Clonality and Persistence of Multiresistant Methicillin-Resistant Coagulase-Negative Staphylococci Isolated from the Staff of a University Veterinary Hospital
Abstract
:1. Introduction
2. Results
2.1. Prevalence and Individual Persistence of mecA Gene
2.2. Species of MRS
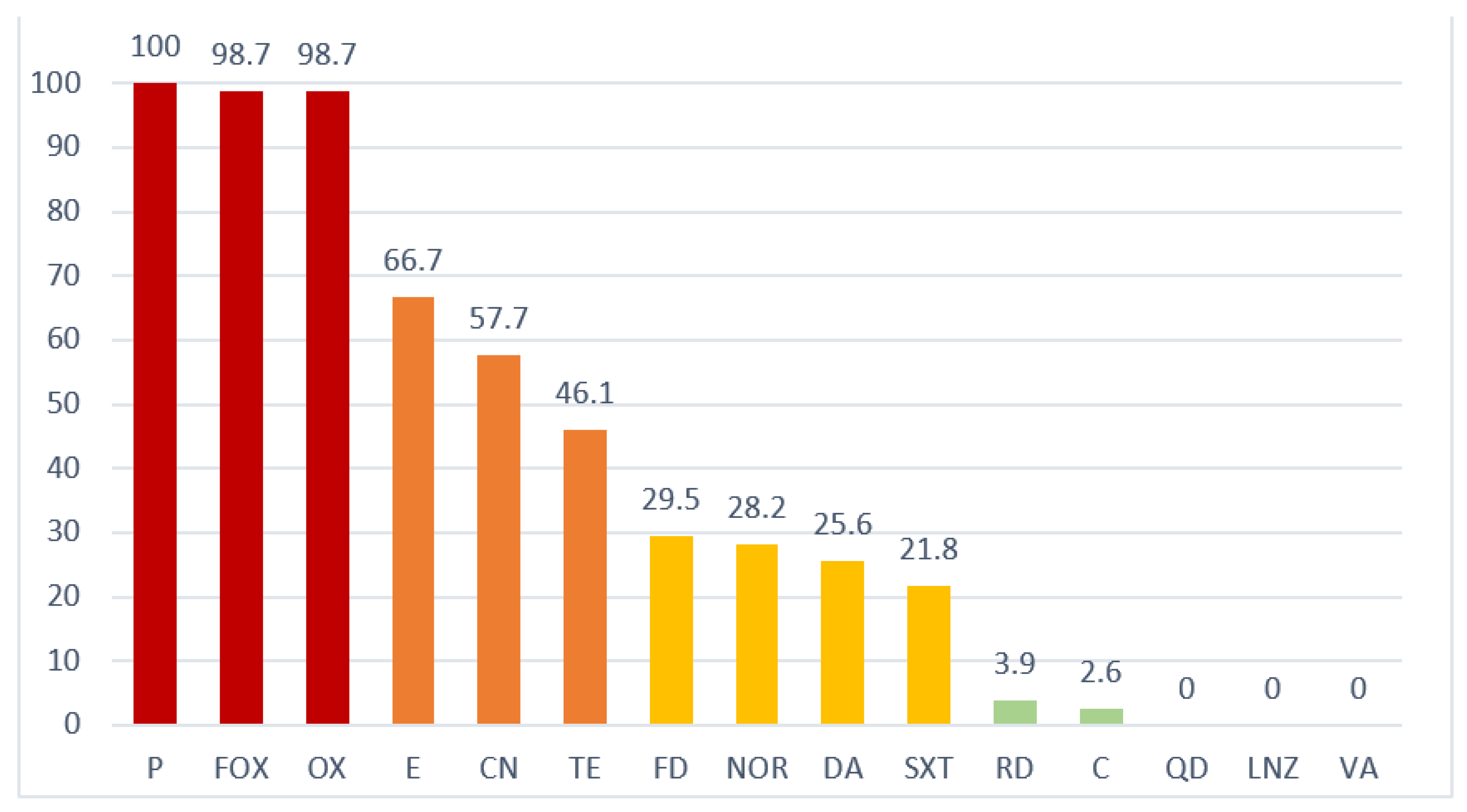
2.3. Antimicrobial Resistance
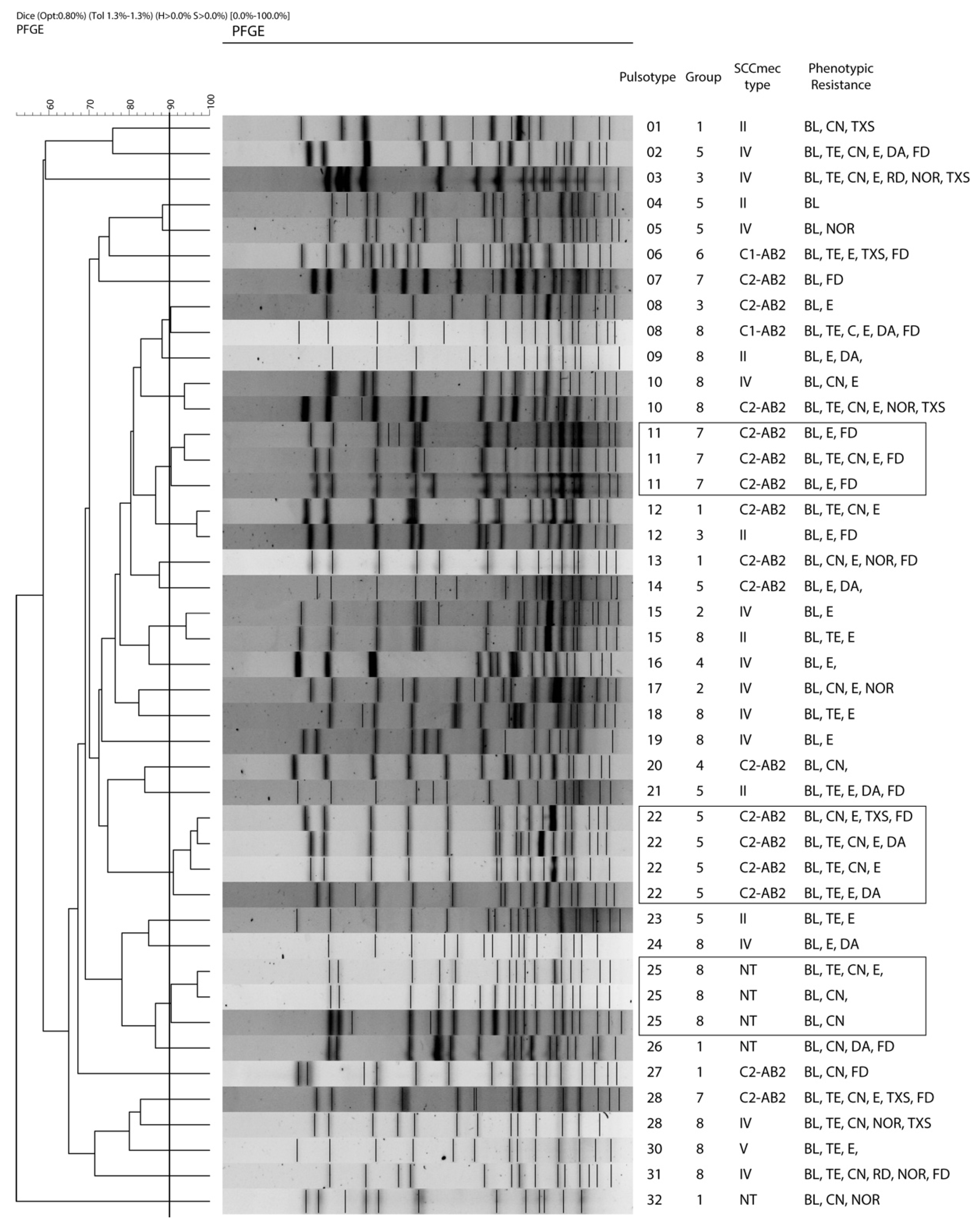
2.4. Clonality and Individual Persistence of S. epidermidis
2.5. SCCmec Typing
3. Discussion
4. Materials and Methods
4.1. Study Design and Population
4.2. Sample Collection
4.3. Bacterial Culture Detection of the mecA Gene and Conservation
4.4. Species Identification
4.5. Antimicrobial Resistance
4.6. Staphylococcal Cassette Chromosome mec (SCCmec) Typing
4.7. Phylogenetic Analysis Using Pulsed-Field Gel Electrophoresis (PFGE)
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Von Eiff, C.; Arciola, C.R.; Montanaro, L.; Becker, K.; Campoccia, D. Emerging staphylococcus species as new pathogens in implant infections. Int. J. Artif. Organs 2006, 29, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Heilmann, C.; Peters, G. Coagulase-negative staphylococci. Clin. Microbiol. Rev. 2014, 27, 870–926. [Google Scholar] [CrossRef] [Green Version]
- Gill, S.R.; Fouts, D.E.; Archer, G.L.; Mongodin, E.F.; DeBoy, R.T.; Ravel, J.; Paulsen, I.T.; Kolonay, J.F.; Brinkac, L.; Beanan, M.; et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 2005, 187, 2426–2438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanssen, A.M.; Ericson Sollid, J.U. Multiple staphylococcal cassette chromosomes and allelic variants of cassette chromosome recombinases in Staphylococcus aureus and coagulase-negative staphylococci from Norway. Antimicrob. Agents Chemother. 2007, 51, 1671–1677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakhundi, S.; Zhang, K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 2018, 31, e00020-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lord, J.; Millis, N.; Jones, R.D.; Johnson, B.; Kania, S.A.; Odoi, A. Patterns of antimicrobial, multidrug and methicillin resistance among Staphylococcus spp. isolated from canine specimens submitted to a diagnostic laboratory in Tennessee, USA: A descriptive study. BMC Vet. Res. 2022, 18, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Sukri, A.; Zaki, H.H.M.; Zin, N.M. Antibiotic Resistance Patterns of Coagulase-Negative Staphylococcus (CoNS) Isolates from a Major Teaching Hospital in Kuala Lumpur, Malaysia. Sains Malays. 2022, 51, 865–872. [Google Scholar] [CrossRef]
- Guggenheim, M.; Zbinden, R.; Handschin, A.E.; Gohritz, A.; Altintas, M.A.; Giovanoli, P. Changes in bacterial isolates from burn wounds and their antibiograms: A 20-year study (1986–2005). Burns 2009, 35, 553–560. [Google Scholar] [CrossRef]
- Ibrahem, S.; Salmenlinna, S.; Virolainen, A.; Kerttula, A.M.; Lyytikainen, O.; Jägerroos, H.; Broas, M.; Vuopio-Varkila, J. Carriage of methicillin-resistant staphylococci and their SCCmec types in a long-term-care facility. J. Clin. Microbiol. 2009, 47, 32–37. [Google Scholar] [CrossRef] [Green Version]
- Widerström, M.; Wiström, J.; Ek, E.; Edebro, H.; Monsen, T. Near absence of methicillin-resistance and pronounced genetic diversity among Staphylococcus epidermidis isolated from healthy persons in northern Sweden. Apmis 2011, 119, 505–512. [Google Scholar] [CrossRef] [PubMed]
- de Souza, C.S.M.; Teixeira, N.B.; de Lourdes Ribeiro de Souza da Cunha, M. Community-associated Staphylococcus aureus (CA-MRSA) in special groups. In The Encyclopedia of Bacteriology Research Developments; Nova Science Publishers: Hauppauge, NY, USA, 2021; Volume 11, pp. 2091–2097. [Google Scholar]
- Thammavongsa, V.; Kim, H.K.; Missiakas, D.; Schneewind, O. Staphylococcal manipulation of host immune responses. Nat. Rev. Microbiol. 2015, 13, 529–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Leekha, S.; Diekema, D.J.; Perencevich, E.N. Seasonality of staphylococcal infections. Clin. Microbiol. Infect. 2012, 18, 927–933. [Google Scholar] [CrossRef] [Green Version]
- Fišarová, L.; Pantůček, R.; Botka, T.; Doškař, J. Variability of resistance plasmids in coagulase-negative staphylococci and their importance as a reservoir of antimicrobial resistance. Res. Microbiol. 2019, 170, 105–111. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus epidermidis—The ‘accidental’ pathogen. Nat. Rev. Microbiol. 2009, 7, 555–567. [Google Scholar] [CrossRef] [Green Version]
- Couto, I.; De Lencastre, H.; Severina, E.; Kloos, W.; Webster, J.A.; Hubner, R.J.; Sanches, I.S.; Tomasz, A. Ubiquitous presence of a mecA homologue in natural isolates of Staphylococcus sciuri. Microb. Drug Resist. 1996, 2, 377–391. [Google Scholar] [CrossRef]
- Abdelmalek, S.M.A.; Qinna, M.W.; Al-Ejielat, R.; Collier, P.J. Methicillin-Resistant Staphylococci (MRS): Carriage and Antibiotic Resistance Patterns in College Students. J. Community Health 2022, 47, 416–424. [Google Scholar] [CrossRef]
- Martins, A.; Riboli, D.F.M.; Camargo, C.H.; Pereira, V.C.; De Almeida Sampaio, R.; De Souza da Cunha, M.D.L.R. Antimicrobial resistance and persistence of Staphylococcus epidermidis clones in a Brazilian university hospital. Diagn. Microbiol. Infect. Dis. 2013, 77, 164–168. [Google Scholar] [CrossRef] [Green Version]
- Garcia, J.F.; Diez, M.J.; Sahagun, A.M.; Diez, R.; Sierra, M.; Garcia, J.J.; Fernandez, M.N. The online sale of antibiotics for veterinary use. Animals 2020, 10, 503. [Google Scholar] [CrossRef] [Green Version]
- Miragaia, M.; Carriço, J.A.; Thomas, J.C.; Couto, I.; Enright, M.C.; De Lencastre, H. Comparison of molecular typing methods for characterization of Staphylococcus epidermidis: Proposal for clone definition. J. Clin. Microbiol. 2008, 46, 118–129. [Google Scholar] [CrossRef] [Green Version]
- Hartman, B.J.; Tomasz, A. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J. Bacteriol. 1984, 158, 513–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, T.; Hiramatsu, K.; Oliveira, D.C.; De Lencastre, H.; Zhang, K.; Westh, H.; O’Brien, F.; Giffard, P.M.; Coleman, D.; Tenover, F.C.; et al. Classification of staphylococcal cassette chromosome mec (SCCmec): Guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 2009, 53, 4961–4967. [Google Scholar] [CrossRef] [Green Version]
- Uehara, Y. Current Status of Staphylococcal Cassette Chromosome mec (SCCmec). Antibiotics 2022, 11, 86. [Google Scholar] [CrossRef] [PubMed]
- Zong, Z.; Peng, C.; Lü, X. Diversity of SCCmec elements in methicillin-resistant coagulase-negative staphylococci clinical isolates. PLoS ONE 2011, 6, e20191. [Google Scholar] [CrossRef]
- Barbier, F.; Ruppé, E.; Hernandez, D.; Lebeaux, D.; Francois, P.; Felix, B.; Desprez, A.; Maiga, A.; Woerther, P.L.; Gaillard, K.; et al. Methicillin-resistant coagulase-negative staphylococci in the community: High homology of SCCmec IVa between Staphylococcus epidermidis and major clones of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 2010, 202, 270–281. [Google Scholar] [CrossRef] [Green Version]
- Saber, H.; Jasni, A.S.; Jamaluddin, T.Z.M.T.; Ibrahim, R. A review of staphylococcal cassette chromosome mec (SCCmec) types in coagulase-negative staphylococci (CoNS) species. Malays. J. Med. Sci. 2017, 24, 7–18. [Google Scholar] [CrossRef]
- Ruppé, E.; Barbier, F.; Mesli, Y.; Maiga, A.; Cojocaru, R.; Benkhalfat, M.; Benchouk, S.; Hassaine, H.; Maiga, I.; Diallo, A.; et al. Diversity of staphylococcal cassette chromosome mec structures in methicillin-resistant Staphylococcus epidermidis and Staphylococcus haemolyticus strains among outpatients from four countries. Antimicrob. Agents Chemother. 2009, 53, 442–449. [Google Scholar] [CrossRef] [Green Version]
- Miragaia, M.; Couto, I.; De Lencastre, H. Genetic diversity among methicillin-resistant Staphylococcus epidermidis (MRSE). Microb. Drug Resist. 2005, 11, 83–93. [Google Scholar] [CrossRef]
- Miragaia, M.; Thomas, J.C.; Couto, I.; Enright, M.C.; De Lencastre, H. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J. Bacteriol. 2007, 189, 2540–2552. [Google Scholar] [CrossRef] [Green Version]
- Jamaluddin, T.Z.M.T.; Kuwahara-Arai, K.; Hisata, K.; Terasawa, M.; Cui, L.; Baba, T.; Sotozono, C.; Kinoshita, S.; Ito, T.; Hiramatsu, K. Extreme genetic diversity of methicillin-resistant Staphylococcus epidermidis strains disseminated among healthy Japanese children. J. Clin. Microbiol. 2008, 46, 3778–3783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, R.J.; Miragaia, M.; Weinberg, A.D.; Lee, C.J.; Rolo, J.; Giacalone, J.C.; Slaughter, M.S.; Pappas, P.; Naka, Y.; Tector, A.J.; et al. Staphylococcus epidermidis colonization is highly clonal across US cardiac centers. J. Infect. Dis. 2012, 205, 1391–1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harbarth, S.; Hawkey, P.M.; Tenover, F.; Stefani, S.; Pantosti, A.; Struelens, M.J. Update on screening and clinical diagnosis of meticillin-resistant Staphylococcus aureus (MRSA). Int. J. Antimicrob. Agents 2011, 37, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Rey Pérez, J.; Zálama Rosa, L.; García Sánchez, A.; Hermoso de Mendoza Salcedo, J.; Alonso Rodríguez, J.M.; Cerrato Horrillo, R.; Zurita, S.G.; Gil Molino, M. Multiple antimicrobial resistance in methicillin-resistant staphylococcus sciuri group isolates from wild ungulates in Spain. Antibiotics 2021, 10, 920. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.E.; Kaleta, E.J.; Arora, A.; Wolk, D.M. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: A fundamental shift in the routine practice of clinical microbiology. Clin. Microbiol. Rev. 2013, 26, 547–603. [Google Scholar] [CrossRef] [Green Version]
- EUCAST. Clinical Breakpoint Bacteria (v. 10). Available online: https://eucast.org/ (accessed on 3 May 2021).
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, T.; Ono, D.; Sato, A. Staphylococcal Cassette Chromosome mec (SCCmec) analysis of MRSA. In Methicillin-Resistant Staphylococcus aureus (MRSA) Protocols, Cutting-Edge Technologies and Advancements, 3rd ed.; Ji, Y., Ed.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2020; pp. 59–78. [Google Scholar]
- Centers for Diseases Control and Prevention (CDC). Oxacillin-Resistant Staphylococcus aureus on PulseNet (OPN). Laboratory Protocol for Molecular Typing of S. aureus by PFGE. Available online: https://www.cdc.gov/mrsa/pdf/ar_mras_PFGE_s_aureus.pdf (accessed on 5 September 2021).
- Tenover, F.C.; Arbeit, R.D.; Goering, R.V.; Mickelsen, P.A.; Murray, B.E.; Persing, D.H.; Swaminathan, B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J. Clin. Microbiol. 1995, 33, 2233–2239. [Google Scholar] [CrossRef] [Green Version]

| Individuals | Samples | Prevalence 2020 | Prevalence 2021 | General Prevalence | Persistent Carriage | Isolates mecA+ |
|---|---|---|---|---|---|---|
| 94 | 183 | 53.2% (50/94) | 31.5% (28/89) | 61.7% (58/94) | 19.1% (18/94) | 78 |
| MRS Species | No. of Isolates | No. of Isolates 2020/21 (Percentage) |
|---|---|---|
| S. epidermidis | 72 (92.3%) | 48/24 |
| S. warneri | 3 (3.8%) | 1/2 |
| S. pseudintermedius | 1 (1.3%) | 0/1 |
| S. haemolyticus | 1 (1.3%) | 1/0 |
| S. sciuri | 1 (1.3%) | 0/1 |
| No. of PTs (No. of Strains in Each) | ID PTs |
|---|---|
| 25 (1) | 1–7, 9, 13, 14, 16–21, 23, 24, 26–32 |
| 4 (2) | 8, 10, 12, 15 |
| 2 (3) | 11, 25 |
| 1 (4) | 22 |
| mec Complex Class | ccr Complex(es) | SCCmec Type (Percentage) | Species (No. of Isolates) |
|---|---|---|---|
| A | AB2 | II (11.5%) | S. epidermidis (8), S. warneri (1) |
| B | AB2 | IV (25.6%) | S. epidermidis (20) |
| C1 | AB2 | UC (9%) | S. epidermidis (6), S. warneri (1) |
| C2 | AB2 | UC (37.2%) | S. epidermidis (28), S pseudintermedius (1) |
| C2 | AB2, C1 | UC (5.1%) | S. epidermidis (3), S. warneri (1) |
| C2 | C1 | V (1.3%) | S. epidermidis (1) |
| NT | NT | NT (10.3%) | S. epidermidis (6), S. haemolyticus (1), S. sciuri (1) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rey, J.; Gil, M.; de Mendoza, J.H.; García, A.; Gaitskell-Phillips, G.; Bastidas-Caldes, C.; Zalama, L. Clonality and Persistence of Multiresistant Methicillin-Resistant Coagulase-Negative Staphylococci Isolated from the Staff of a University Veterinary Hospital. Antibiotics 2022, 11, 811. https://doi.org/10.3390/antibiotics11060811
Rey J, Gil M, de Mendoza JH, García A, Gaitskell-Phillips G, Bastidas-Caldes C, Zalama L. Clonality and Persistence of Multiresistant Methicillin-Resistant Coagulase-Negative Staphylococci Isolated from the Staff of a University Veterinary Hospital. Antibiotics. 2022; 11(6):811. https://doi.org/10.3390/antibiotics11060811
Chicago/Turabian StyleRey, Joaquín, María Gil, Javier Hermoso de Mendoza, Alfredo García, Gemma Gaitskell-Phillips, Carlos Bastidas-Caldes, and Laura Zalama. 2022. "Clonality and Persistence of Multiresistant Methicillin-Resistant Coagulase-Negative Staphylococci Isolated from the Staff of a University Veterinary Hospital" Antibiotics 11, no. 6: 811. https://doi.org/10.3390/antibiotics11060811
APA StyleRey, J., Gil, M., de Mendoza, J. H., García, A., Gaitskell-Phillips, G., Bastidas-Caldes, C., & Zalama, L. (2022). Clonality and Persistence of Multiresistant Methicillin-Resistant Coagulase-Negative Staphylococci Isolated from the Staff of a University Veterinary Hospital. Antibiotics, 11(6), 811. https://doi.org/10.3390/antibiotics11060811