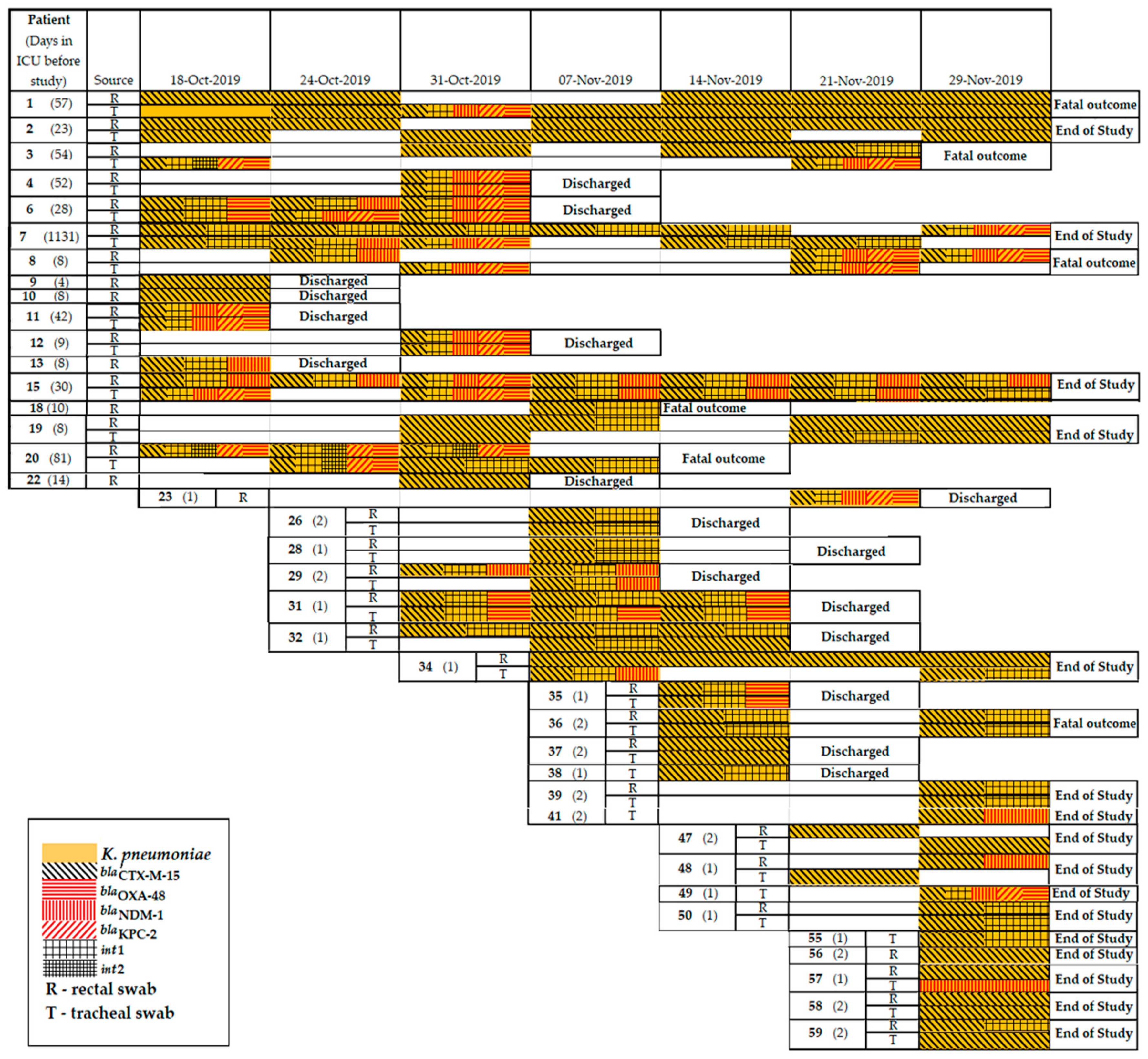
2.5. Resistomes of K. pneumoniae Clinical Isolates
2.6. Resistomes of E. coli Clinical Isolates
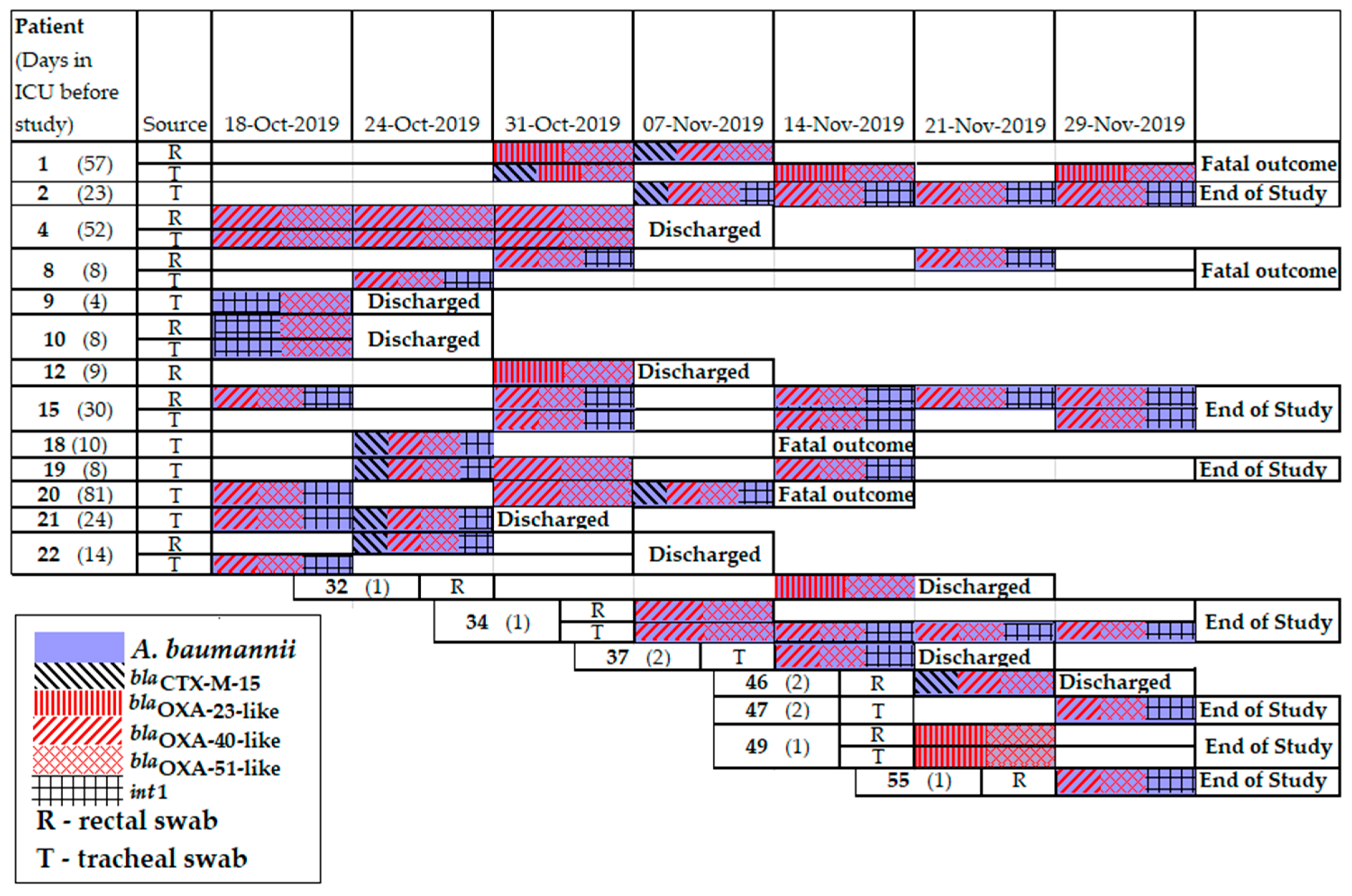
2.7. Resistomes of A. baumannii Clinical Isolates
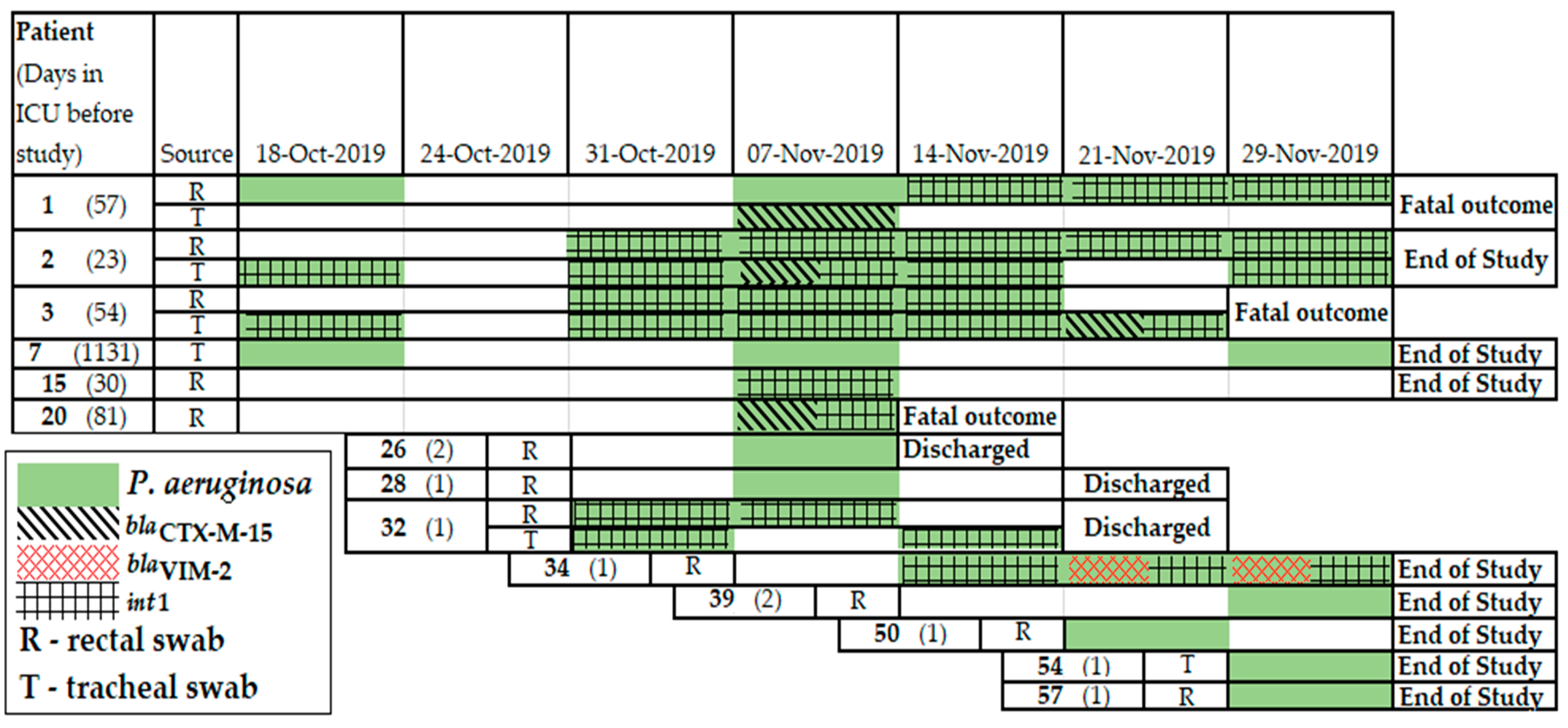
2.8. Resistomes of P. aeruginosa Clinical Isolates
3. Discussion
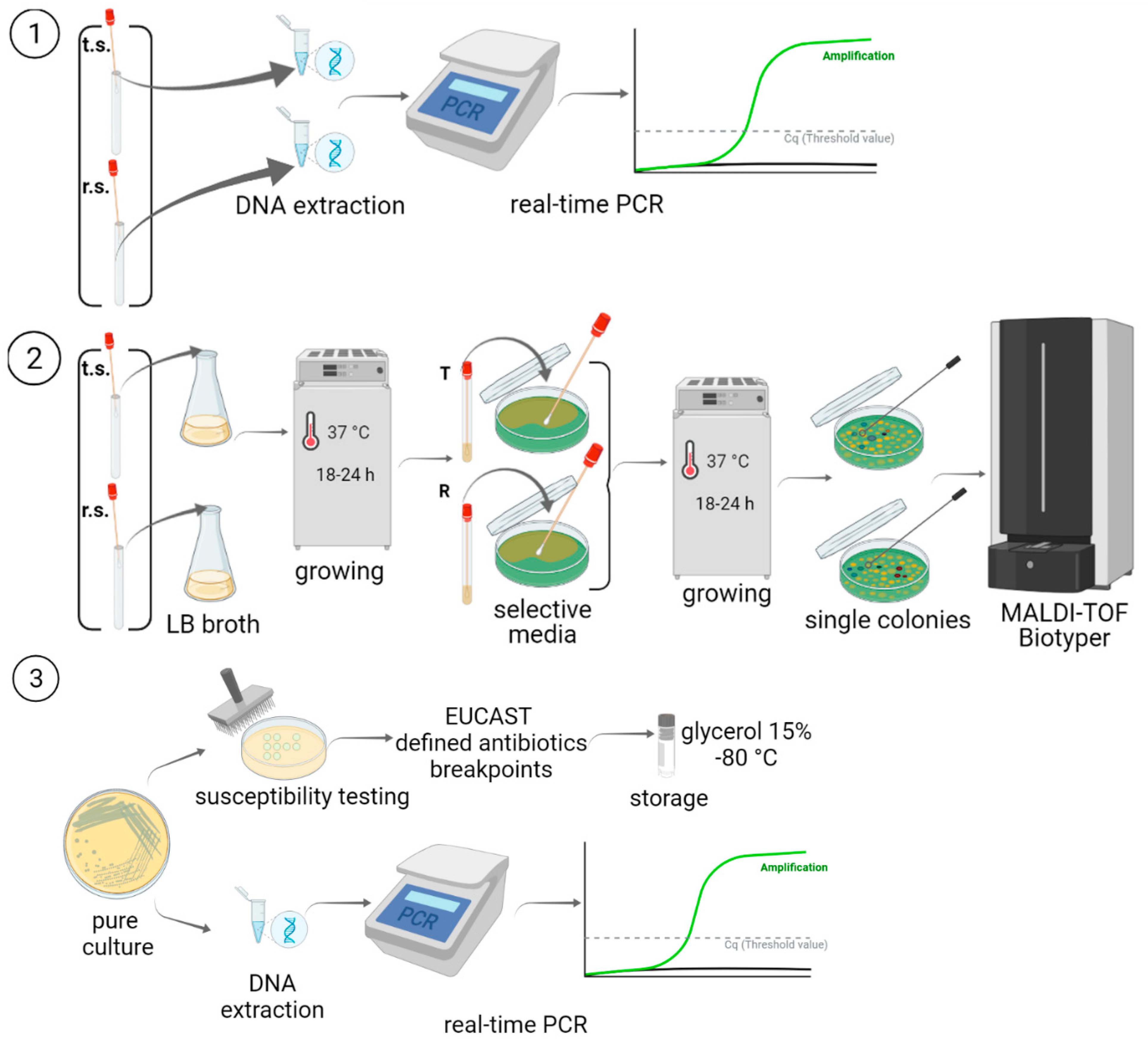
4. Materials and Methods
4.1. Bioethical Requirements
4.2. Study Design
4.3. DNA Extraction and AMR Gene Detection in Clinical Samples
4.4. Isolation and Identification of Gram-Negative Bacteria
4.5. Susceptibility to Antimicrobials
4.6. DNA Extraction and PCR Detection of the Resistance Genes in Clinical Isolates
4.7. Whole-Genome Sequencing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jian, Z.; Zeng, L.; Xu, T.; Sun, S.; Yan, S.; Yang, L.; Huang, Y.; Jia, J.; Dou, T. Antibiotic resistance genes in bacteria: Occurrence, spread, and control. J. Basic Microbiol. 2021, 61, 1049–1070. [Google Scholar] [CrossRef] [PubMed]
- Qadri, H.; Shah, A.H.; Mir, M. Novel Strategies to Combat the Emerging Drug Resistance in Human Pathogenic Microbes. Curr. Drug Targets 2021, 22, 1424–1436. [Google Scholar] [CrossRef] [PubMed]
- Brink, A.J. Epidemiology of carbapenem-resistant Gram-negative infections globally. Curr. Opin. Infect. Dis. 2019, 32, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.V.; Reed, E.E.; Herman, D.D.; Magrum, B.; Beatty, J.J.; Stevenson, K.B. Antimicrobial Stewardship in the ICU. Semin. Respir. Crit. Care Med. 2022, 43, 131–140. [Google Scholar] [CrossRef]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef]
- Miao, M.; Wen, H.; Xu, P.; Niu, S.; Lv, J.; Xie, X.; Mediavilla, J.R.; Tang, Y.W.; Kreiswirth, B.N.; Zhang, X.; et al. Genetic Diversity of Carbapenem-Resistant Enterobacteriaceae (CRE) Clinical isolates from a tertiary hospital in Eastern China. Front. Microbiol. 2019, 9, 3341. [Google Scholar] [CrossRef]
- Mittal, G.; Gaind, R.; Kumar, D.; Kaushik, G.; Gupta, K.B.; Verma, P.K.; Deb, M. Risk factors for fecal carriage of carbapenemase producing Enterobacteriaceae among intensive care unit patients from a tertiary care center in India. BMC Microbiol. 2016, 16, 138. [Google Scholar] [CrossRef]
- Yan, L.; Sun, J.; Xu, X.; Huang, S. Epidemiology and risk factors of rectal colonization of carbapenemase-producing Enterobacteriaceae among high-risk patients from ICU and HSCT wards in a university hospital. Antimicrob. Resist. Infect. Control 2020, 9, 155. [Google Scholar] [CrossRef]
- Linh, T.D.; Thu, N.H.; Shibayama, K.; Suzuki, M.; Yoshida, L.; Thai, P.D.; Anh, D.D.; Duong, T.N.; Trinh, H.S.; Thom, V.P.; et al. Expansion of KPC-producing Enterobacterales in four large hospitals in Hanoi, Vietnam. J. Glob. Antimicrob. Resist. 2021, 27, 200–211. [Google Scholar] [CrossRef]
- Zhen, X.; Stålsby Lundborg, C.; Sun, X.; Gu, S.; Dong, H. Clinical and economic burden of carbapenem-resistant infection or colonization caused by Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii: A multicenter study in China. Antibiotics 2020, 9, 514. [Google Scholar] [CrossRef]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef]
- Zaha, D.C.; Bungau, S.; Aleya, S.; Tit, D.M.; Vesa, C.M.; Popa, A.R.; Pantis, C.; Maghiar, O.A.; Bratu, O.G.; Furau, C.; et al. What antibiotics for what pathogens? The sensitivity spectrum of isolated strains in an intensive care unit. Sci. Total Environ. 2019, 687, 118–127. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Surveillance of Antimicrobial Resistance in Europe–Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net) 2017; ECDC: Stockholm, Sweden, 2018.
- Arhoune, B.; Oumokhtar, B.; Hmami, F.; Barguigua, A.; Timinouni, M.; El Fakir, S.; Chami, F.; Bouharrou, A. Rectal carriage of extended-spectrum β-lactamase- and carbapenemase-producing Enterobacteriaceae among hospitalised neonates in a neonatal intensive care unit in Fez, Morocco. J. Glob. Antimicrob. Resist. 2017, 8, 90–96. [Google Scholar] [CrossRef]
- Hammami, S.; Dahdeh, C.; Mamlouk, K.; Ferjeni, S.; Maamar, E.; Hamzaoui, Z.; Saidani, M.; Ghedira, S.; Houissa, M.; Slim, A.; et al. Rectal carriage of extended-spectrum beta-lactamase and carbapenemase producing gram-negative bacilli in intensive care units in Tunisia. Microb. Drug Resist. 2017, 23, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Aruhomukama, D.; Najjuka, C.F.; Kajumbula, H.; Okee, M.; Mboowa, G.; Sserwadda, I.; Mayanja, R.; Joloba, M.L.; Kateete, D.P. blaVIM- and blaOXA-mediated carbapenem resistance among Acinetobacter baumannii and Pseudomonas aeruginosa isolates from the Mulago hospital intensive care unit in Kampala, Uganda. BMC Infect. Dis. 2019, 19, 853. [Google Scholar] [CrossRef] [PubMed]
- Atterby, C.; Osbjer, K.; Tepper, V.; Rajala, E.; Hernandez, J.; Seng, S.; Holl, D.; Bonnedahl, J.; Börjesson, S.; Magnusson, U.; et al. Carriage of carbapenemase- and extended-spectrum cephalosporinase-producing Escherichia coli and Klebsiella pneumoniae in humans and livestock in rural Cambodia; gender and age differences and detection of blaOXA-48 in humans. Zoonoses Public Health 2019, 66, 603–617. [Google Scholar] [CrossRef]
- Liu, X.; Li, X.; Yang, A.W.; Tang, B.; Jian, Z.J.; Zhong, Y.M.; Li, H.L.; Li, Y.M.; Yan, Q.; Liang, X.H.; et al. Community fecal carriage and molecular epidemiology of extended-spectrum β-lactamase- and carbapenemase-producing Escherichia coli from healthy children in the Central South China. Infect. Drug Resist. 2022, 15, 1601–1611. [Google Scholar] [CrossRef]
- Siikamaki, H.; Kivela, P.; Fotopoulos, M.; Ollgren, J.; Kantele, A. Illness and injury of travellers abroad: Finnish nationwide data from 2010 to 2012, with incidences in various regions of the world. Eurosurveillance 2015, 20, 15–26. [Google Scholar] [CrossRef]
- Fournier, S.; Monteil, C.; Lepainteur, M.; Richard, C.; Brun-Buisson, C.; Jarlier, V. Long-term control of carbapenemase-producing Enterobacteriaceae at the scale of a large French multihospital institution: A nine-year experience, France, 2004 to 2012. Eurosurveillance 2014, 19, 20802. [Google Scholar] [CrossRef]
- Ershova, K.; Savin, I.; Kurdyumova, N.; Wong, D.; Danilov, G.; Shifrin, M.; Alexandrova, I.; Sokolova, E.; Fursova, N.; Zelman, V.; et al. Implementing an infection control and prevention program decreases the incidence of healthcare-associated infections and antibiotic resistance in a Russian neuro-ICU. Antimicrob. Resist. Infect. Control 2018, 7, 94. [Google Scholar] [CrossRef]
- Schneider, A.; Coope, C.; Michie, S.; Puleston, R.; Hopkins, S.; Oliver, I. Implementing a toolkit for the prevention, management and control of carbapenemase-producing Enterobacteriaceae in English acute hospitals trusts: A qualitative evaluation. BMC Health Serv. Res. 2019, 19, 689. [Google Scholar] [CrossRef] [PubMed]
- Jamal, A.J.; Garcia-Jeldes, F.; Baqi, M.; Borgia, S.; Johnstone, J.; Katz, K.; Kohler, P.; Muller, M.P.; McGeer, A.J. Investigators of the Toronto Invasive Bacterial Diseases Network Infection prevention and control practices related to carbapenemase-producing Enterobacteriaceae (CPE) in acute-care hospitals in Ontario, Canada. Infect. Control Hosp. Epidemiol. 2019, 40, 1006–1012. [Google Scholar] [CrossRef] [PubMed]
- Solomennyĭ, A.P.; Maksimov, A.; Saralov, A.I.; Iafaev, R.; Goncharov, A.E.; Keropian, E.A.; Multykh, I.G. Emergence of the integron-positive multi-drug resistant strain of Acinetobacter baumanii in Russian hospitals. Zhurnal Mikrobiol. Epidemiol. Immunobiol. 2008, 4, 89–91. [Google Scholar]
- Fursova, N.K.; Astashkin, E.I.; Knyazeva, A.I.; Kartsev, N.N.; Leonova, E.S.; Ershova, O.N.; Alexandrova, I.A.; Kurdyumova, N.V.; Sazikina, S.Y.; Volozhantsev, N.V.; et al. The spread of blaOXA-48 and blaOXA-244 carbapenemase genes among Klebsiella pneumoniae, Proteus mirabilis and Enterobacter spp. isolated in Moscow, Russia. Ann. Clin. Microbiol. Antimicrob. 2015, 14, 46. [Google Scholar] [CrossRef]
- Shamaeva, S.K.; Portnyagina, U.S.; Edelstein, M.V.; Kuzmina, A.A.; Maloguloval, S.; Varfolomeeva, N.A. Results of monitoring metallo-beta-lactamase-producing strains of Pseudomonas aeruginosa in a multi-profile hospital. Wiad. Lek. 2015, 68, 546–548. [Google Scholar] [PubMed]
- Yakovlev, S.V.; Suvorova, M.P.; Beloborodov, V.B.; Basin, E.E.; Eliseev, E.V.; Kovelenov, S.V.; Portyagina, U.S.; Rog, A.A.; Rudnov, V.A.; Barkanova, O.N. Multicentre study of the prevalence and clinical value of hospital-acquired infections in emergency hospitals of Russia: ERGINI study team. Antibiot. Khimioter. 2016, 61, 32–42. [Google Scholar]
- Pisanenko, D.N.; Gasrataliev, V.E.; Gorshkova, T.N.; Ivaneeva, M.V.; Abramov, D.V.; Danilov, A.A.; Galitsky, T.V.; Ruina, O.V.; Stroganov, A.B.; Atduev, V.A. Microbiological analysis as effective tool for optimization of empirical antibiotic therapy in the urological clinic. Urologiia 2018, 6, 45–51. [Google Scholar] [CrossRef]
- Rafalskiy, V.; Pushkar, D.; Yakovlev, S.; Epstein, O.; Putilovskiy, M.; Tarasov, S.; Glazunov, A.; Korenev, S.; Moiseeva, E.; Gorelysheva, N. Distribution and antibiotic resistance profile of key Gram-negative bacteria that cause community-onset urinary tract infections in the Russian Federation: RESOURCE multicentre surveillance 2017 study. J. Glob. Antimicrob. Resist. 2020, 21, 188–194. [Google Scholar] [CrossRef]
- Kuzina, E.S.; Novikova, T.S.; Solomentsev, V.I.; Sizova, A.A.; Astashkin, E.I.; Kolupaeva, N.V.; Potapov, V.D.; Fursova, N.K. Carriage of capsular serotype K1 Klebsiella pneumoniae sequence type 23 strains in healthy microbiology laboratory staff in Russia. Microbiol. Resour. Announc. 2021, 10, e0052721. [Google Scholar] [CrossRef]
- Zaha, D.C.; Bungau, S.; Uivarosan, D.; Tit, D.M.; Maghiar, T.A.; Maghiar, O.; Pantis, C.; Fratila, O.; Rus, M.; Vesa, C.M. Antibiotic consumption and microbiological epidemiology in surgery departments: Results from a single study center. Antibiotics 2020, 9, 81. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Alqahtani, M.; Tickler, I.A.; Al Deesi, Z.; AlFouzan, W.; Al Jabri, A.; Al Jindan, R.; Al Johani, S.; Alkahtani, S.A.; Al Kharusi, A.; Mokaddas, E.; et al. Molecular detection of carbapenem resistance genes in rectal swabs from patients in Gulf Cooperation Council hospitals. J. Hosp. Infect. 2021, 112, 96–103. [Google Scholar] [CrossRef]
- Schechner, V.; Kotlovsky, T.; Kazma, M.; Mishali, H.; Schwartz, D.; Navon-Venezia, S.; Schwaber, M.J.; Carmeli, Y. Asymptomatic rectal carriage of blaKPC producing carbapenem-resistant Enterobacteriaceae: Who is prone to become clinically infected? Clin. Microbiol. Infect. 2013, 19, 451–456. [Google Scholar] [CrossRef]
- Madueño, A.; González-García, J.; Alonso Socas, M.; Miguel Gómez, M.A.; Lecuona, M. Clinical features and outcomes of bacteraemia due to OXA-48-like carbapenemase-producing Klebsiella pneumoniae in a tertiary hospital. Enferm. Infecc. Microbiol. Clin. (Engl. Ed.) 2018, 36, 498–501. [Google Scholar] [CrossRef]
- Lin, H.Y. Why does the sepsis induced by severe COVID-19 have different clinical features from sepsis induced by CrKP? Chin. J. Traumatol. 2022, 25, 25–26. [Google Scholar] [CrossRef]
- Tfifha, M.; Ferjani, A.; Mallouli, M.; Mlika, N.; Abroug, S.; Boukadida, J. Carriage of multidrug-resistant bacteria among pediatric patients before and during their hospitalization in a tertiary pediatric unit in Tunisia. Libyan. J. Med. 2018, 13, 1419047. [Google Scholar] [CrossRef]
- Fernández-Martínez, E.; Mapango, E.A.; Martínez-Fernández, M.C.; Valle-Barrio, V. Family-centred care of patients admitted to the intensive care unit in times of COVID-19: A systematic review. Intensive Crit. Care Nurs. 2022, 70, 103223. [Google Scholar] [CrossRef]
- Kumar, A.; Mohapatra, S.; Bir, R.; Tyagi, S.; Bakhshi, S.; Mahapatra, M.; Gautam, H.; Sood, S.; Das, B.K.; Kapil, A. Intestinal colonization due to carbapenem–resistant Enterobacteriaceae among hematological malignancy patients in India: Prevalence and molecular charecterisation. Indian J. Hematol. Blood Transfus. 2022, 38, 1–7. [Google Scholar] [CrossRef]
- Ramadan, R.A.; Gebriel, M.G.; Kadry, H.M.; Mosallem, A. Carbapenem-resistant Acinetobacter baumannii and Pseudomonas aeruginosa: Characterization of carbapenemase genes and E-test evaluation of colistin-based combinations. Infect. Drug Resist. 2018, 11, 1261–1269. [Google Scholar] [CrossRef]
- Hassan, R.M.; Salem, S.T.; Hassan, S.; Hegab, A.S.; Elkholy, Y.S. Molecular characterization of carbapenem-resistant Acinetobacter baumannii clinical isolates from Egyptian patients. PLoS ONE 2021, 16, e0251508. [Google Scholar] [CrossRef]
- Khurshid, M.; Rasool, M.H.; Ashfaq, U.A.; Aslam, B.; Waseem, M.; Ali, M.A.; Almatroudi, A.; Rasheed, F.; Saeed, M.; Guo, Q.; et al. Acinetobacter baumannii Sequence types harboring genes encoding aminoglycoside modifying enzymes and 16SrRNA methylase; a multicenter study from Pakistan. Infect. Drug Resist. 2020, 13, 2855–2862. [Google Scholar] [CrossRef]
- Al-Agamy, M.H.; Jeannot, K.; El-Mahdy, T.S.; Samaha, H.A.; Shibl, A.M.; Plésiat, P.; Courvalin, P. Diversity of Molecular Mechanisms Conferring Carbapenem Resistance to Pseudomonas aeruginosa Isolates from Saudi Arabia. Can. J. Infect. Dis. Med. Microbiol. 2016, 2016, 4379686. [Google Scholar] [CrossRef]
- Kateete, D.P.; Nakanjako, R.; Namugenyi, J.; Erume, J.; Joloba, M.L.; Najjuka, C.F. Carbapenem resistant Pseudomonas aeruginosa and Acinetobacter baumannii at Mulago hospital in Kampala, Uganda (2007–2009). Springerplus 2016, 5, 1308. [Google Scholar] [CrossRef]
- Sun, S.; Chen, K.; Kong, X.; Tian, W.; Niu, S. Genetic diversity and in vitro activity of aztreonam/avibactam and ceftazidime/avibactam against carbapenem–resistant Enterobacterales: A multi–center study in Southwest China. Infect. Drug Resist. 2022, 15, 2243–2251. [Google Scholar] [CrossRef]
- Depka, D.; Mikucka, A.; Bogiel, T.; Rzepka, M.; Zawadka, P.; Gospodarek–Komkowska, E. Conventional and real–time PCR targeting blaOXA genes as reliable methods for a rapid detection of carbapenem–resistant Acinetobacter baumannii clinical strains. Antibiotics 2022, 11, 455. [Google Scholar] [CrossRef]
- Haider, M.H.; McHugh, T.D.; Roulston, K.; Arruda, L.B.; Sadouki, Z.; Riaz, S. Detection of carbapenemases blaOXA48-blaKPC-blaNDM-blaVIM and extended-spectrum-β-lactamase blaOXA1-blaSHV-blaTEM genes in Gram–negative bacterial isolates from ICU burns patients. Ann. Clin. Microbiol. Antimicrob. 2022, 21, 18. [Google Scholar] [CrossRef]
- Chainier, D.; Barraud, O.; Masson, G.; Couve-Deacon, E.; François, B.; Couquet, C.Y.; Ploy, M.C. Integron digestive carriage in human and cattle: A “One Health” cultivation-independent approach. Front. Microbiol. 2017, 8, 1891. [Google Scholar] [CrossRef]
- Moazami Goudarzi, S.; Eftekhar, F. Multidrug resistance and integron carriage in clinical isolates of Pseudomonas aeruginosa in Tehran, Iran. Turk. J. Med. Sci. 2015, 45, 789–793. [Google Scholar] [CrossRef]
- Chairat, S.; Ben Yahia, H.; Rojo-Bezares, B.; Sáenz, Y.; Torres, C.; Ben Slama, K. High prevalence of imipenem-resistant and metallo-beta-lactamase-producing Pseudomonas aeruginosa in the Burns Hospital in Tunisia: Detection of a novel class 1 integron. J. Chemother. 2019, 31, 120–126. [Google Scholar] [CrossRef]
- Farshadzadeh, Z.; Hashemi, F.B.; Rahimi, S.; Pourakbari, B.; Esmaeili, D.; Haghighi, M.A.; Majidpour, A.; Shojaa, S.; Rahmani, M.; Gharesi, S.; et al. Wide distribution of carbapenem resistant Acinetobacter baumannii in burns patients in Iran. Front. Microb. 2015, 6, 1146. [Google Scholar] [CrossRef]
- Bagheri-Nesami, M.; Rezai, M.S.; Ahangarkani, F.; Rafiei, A.; Nikkhah, A.; Eslami, G.; Shafahi, K.; Hajalibeig, A.; Khajavi, R. Multidrug and co-resistance patterns of non-fermenting Gram-negative bacilli involved in ventilator-associated pneumonia carrying class 1 integron in the North of Iran. Germs 2017, 7, 123–131. [Google Scholar] [CrossRef]
- Hastak, P.; Cummins, M.L.; Gottlieb, T.; Cheong, E.; Merlino, J.; Myers, G.; Djordjevic, S.P.; Roy Chowdhury, P. Genomic profiling of Escherichia coli isolates from bacteraemia patients: A 3-year cohort study of isolates collected at a Sydney teaching hospital. Microb. Genom. 2020, 6, e000371. [Google Scholar] [CrossRef]
- Malekjamshidi, M.R.; Zandi, H.; Eftekhar, F. Prevalence of extended-spectrum β-lactamase and integron gene carriage in multidrug-resistant Klebsiella species isolated from outpatients in Yazd. Iran. J. Med. Sci. 2020, 45, 23–31. [Google Scholar] [CrossRef]
- Fursova, A.D.; Fursov, M.V.; Astashkin, E.I.; Novikova, T.S.; Fedyukina, G.N.; Kislichkina, A.A.; Alexandrova, I.A.; Ershova, O.N.; Dyatlov, I.A.; Fursova, N.K. Early response of antimicrobial resistance and virulence genes expression in classical, hypervirulent, and hybrid hvKp-MDR Klebsiella pneumoniae on antimicrobial stress. Antibiotics 2021, 11, 7. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. In CLSI Supplement M100–S29, 29th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]
- Priamchuk, S.D.; Fursova, N.K.; Abaev, I.V.; Kovalev, I.; Shishkova, N.A.; Pecherskikh, E.I.; Korobova, O.V.; Astashkin, E.I.; Pachkunov, D.M.; Kruglov, A.N.; et al. Genetic determinants of antibacterial resistance among nosocomial Escherichia coli, Klebsiella spp., and Enterobacter spp. isolates collected in Russia within 2003–2007. Antibiot. Khimioter. 2010, 55, 3–10. (In Russian) [Google Scholar]
- Eckert, C.; Gautier, V.; Arlet, G. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 2005, 57, 14–23. [Google Scholar] [CrossRef]
- Edelstein, M.; Pimkin, M.; Palagin, I.; Stratchounski, L. Prevalence and molecular epidemiology of CTX-M extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob. Agents Chemother. 2003, 47, 3724–3732. [Google Scholar] [CrossRef]
- Poirel, L.; Bonnin, R.A.; Nordmann, P. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob. Agents Chemother. 2011, 56, 559–562. [Google Scholar] [CrossRef]
- Rasheed, J.K.; Biddle, J.W.; Anderson, K.F.; Washer, L.; Chenoweth, C.; Perrin, J.; Newton, D.W.; Patel, J.B. Detection of the Klebsiella pneumoniae carbapenemase type 2 carbapenem-hydrolyzing enzyme in clinical isolates of Citrobacter freundii and K. oxytoca carrying a common plasmid. J. Clin. Microbiol. 2008, 46, 2066–2069. [Google Scholar] [CrossRef][Green Version]
- Hujer, K.M.; Hujer, A.M.; Hulten, E.A.; Bajaksouzian, S.; Adams, J.M.; Donskey, C.J.; Ecker, D.J.; Massire, C.; Eshoo, M.W.; Sampath, R.; et al. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob. Agents Chemother. 2006, 50, 4114–4123. [Google Scholar] [CrossRef]
- Yang, J.; Chen, Y.; Jia, X.; Luo, Y.; Song, Q.; Zhao, W.; Wang, Y.; Liu, H.; Zheng, D.; Xia, Y.; et al. Dissemination and characterization of NDM-1-producing Acinetobacter pittii in an intensive care unit in China. Clin. Microbiol. Infect. 2012, 18, E506–E513. [Google Scholar] [CrossRef]
- Machado, E.; Cantón, R.; Baquero, F.; Galán, J.-C.; Rollán, A.; Peixe, L.; Coque, T.M. Integron content of extended-spectrum-β- lactamase-producing Escherichia coli strains over 12 years in a single hospital in Madrid, Spain. Antimicrob. Agents Chemother. 2005, 49, 1823–1829. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
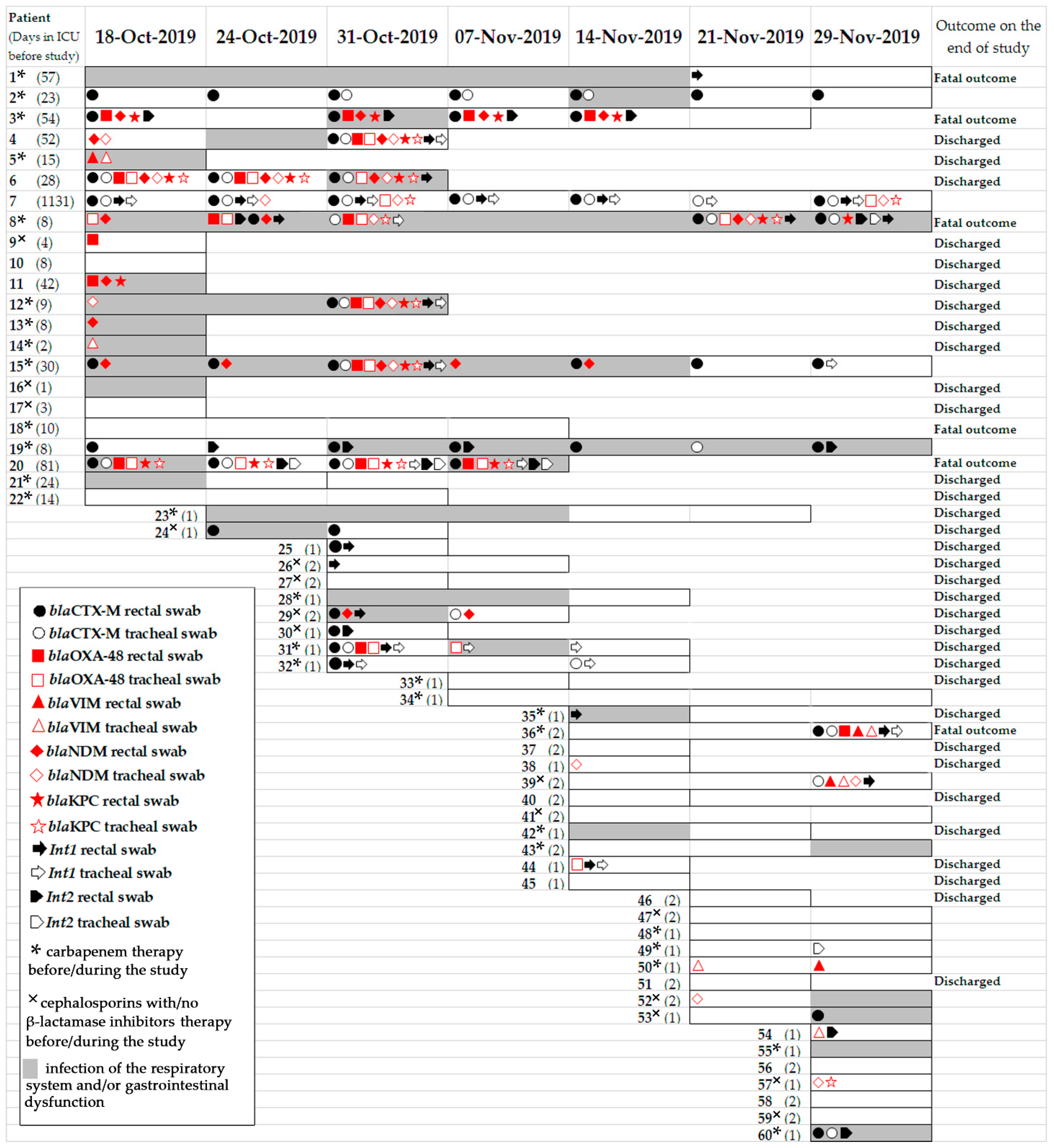
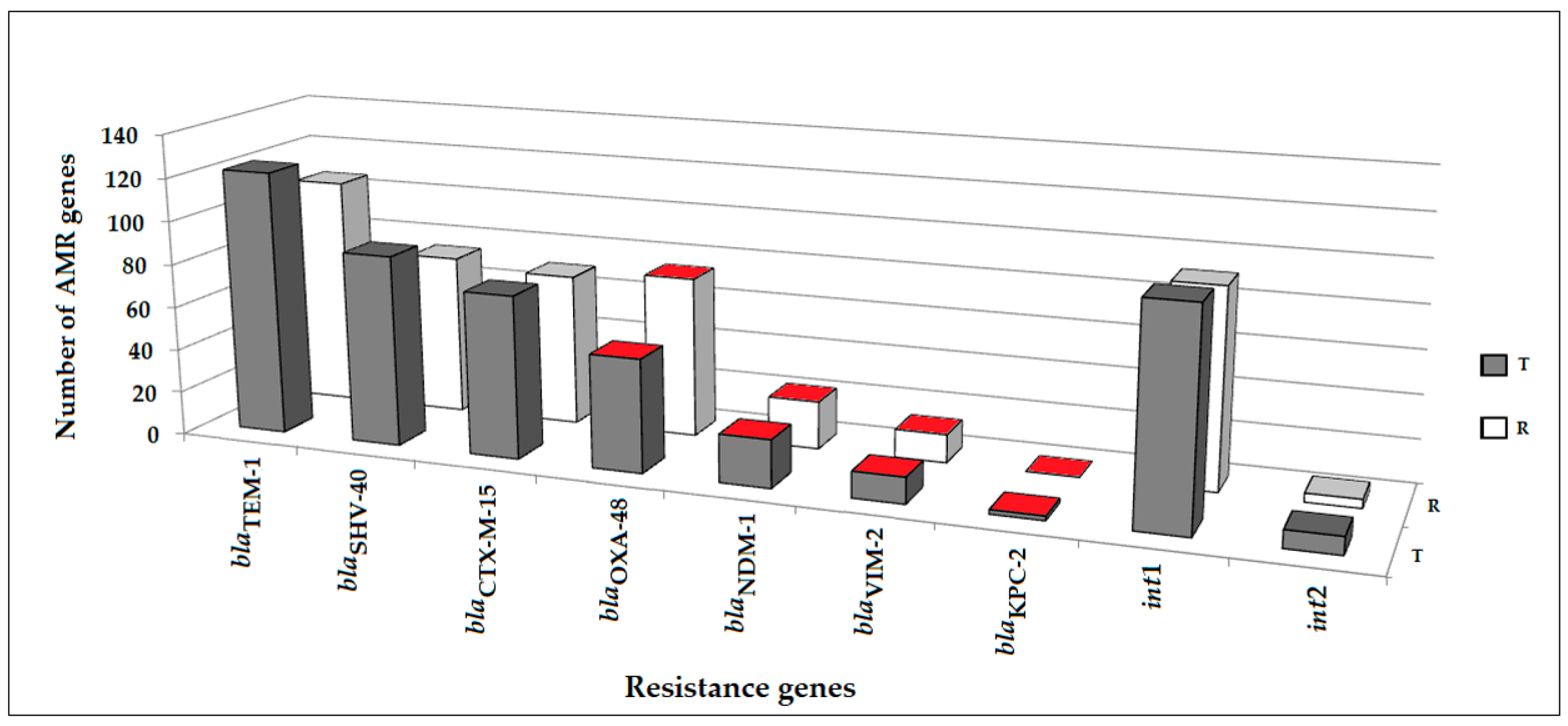
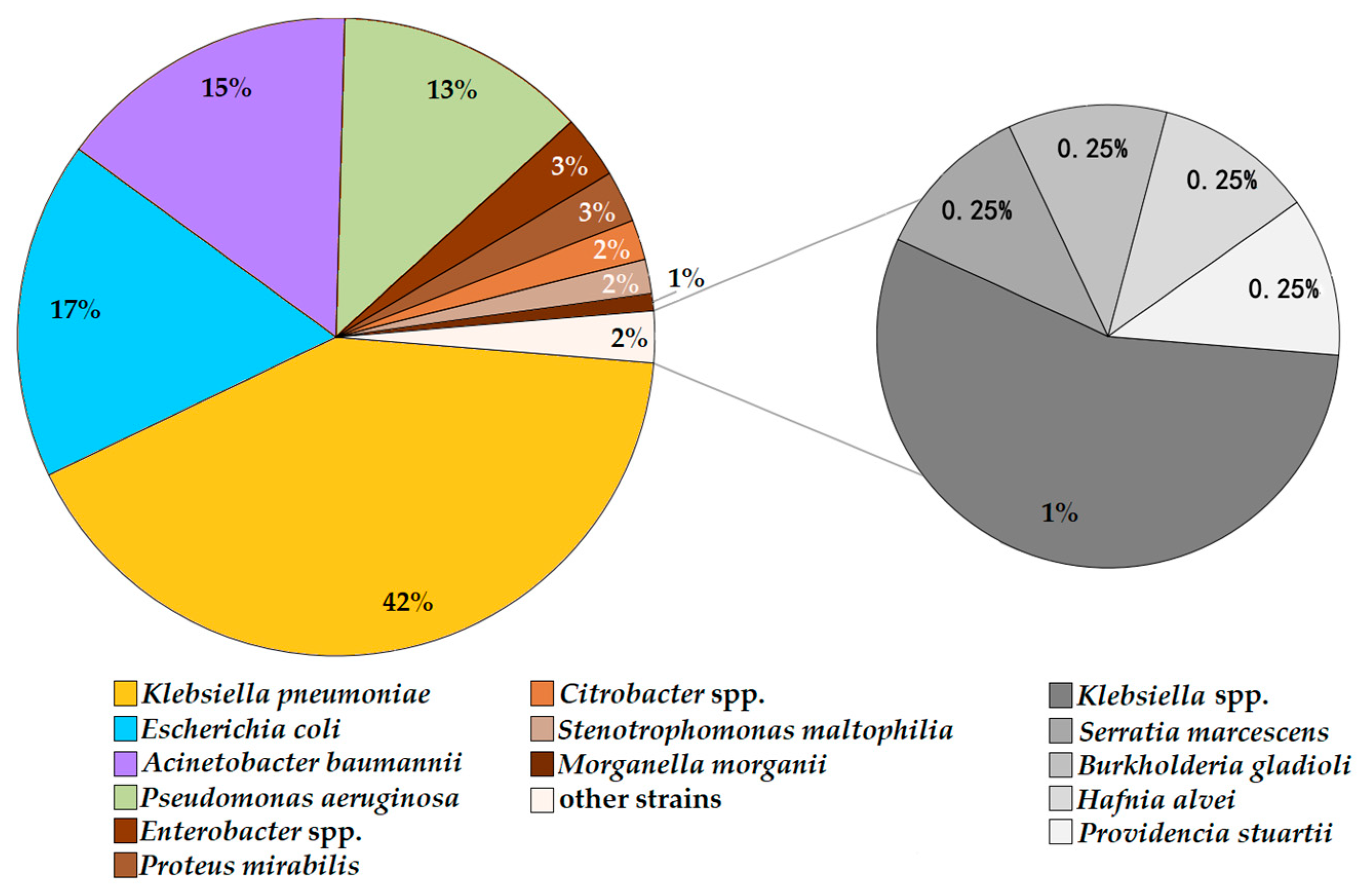
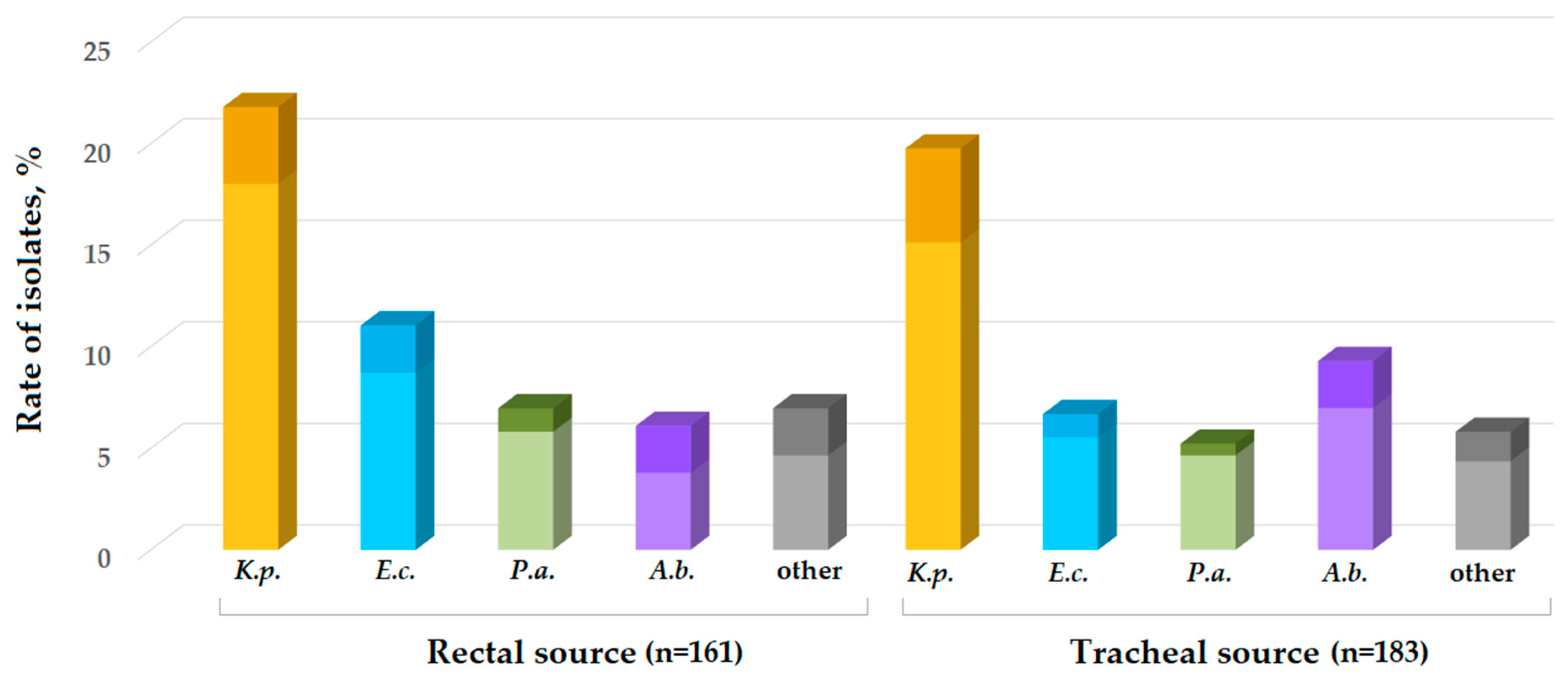

| Item | Dates of Point-Prevalence Surveys/Number of Patients Per Survey | ||||||
|---|---|---|---|---|---|---|---|
| 18 October 2019 | 24 October 2019 | 31 October 2019 | 07 October 2019 | 14 October 2019 | 21 November 2019 | 29 November 2019 | |
| Total patients | 22 | 16 | 23 | 17 | 23 | 21 | 24 |
| Respiratory infection | 8 | 6 | 6 | 6 | 5 | 2 | 6 |
| Gastrointestinal dysfunction | 4 | 2 | 4 | 2 | 1 | 0 | 1 |
| Carbapenem therapy | 13 | 10 | 13 | 13 | 15 | 14 | 13 |
| Cephalosporins with/no β-lactamase inhibitors therapy | 3 | 1 | 5 | 2 | 2 | 5 | 7 |
| Admission to the ICU | 22 | 2 | 8 | 2 | 11 | 8 | 7 |
| Fatal outcome | 0 | 0 | 0 | 0 | 2 | 1 | 3 |
| Discharged | 0 | 8 | 1 | 8 | 3 | 9 | 1 |
| Trends in Carbapenemase Gene Content | Patients |
|---|---|
| Patients carried carbapenemase genes both in r.s. and t.s. simultaneously | 4, 5, 6, 8, 12, 15, 20, 31, 36, 39 |
| Patients positive on carbapenemase genes during the study | 3, 5, 6, 8, 9, 11, 13, 14, 20, 38, 44, 50, 54, 57 |
| Patients became positive for carbapenemase genes during the study | 7, 36, 39 |
| Patients became negative for carbapenemase genes during the study | 15, 31, 52 |
| Patients positive on blaNDM+blaKPC+ blaOXA-48+blaCTX-M+int1 | r.s. 3, 4, 6, 8, 11,12, 15 t.s. 4, 6, 7, 8, 12, 15 |
| Patients positive on blaNDM+blaKPC | r.s. 6 t.s. 57 |
| Patients positive on blaKPC+blaOXA-48 | r.s. 8, 20 t.s. 20 |
| Patients positive on blaVIM+blaOXA-48 | r.s. 36 |
| Patients positive on blaVIM+blaNDM | t.s. 36 |
| Patients positive on blaOXA-48+blaNDM+blaKPC | r.s. 4, 6, 8, 12, 11, 15 t.s. 4, 6, 7, 8, 12, 15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuzina, E.S.; Novikova, T.S.; Astashkin, E.I.; Fedyukina, G.N.; Kislichkina, A.A.; Kurdyumova, N.V.; Savin, I.A.; Ershova, O.N.; Fursova, N.K. Rectal and Tracheal Carriage of Carbapenemase Genes and Class 1 and 2 Integrons in Patients in Neurosurgery Intensive Care Unit. Antibiotics 2022, 11, 886. https://doi.org/10.3390/antibiotics11070886
Kuzina ES, Novikova TS, Astashkin EI, Fedyukina GN, Kislichkina AA, Kurdyumova NV, Savin IA, Ershova ON, Fursova NK. Rectal and Tracheal Carriage of Carbapenemase Genes and Class 1 and 2 Integrons in Patients in Neurosurgery Intensive Care Unit. Antibiotics. 2022; 11(7):886. https://doi.org/10.3390/antibiotics11070886
Chicago/Turabian StyleKuzina, Ekaterina S., Tatiana S. Novikova, Evgeny I. Astashkin, Galina N. Fedyukina, Angelina A. Kislichkina, Natalia V. Kurdyumova, Ivan A. Savin, Olga N. Ershova, and Nadezhda K. Fursova. 2022. "Rectal and Tracheal Carriage of Carbapenemase Genes and Class 1 and 2 Integrons in Patients in Neurosurgery Intensive Care Unit" Antibiotics 11, no. 7: 886. https://doi.org/10.3390/antibiotics11070886
APA StyleKuzina, E. S., Novikova, T. S., Astashkin, E. I., Fedyukina, G. N., Kislichkina, A. A., Kurdyumova, N. V., Savin, I. A., Ershova, O. N., & Fursova, N. K. (2022). Rectal and Tracheal Carriage of Carbapenemase Genes and Class 1 and 2 Integrons in Patients in Neurosurgery Intensive Care Unit. Antibiotics, 11(7), 886. https://doi.org/10.3390/antibiotics11070886






