Abstract
Colistin, a last-resort antibiotic, is used to treat infections caused by multi-drug-resistant Gram-negative bacteria. Colistin resistance can emerge by acquiring the mobile colistin gene, mcr-1, usually plasmid borne. Studies on mcr-1 and its transmissibility are limited in the Middle East and North Africa (MENA) region. Here, we investigated the occurrence of mcr-1 in 18 previously collected Escherichia coli isolates collected from chicken samples in Qatar; whole-genome sequencing was performed to determine the location (plasmid-borne and chromosomal) of mcr-1 in the isolates. Additionally, we assessed the transmissibility of plasmid-borne mcr-1 and its cost on fitness in E. coli biofilms. Our results showed that the E. coli isolates belonged to different sequence types, indicating that mcr-1 was occurring in strains with diverse genetic backgrounds. In silico analysis and transformation assays showed that all the isolates carried mcr-1 on plasmids that were mainly IncI2 types. All the mcr-1 plasmids were found to be transmissible by conjugation. In biofilms, a significant reduction in the number of CFU (≈0.055 logs CFU/mL) and colistin resistance (≈2.19 log CFU/mL) was observed; however, the reduction in resistance was significantly larger, indicating that the plasmids incur a high fitness cost. To our knowledge, this is the first study that investigates mcr-1 transmissibility and persistence in Qatar. Our findings highlight that mcr has the potential to spread colistin resistance to potentially disparate strains and niches in Qatar, posing a risk that requires intervention.
1. Introduction
Colistin (polymyxin E) is a cationic polypeptide antibiotic. Colistin was approved in 1959 to treat Gram-negative bacterial infections but was withdrawn in the 1980s due to its nephrotoxicity and neurotoxicity [1,2]. However, the emergence of multi-drug-resistant (MDR) bacteria and the scarcity of new antibiotics resulted in the reintroduction of colistin in the 2000s as a last-resort antibiotic to treat complicated MDR infections [3,4,5].
Resistance to colistin was associated with chromosomal mutations that lead to lipopolysaccharide modifications [6]. However, in 2015, the plasmid-borne mobile colistin resistance gene (mcr-1) was first reported in an Escherichia coli isolated from a pig in China [7]. mcr-1 encodes for a phosphoethanolamine transferase, which modifies lipid A by increasing its positive charge, resulting in resistance to colistin [7]. Since the discovery of mcr-1, multiple mcr genes and variants have been identified (mcr-2 to mcr-10) [8,9,10,11,12,13,14,15,16]. However, an analysis of 1386 mcr carrying Gram-negative bacterial genomes from the NCBI database found that E. coli (952 isolates, ≈68.7%) were the most common carriers of mcr-1 so far [17]. To date, mcr-1 has been reported in isolates belonging to different species from diverse sources, including animals, humans, and the environment [17,18,19].
The wide and relatively rapid dissemination of mcr-1 can be attributed to the robust plasmids that carry this gene. Previous studies have reported several mcr-1-carrying plasmids with varying sizes (58–256 kb) and Inc types, including IncI2, IncHI2, IncX4, IncP, IncY, IncF, IncFI, IncFII, IncFIB, INcK2, IncN, and IncQ. However, IncI2, IncHI2, and IncX4 appear to be the most common carriers of mcr-1 [7,18,20,21,22,23,24,25]. Notably, a single E. coli isolate can harbor two different mcr-1-carrying plasmids simultaneously, increasing the possibility of the transmission of this gene [26]. The mobility of mcr-1 poses an apparent public health concern associated with the spread of colistin resistance, rendering this critically important antibiotic ineffective. Additionally, the latter is complicated further because mcr-1-carrying plasmids often harbor other antimicrobial resistance (AMR) genes, including those encoding resistance to -lactams, fluoroquinolones, and tetracyclines, among others [20].
Many of the mcr genes were first reported in animals, highlighting their importance as a reservoir for colistin resistance [8,9,10,11,12,13,14,15,16,20,27]. It was hypothesized that the dissemination of mcr-1 among livestock-associated isolates was accelerated by using colistin as a growth promoter and/or prophylaxis in the treatment of livestock [18,19,20]. Therefore, it is important to closely monitor the emergence of mcr and associated plasmids and strains in livestock to devise appropriate control strategies and reduce the dissemination of colistin resistance via the food chain and/or environment. We previously reported colistin resistance in samples from humans, broiler chickens, and chicken carcasses in Qatar [28,29,30]. However, our previous analysis focused primarily on detecting mcr using PCR analysis. Here, the mcr-1-harboring isolates from broiler chickens (fecal matter) and chicken carcasses were subjected to an in-depth investigation to determine (1) the location of mcr-1 (plasmid-borne or chromosomal), (2) the transmissibility of plasmid-borne mcr-1, (3) the stability of existence of mcr-1 in the isolates after biofilms formation, and (4) the plasmid types and other properties (sequence type, acquired resistome, virulence) of the strains that carried this gene.
2. Results
2.1. Antibiotic Susceptibility Profiles of the mcr-1-Carrying E.coli
All 18 isolates were resistant to colistin with MICs ranging between 3 and 12 g/mL (median = 6 g/mL; Table 1). The majority of the isolates (17 of 18, ≈94.4%) were identified as MDR due to the resistance to the ≥3 antibiotic classes [30]. However, none of isolates were extended-spectrum -lactamase or carbapenemase producers because they were sensitive to -lactam/-lactamase inhibitor combinations; second-, third-, and fourth-generation cephalosporins; and carbapenemases (Figure 1). Resistances to sulfamethoxazole–trimethoprim, ciprofloxacin, tetracycline, and fosfomycin were highly detected at 94.44%, 66.7%, 50%, and 44%, respectively.

Table 1.
Colistin minimum inhibitory concentrations (MICs), genomic analysis, and properties of the mcr-1-harbouring E. coli isolates.
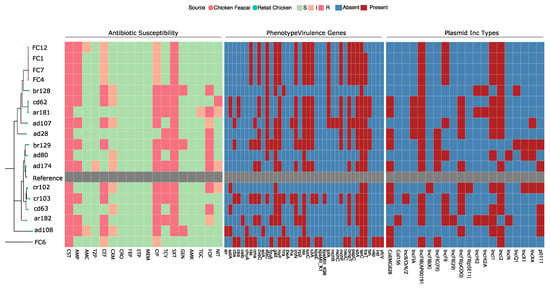
Figure 1.
Phylogenetic tree, antimicrobial susceptibility profiles, virulence factors, and plasmid incompatibility (Inc) types of the 18 mcr-1-carrying Escherichia coli isolated from retail chicken carcasses and broiler chicken fecal samples. E. coli K-12 MG1655 (GenBank: CP014225.1) was used as a reference strain for the phylogeny and indicated in grey. The antibiotics tested were ampicillin (AMP), amoxicillin-clavulanic acid (AMC), piperacillin-tazobactam (TZP), cephalothin (CEF), cefuroxime (CXM), ceftriaxone (CRO), cefepime (FEP), ertapenem (ETP), meropenem (MEM), ciprofloxacin (CIP), tetracycline (TCY), sulfamethoxazole-trimethoprim (SXT), gentamicin (GEN), amikacin (AMK), fosfomycin (FOF), nitrofurantoin (NIT), and colistin (CST). S, I, and R correspond to susceptible, intermediate, and resistant in the antibiotic susceptibility profiles, respectively. For the virulence factors and plasmid Inc types, red indicates presence, and blue indicates the absence from the genome assembly.
2.2. Genomic Features and Diversity
The isolates’ genomic features, including phylogroups, sequence types (ST), antibiotic resistance genes (ARG), plasmid Inc types, FimH type, and virulence genes, are listed in Table 1 and shown in Figure 1. The isolates belonged to 12 STs, with four isolates (22%) belonging to ST 602, two to ST295, and another two to ST48. The remaining isolates belonged to ST10, ST1011, ST155, ST224, ST3270, ST34, ST355, ST6448, and ST744. Lastly, one isolate (ar182) was not assigned an ST because the housekeeping gene alleles did not match any known ST in the database. Notably, the four isolates (FC1, FC4, FC7, and FC12) that belonged to ST 602 were isolated from fecal samples collected from chickens on the same farm. These isolates also had similar ARGs, virulence factors, and plasmid types.
As for phylogroups (Figure 1), most of the isolates (n = 9; 50%) belonged to phylogroup B1, followed by phylogroup A (n = 7; 38.9%), and an isolate belonged to each of the phylogroups B2 and E, respectively.
The FimH types FimH86 and FimH54 were the most common, with four isolates each. Two isolates had FimH39, and the remaining isolates belonged to individual FimH types. Analysis of the plasmid Inc types showed that all isolates carried at least four plasmids and at most nine (median = 6.5).
2.3. Location of the mcr-1
The location of the mcr-1 was analyzed using mlplasmids and MGEfinder (Table 2) [31,32]. The mlplasmids showed that most isolates (83.3%) carried mcr-1 on a plasmid. However, analysis with MGEfinder showed that only 66.67% of the isolates harbored the gene on a plasmid. The discrepancies were in three cases where mlplasmids flagged the gene to be on plasmid, while MGEfinder assigned it as chromosomal. In another case, the opposite occurred between the two programs. However, there was a concordance of 83.3%. Moreover, the results of the MGEfinder analysis showed that all isolates that had plasmid-harbored mcr-1 carried the gene on an IncI2 plasmid, except for two isolates that carried mcr-1 on IncX4.

Table 2.
Results of the in silico analysis of the mcr-1 gene location.
Plasmids were extracted from each isolate and transformed into a chemically competent colistin-sensitive E. coli to verify the in silico results and assess the transmissibility of the mcr-1. Successful transformation was evaluated by colonies growing on colistin-supplemented media, and the transformation resulted in colonies in all cases (Figure 2). All transformants were colistin-resistant, confirming that mcr-1 was carried on plasmids in all the isolates. DNA was extracted from the colistin-resistant transformants and by mcr-1-specific PCR analysis for further verification. All the transformants showed positive amplification of mcr-1 (Figure 2).
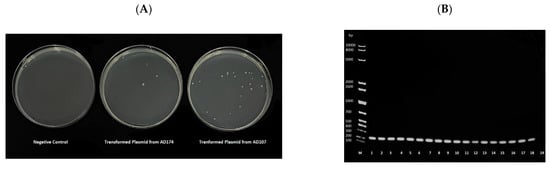
Figure 2.
(A) Representative image of the One ShotTM OmniMAXTM 2 T1R E. coli growth in Muller–Hinton media supplemented with 4 μg/mL of colistin following heat-shock transformation. The negative control underwent the heat-shock method without adding plasmid. (B) Agarose gel electrophoresis of the mcr-1 PCR product from DNA extracts of the transformants. The first well, labelled M, contained a 1 kb Ladder. Plasmids extracted from each sample were transformed into a chemically competent E. coli. The origin of the DNA in each well were as follows, 1: ad28, 2: ad80, 3: ad107, 4: ad108, 5: ad174, 6: ar181, 7: ar182, 8: br128, 9: br129, 10: cd62, 11: cd63, 12: cr102, 13: cr103, 14: FC1, 15: FC4, 16: FC6, 17: FC7, 18: FC12, 19: PCR negative control.
2.4. Trasnmissibility of the mcr-1 via Conjugation
To further assess the transmissibility of mcr-carrying plasmids, conjugation experiments were performed. Transconjugants were obtained for all donors. Hence, conjugation was successful in all cases (Figure 3). For confirmation, DNA was extracted from the transconjugants that grew on MH media containing colistin and streptomycin and subjected to mcr-1-specific PCR analysis. Transconjugants from all donors showed positive amplification of mcr-1 (Figure 3).
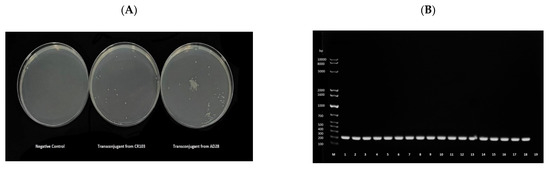
Figure 3.
(A) Representative image of the trans-conjugant E. coli K12 growth in Muller–Hinton media supplemented with 2 μg/mL of colistin and 2000 μg/mL of streptomycin following conjugation. The negative control underwent the conjugation method without adding donor cells. (B) Agarose gel electrophoresis of the mcr-1 PCR product from DNA extracts of the transconjugants. The first well, labeled M, contains a 1 kb Ladder. mcr-1 positive E. coli isolates (donors) underwent conjugation with E. coli K12, strain IM93b (recipient). The origin of the DNA in each well were as follows, 1: ad28, 2: ad80, 3: ad107, 4: ad108, 5: ad174, 6: ar181, 7: ar182, 8: br128, 9: br129, 10: cd62, 11: cd63, 12: cr102, 13: cr103, 14: FC1, 15: FC4, 16: FC6, 17: FC7, 18: FC12, 19: PCR negative control.
2.5. Presistence of the mcr-1 Plasmids in Biofilms
All isolates showed an ability to form biofilms as measured by the absorbance at 570 nm for the crystal violet dye (see Figure S1a,b). A Wilcoxon signed-rank test indicated that the log CFU/mL from the biofilms was significantly lower on day 6 compared to day 3 on both MH agar without colistin (p = 0.033) and MH with colistin (p = 7.6 × 10−6) (Figure 4). However, the reduction in the colistin-supplemented medium was significantly larger than the reduction in the non-supplemented medium (p = 7.6 × 10−6; 0.055 log CFU/mL vs. 2.19 log CFU/mL), suggesting a partial loss of mcr-1-carrying plasmids in a portion of the biofilm E. coli population.
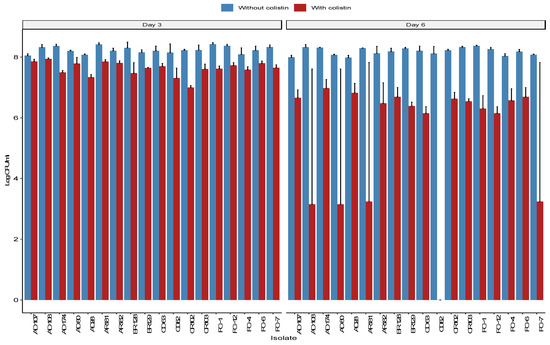
Figure 4.
Log bacterial concentration (CFU/mL) was obtained from each sample on days 3 and 6 of the biofilm formation. Each count was performed on two media, one in Mueller–Hinton (MH) agar (blue) and one in MH supplemented with 4 μg/mL of colistin (red). The left side shows the bacterial concentration on day 3, and the right side represents the bacteria concentrations on day 6.
3. Discussion
Several studies have shown that the prevalence of mcr-carrying colistin-resistant E. coli is usually higher in livestock than in humans [33]. For example, studies in Vietnam found that 97.2% of the domestic chickens tested carried mcr-1-positive E. coli with evidence of clonal relationships [34]. Similarly, in Lebanon, approximately 98% of the colistin-resistant E. coli retrieved from feces of broiler chicken were mcr-1-positive [35]. Additionally, there are increasing reports of acquiring mcr by strains that carry other antibiotic resistance determinates such as ESBL- or pAMP-C-encoding genes [36,37]. These observations and the mobility of mcr highlight the importance of monitoring and analyzing the epidemiology and transmissibility of mcr in livestock and associated products in different countries. This is imperative to reduce the risk of spreading colistin resistance via animal farming, the food chain, and the environment, as well as to control potentially complicated infections. In previous studies, 270 E. coli from retail chicken carcasses and 172 from fecal samples from broiler chicken farms were isolated in Qatar [28,38]. In those studies, the prevalence of mcr-1-carrying E. coli was 31.9% in retail chicken carcasses and 15.6% in the feces of broiler chickens. However, the isolates were not investigated further. On the basis of these observations and studies, 18 mcr-1-positive chicken-associated E. coli were randomly selected (13 from retail chicken and 5 from broiler chicken) for an in-depth analysis to understand further the emergence and potential spread of mcr in Qatar.
Most of the isolates included in this study showed an MDR phenotype. However, none of the isolates were likely ESBL producers as none of them were resistant to third- or fourth-generation cephalosporins, and they were not carbapenem resistant. Interestingly, the majority of the isolates were ciprofloxacin resistant. Ciprofloxacin and colistin are frequently used as growth promoters and prophylactic agents [39]. The latter might have contributed to the observed resistance in these isolates. However, data on antibiotic use in these animals were not available. Regardless, the over-reliance on antibiotics in animal farming might drive the selection of more resistant strains that can pose a risk for both animals and humans.
Phenotypic and genomic analyses revealed a notable diversity between the isolates (Figure 1 and Table 1). Isolates belonging to Phylogroup B1 were the most common (Table 1). This was expected because a high proportion of group B1 is common in mammals and birds [40,41]. One fecal isolate (FC6) belonged to phylogroup B2, common in humans and associated with inflammatory bowel disease and urinary tract infections [41,42]. However, the most frequently detected STs were ST602 (22%), while the remaining isolates were mainly assigned disparate STs. All four isolates that belonged to ST602 were closely related and were obtained from feces of broiler chickens on the same farm, highlighting the possibility of clonal transmission. These four isolates carried identical ARGs, virulence genes, and plasmid replicons. Notably, ST602 is associated with the global spread of AMR in humans and food-producing animals, particularly poultry [43]. However, detecting different and diverse STs is consistent with other studies that found that the mcr-1-positive E. coli were not necessarily clonal [44,45].
Each mcr-1-carrying E. coli isolate in this study carried between 4 and 9 plasmid Inc types (median = 6.5). The majority of the isolates (n = 10) harbored the IncI2 plasmid, one of the most prevalent mcr-carrying plasmids detected in poultry, human, and environmental samples [33]. The bioinformatics analysis also revealed that two isolates had mcr-1 on IncX4 plasmids (Table 2), which are also associated with the global dissemination of mcr-mediated colistin resistance [46]. The mcr in the remaining isolates was not assigned to a plasmid Inc type and had a low probability of being on a plasmid according to in silico analysis. However, there were discrepancies between the two algorithms used to identify the location of mcr-1. This discrepancy might have resulted from limitations associated with short-read WGS for plasmids or because the flanking regions of the mcr-1 did not match a known plasmid sequence. The latter is very interesting because mcr-1 in all the isolates was plasmid-born using heat-shock and conjugation assays, suggesting the potential to detect unknown mcr-1-carrying plasmid types. To date, more than 12 mcr-1-carrying plasmid types have been identified [26]. This plasmid diversity contributes to mcr-1 spread; however, the exact role and diversity of the plasmids still require further investigation.
Whether the gene is carried on a chromosome or a plasmid might result in different transmission dynamics. The mcr-1 is typically flanked by ISApl1 transposable elements [47]. ISApl1 is thought to be the element that mobilizes the gene on plasmids and allows for it to be integrated into chromosomes [44,47]. A gene on a chromosome can be transmitted vertically within a clonal lineage, while carrying it on a plasmid (in addition to mobilization through ISApl1) will enable it to be transferred horizontally between clones and to other bacterial genera. Therefore, a transformation experiment was performed to validate in silico analysis. The transformation was successful for plasmids extracted from all the isolates, as evidenced by growth on colistin-supplemented media and subsequent detection of mcr-1 in the transformants (Figure 2).
Similarly, mating in experiments was conducted to evaluate further if mcr-1 was transmissible plasmid borne. Conjugation was observed with all 18 isolates as donors, as evidenced by the growth of transconjugants on media supplemented with colistin and streptomycin and subsequent detection of mcr-1 in the transconjugants (Figure 3). The natural transformation has been observed in E. coli, albeit at low levels and seemingly environmentally dependent [48,49,50]. For example, a 1996 study that assessed natural transformation in E. coli in environmental water samples showed that the transformation efficiency was higher in samples taken downstream of wastewater effluent than elsewhere [48]. Additionally, conjugation was formerly shown for IncI2 and IncX plasmids [5,7,13,51]. Regardless, the data in this study indicate that mcr is transmissible and, as such, poses a problem that requires immediate action to identify and limit the factors that are driving the emergence of mcr in livestock and associated products in Qatar. Further investigations are necessary to examine the frequency of conjugation and whether it leads to similar resistance levels in the recipient strain and to track mcr-1-carrying plasmid transmission to other niches and bacterial hosts.
AMR dissemination continues to be a significant public health challenge and has been receiving increased research interest. However, ARGs are not the only mechanism of resistance/persistence possessed by bacteria. Biofilm formation has been shown to increase antimicrobial tolerance and resistance and promote ARG transfer [52]. However, dogmatically mcr-1-carrying plasmids might be unstable and can incur a high fitness cost [53]. It was hypothesized that when a more cost-efficient form of resistance exists, the bacteria will shed high-cost plasmids. However, in reality, this remains not well investigated for mcr-1-carrying plasmids. Therefore, a biofilm formation assay was performed to test this hypothesis. There was a significant reduction in biofilm-associated bacterial density between days 3 and 6 on MH agar without and with colistin (p = 0.033 and p = 7.6 × 10−6, respectively). The reduction in bacterial density on day 6 was incompatible with the results of the crystal violet assay. The discrepancy was likely due to the differences between the two methods. CFU plate counts is a direct method and accounts only for viable cells, while the crystal violet assay is indirect and does not distinguish between viable and non-viable cells. The reduction in bacterial CFU on the MH media was likely due to the bacteria reaching a growth plateau and resource limitation. However, the decline in colistin resistance was significantly higher than the reduction in bacterial CFU (p = 7.6 × 10−6; 0.055 log CFU/mL vs. 2.19 log CFU/mL), indicating that mcr-1-carrying E. coli persistence in biofilms is reduced after six days. This was likely due to a combination of factors, including energy conservation, protection in biofilms (tolerance/resistance to stressors such as antibiotics), and resource limitations. The results are consistent with experiments performed with Klebsiella pneumoniae using an mcr-1 recombinant plasmid and with the IncX4 plasmid in E. coli [46,53]. The majority of the isolates in this study carried the gene on IncI2 plasmids, indicating that a similar fitness cost existed for these plasmids. Despite the reduction in resistant CFUs, it should be noted that a significant number of resistant and mcr-1-carrying bacteria persisted in the 6-day-old biofilms. This might increase the potential of mcr-1 transmission, even under unfavorable survival conditions. Further studies are required to assess the duration of mcr persistence and its transmissibility in single- and multi-species biofilms, respectively.
4. Materials and Methods
4.1. Bacterial Isolates
The E. coli strains investigated in this study were isolated between September 2016 and April 2018 and are described in previously published studies [28,38]. Briefly, E. coli were isolated from chicken carcasses collected from three major hypermarkets in Qatar and from chicken fecal samples from multiple broiler farms. The isolates were collected using a stratified random sampling approach under the supervision of the Ministry of Public Health (MoPH) and the Ministry of Municipality (MM). All the isolates were stored at −80 °C. Eighteen isolates that tested positive for mcr-1 by PCR were randomly selected for this study.
4.2. Antimicrobial Susceptibility Testing
The susceptibility of the mcr-1-positive E. coli to 17 antibiotics was assessed using E-test strips. Briefly, each isolate was grown on blood agar plates (BD-Medysinal FZCO, Dubai, UAE) overnight. Colonies from each plate were suspended in a phosphate buffer solution to achieve a 0.5 McFarland (McF) as measured by DensiCHEK Plus (bioMérieux, Marciy-L’Étoile, France), and the suspension was then spread onto Mueller–Hinton (MH) agar plates (HiMedia, Maharashtra, India). The E-test strips for each antibiotic (Liofilchem, Roseto delgi Abruzzi, Italy) were applied to the agar surface, and the plates were incubated at 37 °C overnight. The antibiotics tested were ampicillin (AMP), amoxicillin-clavulanic acid (AMC), piperacillin-tazobactam (TZP), cephalothin (KF), cefuroxime (CXM), ceftriaxone (CRO), cefepime (FEP), ertapenem (ETP), meropenem (MEM), ciprofloxacin (CIP), tetracycline (TCY), sulfamethoxazole-trimethoprim (SXT), gentamicin (GEN), amikacin (AMK), fosfomycin (FOF), nitrofurantoin (NIT), and colistin (CST). The minimum inhibitory concentrations (MICs) were interpreted following the Clinical and Laboratory Standards Institute (CLSI) [54]. E. coli strains ATCC 25922 and ATCC 35218 were used as controls.
4.3. Whole-Genome Sequencing
Genomic DNA was extracted from the isolates using the QIAamp® UCP Pathogen mini kit (Qiagen, Hilden, Germany) and following the manufacturer’s protocol. The DNA was then quantified with the Qubit dsDNA high-sensitivity assay (Thermo Fisher, Waltham, MA, USA). Whole-genome sequencing was performed on the BIGSEQ-500 (Beijing Genomics Institute, Shenzhen, China). Briefly, the genomic DNA was randomly fragmented with the Covaris instrument (Covaris LLC., Woburn, MA, USA), and 200–400 bp fragments were selected using the Agencourt AMPure XP beads (Beckman Coulter, Brea, CA, USA). The fragments were then end-repaired and adenylated, and adaptors were ligated. PCR was then performed to amplify the fragments, which were purified using the Agencourt AMPure XP beads. Lastly, the DNA was circularized and then sequenced using DNA nano-balls (DNB), resulting in 150 bp paired reads.
4.4. Bioinformatics Analysis
The quality of the raw reads was assessed using Fastqc (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/, accessed on 1 November 2021), and trimming was performed using TrimGalore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/, accessed on 1 November 2021) to remove adaptors and low-quality reads. Contamination was checked using kraken2 v2.1.1 [55]. The trimmed reads were assembled using SPAdes v3.13.0 and optimized with Unicycler v0.4.9 [56,57]. The final assemblies (n = 18) are available on the NCBI website under BioProject PRJNA786273 (The genome accessions are listed in Table S1).
Core genome alignment of the isolates with E. coli K-12 MG1655 (GenBank accession: CP014225.1) as the reference strain was performed using the RedDog read mapping pipeline (https://github.com/katholt/reddog-nf, accessed on 1 November 2021). The maximum-likelihood phylogenetic tree was constructed using FastTree [58,59]. Phylotyping was performed using ClermonTyping v20.03 [60]. Multi-locus sequence typing (MLST) and antimicrobial resistance gene (ARG) identification were performed using SRST2 v0.2.0 [61]. The incompatibility (Inc) types of the plasmids carried by the isolates were identified using PlasmidFinder v2.1.6 [62]. Virulence genes were identified with VirulenceFinder v2.0, and the FimH types were determined with FimTyper v1.0 [63,64]. Lastly, the locations (chromosomal or on a plasmid) of the mcr-1 in each isolate were inferred using mlplasmids v1.0.0 and MGEfinder v1.0.3 [31,32].
4.5. Plasmid Extraction and Screening for mcr-1 Using PCR Analysis
The 18 colistin-resistant isolates were sub-cultured in Luria–Bertani (LB) broth and grown overnight (Merck, Dramstadt, Germany). Then, plasmid DNA was extracted using the QIAprep® spin miniprep kit (Qiagen, Hilden, Germany) as described in the manufacturer’s instructions. The concentration and purity of the eluted plasmid DNA were determined using the NanoDrop™ 2000c (Thermo Fisher Scientific, Waltham, MA, USA).
The presence of mcr-1 in the plasmid extracts was confirmed with PCR. The PCR reaction contained 0.5 M of each primer (MCR1_22697 F1: cacttatggcacggtctatga, MCR1_22810 R1: cccaaaccaatgatacgcat) [65], 30 ng of DNA, 12.5 L of A PCR master mix (Hot star Taq Plus, Qiagen, Hilden, Germany), 1x of gel loading dye (CoralLoad, Qiagen, Hilden, Germany), and DEPC-treated water up to a volume of 25 L. The reactions were amplified in a Thermal Cycler using the following program: denaturation at 95 °C for 15 min; 25 cycles of 95 °C for 30 min, 58 °C for 90 min, and 72 °C for 1 min; and a final extension at 72 °C for 10 min. The amplified PCR products were subjected to electrophoresis in a 1.2% agarose gel (AgaroseLE, Ambion®, Austin, TX, USA) stained with ethidium bromide (Promega, Madison, WI, USA). The gel was visualized using a Molecular Imager® Gel Doc™ XR System 170–8170 (Bio-rad, Hercules, CA, USA).
4.6. Assessing mcr-1 Transmissibility Using the Heat Shock Method
The extracted plasmids were transformed into One shot™ OmniMAX™ 2 T1R chemically competent E. coli cells (Thermo Fisher Scientific, Waltham, MA, USA) by heat shock [66,67]. Briefly, 50 L of the chemically competent E. coli were mixed with 3 L of the extracted plasmid and incubated on ice for 1 h. The mixture was then heat-shocked by placing it in a 42 °C water bath for 45 s, which was followed by incubation on ice for 2 min. After heat shock, 1 mL of freshly prepared LB broth was added, and the mixture was incubated at 37 °C in a shaking incubator at 200 rpm for 1 h. The mixture was centrifuged at 3000 rpm for 5 min, and 0.9 mL of the supernatant was removed. The pellet was resuspended in the remaining solution. Moreover, transformants were selected by spreading the suspension on MH agar plates supplemented with 4 g/mL of colistin. Transformant colonies were subcultured in LB broth overnight and subjected to plasmid extraction and mcr-1 PCR analysis to confirm the transmission of mcr-1-carrying plasmids. Chemically competent E. coli with 3 L sterile deionized water were used as a negative control.
4.7. Assessing mcr-1 Transmissibility via Conjugation Assays
Conjugation experiments were conducted to assess further the transmissibility of the mcr-1-carrying plasmids. The mcr-1-positive isolates were used as donors, and the streptomycin-resistant E. coli K-12 strain IM93B (BEI Resources, Manaas, VA, USA) as the recipient. Briefly, all isolates were inoculated into 4 mL of LB broth and incubated for 5 h at 37 °C with shaking at 200 rpm. The donor strain was then mixed with the recipient strain at a proportion of 1:3 and incubated for 5 h at 37 °C in a shaking incubator at 200 rpm. The mixture was spread on MH plates containing 2000 μg/mL streptomycin and 2 μg/mL colistin and incubated at 37 °C for 24 h to select transconjugants. DNA was extracted from the transconjugants and screened by PCR analysis for mcr-1 as described above to verify that the conjugation occurred. As a control, the recipient and donor were separately plated on a streptomycin plus colistin plate.
4.8. Assessing the Persistence of mcr-1 Plasmids in Biofilms
The persistence of mcr-1 plasmids in the biofilm was assessed using the crystal violet biofilm assay as described by Hassan et al. (2020) [46]. Briefly, mcr-1-positive E. coli were grown in LB broth. The cultures were diluted 100-fold and incubated at 37 °C for 2 h in a shaking incubator at 200 rpm. The optical density at 600 nm (OD600) was adjusted to 0.05. Next, 2 mL aliquots of the cultures were transferred to 5 mL sterile borosilicate glass vials. One set of duplicate vials for each isolate was incubated for 3 days and another for 6 days in an incubator at 37 °C. Then, non-adherent bacterial cells were removed by washing with 2 mL of sterile distilled water. The experiment was performed twice, once to assess the biofilm formation and the second to assess the persistence of mcr-1-carrying plasmids.
To assess biofilm formation, the glass vials were air-dried after washing the non-adherent cells and stained with 2 mL of crystal violet solution (Anamol Laboratories Pvt Ltd., Maharashatra, India) for 15 min at room temperature. The vials were then washed with 2 mL sterile distilled water several times and dried to remove the excess crystal violet. Then, 2 mL of acetic acid (Fisher Scientific, Waltham, MA, USA) was added to each vial, and they were incubated for 1 h at room temperature. The optical density of the suspension was measured at 570 nm using a LabQuest spectrophotometer (Vernier, Beaverton, OR, USA). Vials containing only LB broth were used as negative controls.
To assess the mcr-1-carrying plasmids, the biofilms were resuspended in 1 mL of LB broth after removing the non-adherent bacterial then serially diluted components (10-fold). Next, 100 L of each dilution was spread on MH agar plates without and with 4 g/mL of colistin. The plates were incubated at 37 °C overnight, and colony-forming units (CFU) were counted.
4.9. Data Analysis
Statistical analyses were performed using R v4.1.0 [68]. Figures were generated with ggplot v3.3.5, ggpubr v0.4.0, ggtree v3.0.4, and aplot v0.1.1 [69,70,71]. The differences in growth in biofilm assays were assessed using a Wilcoxon signed-rank test as the data did not follow a normal distribution.
5. Conclusions
Antibiotic use in humans and animals facilitates AMR emergence and dissemination and the interactions between humans, animals, and the environment. Consequently, the One Health approach is instrumental in tackling AMR, locally and globally. Colistin resistance, particularly mcr-1-mediated resistance, has been reported in different hosts and niches and constitutes a prime target for One-Health-based intervention. A few studies have previously reported mcr-1-mediated colistin resistance in Qatar’s humans, animals, and food products. In this study, mcr-1-carrying E. coli were subjected to a rigorous analysis that revealed the mcr-1 (1) was plasmid-borne and transmissible, (2) occurred in diverse genetic backgrounds along with other important ARGs, (3) persists in biofilms at a cost, and (4) can pose a significant problem if not tackled appropriately. Consequently, there is a need to continue investigations and identify and monitor the factors that drive the emergence and spread of mcr in Qatar. This will be critical to developing a suite of interventions under a One Health approach.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/antibiotics11060774/s1, Figure S1: (a) Biofilm growth over six days. The figure shows the absorbance at 570 nm of the crystal violet dye as a proxy for biofilm growth. Measurements were taken in duplicates on day 0 (red), day 3 (green), and day 6 (blue) for each isolate. (b) Representative image of bacterial biofilm growth. #1: mcr-positive E. coli growing in LB broth of day 0. #2: mcr-positive E. coli growing in LB broth of day 3. #3: Bacterial suspension of day 3 after being stained with crystal violet stain and measured at 570 nm using a spectrophotometer. #4: mcr-positive E. coli growing in LB broth of day 6. #5: Bacterial suspension of day 6 after being stained with crystal violet stain and measured at 570 nm using a spectrophotometer. Table S1: Genome accessions and sample sources.
Author Contributions
Conceptualization, N.O.E.; methodology, N.O.E., A.A.J. and H.A.M. software, N.O.E. and H.A.M.; validation, N.O.E., H.A.M. and I.I.K.; formal analysis, H.A.M. and N.O.E.; resources, N.O.E.; data curation, N.O.E. and H.A.M.; writing—original draft preparation, H.A.M. and A.A.J.; writing—review and editing, N.O.E. and I.I.K.; supervision, N.O.E.; project administration, N.O.E.; funding acquisition, N.O.E. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Biomedical Research Centre, Qatar University, grant number (BRC-2022-ID-01) to Nahla O. Eltai.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The genome assemblies included in this study are available at the NCBI website (https://www.ncbi.nlm.nih.gov/bioproject, accessed on 1 November 2021) under BioProject PRJNA786273. The assembly accessions are listed in Table S1.
Acknowledgments
The authors would like to acknowledge the administrative staff for assisting in all the paperwork.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the study’s design; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Ross, S.; Puig, J.R.; Zaremba, E.A. Colistin: Some preliminary laboratory and clinical observations in specific gastroenteritis in infants and children. Antibiot Annu. 1960, 7, 89–100. [Google Scholar]
- Akajagbor, D.S.; Wilson, S.L.; Shere-Wolfe, K.D.; Dakum, P.; Charurat, M.E.; Gilliam, B.L. Higher Incidence of Acute Kidney Injury With Intravenous Colistimethate Sodium Compared With Polymyxin B in Critically Ill Patients at a Tertiary Care Medical Center. Clin. Infect. Dis. 2013, 57, 1300–1303. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Kasiakou, S.K.; Saravolatz, L.D. Colistin: The Revival of Polymyxins for the Management of Multidrug-Resistant Gram-Negative Bacterial Infections. Clin. Infect. Dis. 2005, 40, 1333–1341. [Google Scholar] [CrossRef] [PubMed]
- Velkov, T.; Thompson, P.E.; Nation, R.L.; Li, J. Structure−Activity Relationships of Polymyxin Antibiotics. J. Med. Chem. 2009, 53, 1898–1916. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Brunel, J.-M.; Dubus, J.-C.; Reynaud-Gaubert, M.; Rolain, J.-M. Colistin: An update on the antibiotic of the 21st century. Expert Rev. Anti-Infect. Ther. 2012, 10, 917–934. [Google Scholar] [CrossRef] [PubMed]
- Olaitan, A.O.; Morand, S.; Rolain, J.M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Xavier, B.B.; Lammens, C.; Ruhal, R.; Kumar-Singh, S.; Butaye, P.; Goossens, H.; Malhotra-Kumar, S. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 2016, 7, 21. [Google Scholar] [CrossRef]
- Yin, W.; Li, H.; Shen, Y.; Liu, Z.; Wang, S.; Shen, Z.; Zhang, R.; Walsh, T.R.; Shen, J.; Wang, Y. Novel Plasmid-Mediated Colistin Resistance Gene mcr-3 in Escherichia coli. mBio 2017, 8, e00543-17. [Google Scholar] [CrossRef]
- Carattoli, A.; Villa, L.; Feudi, C.; Curcio, L.; Orsini, S.; Luppi, A.; Pezzotti, G.; Magistrali, C.F. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Eurosurveillance 2017, 22, 30589. [Google Scholar] [CrossRef]
- Borowiak, M.; Fischer, J.; Hammerl, J.A.; Hendriksen, R.S.; Szabo, I.; Malorny, B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J. Antimicrob. Chemother. 2017, 72, 3317–3324. [Google Scholar] [CrossRef] [PubMed]
- AbuOun, M.; Stubberfield, E.J.; Duggett, N.A.; Kirchner, M.; Dormer, L.; Nunez-Garcia, J.; Randall, L.P.; Lemma, F.; Crook, D.W.; Teale, C.; et al. mcr-1 and mcr-2 (mcr-6.1) variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J. Antimicrob. Chemother. 2018, 73, 2904. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-Q.; Li, Y.-X.; Lei, C.-W.; Zhang, A.; Wang, H.-N. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2018, 73, 1791–1795. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, Y.; Zhou, Y.; Li, J.; Yin, W.; Wang, S.; Zhang, S.; Shen, J.; Shen, Z.; Wang, Y. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microbes Infect. 2018, 7, 122. [Google Scholar] [CrossRef]
- Carroll, L.M.; Gaballa, A.; Guldimann, C.; Sullivan, G.; Henderson, L.O.; Wiedmann, M. Identification of Novel Mobilized Colistin Resistance Gene mcr-9 in a Multidrug-Resistant, Colistin-Susceptible Salmonella enterica Serotype Typhimurium Isolate. mBio 2019, 10, e00853-19. [Google Scholar] [CrossRef]
- Wang, C.; Feng, Y.; Liu, L.; Wei, L.; Kang, M.; Zong, Z. Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 2020, 9, 508–516. [Google Scholar] [CrossRef]
- Khedher, M.B.; Baron, S.A.; Riziki, T.; Ruimy, R.; Raoult, D.; Diene, S.M.; Rolain, J.-M. Massive analysis of 64,628 bacterial genomes to decipher water reservoir and origin of mobile colistin resistance genes: Is there another role for these enzymes? Sci Rep. 2020, 10, 5970. [Google Scholar] [CrossRef]
- Feng, Y. Transferability of MCR-1/2 Polymyxin Resistance: Complex Dissemination and Genetic Mechanism. ACS Infect. Dis. 2018, 4, 291–300. [Google Scholar] [CrossRef]
- El-Sayed Ahmed, M.A.E.G.; Zhong, L.L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.B. Colistin and its role in the Era of antibiotic resistance: An extended review (2000–2019). Emerg. Microbes Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef]
- Lima, T.; Domingues, S.; Da Silva, G.J. Plasmid-Mediated Colistin Resistance in Salmonella enterica: A Review. Microorganisms 2019, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Feng, Y.; Lü, X.; McNally, A.; Zong, Z. IncP Plasmid Carrying Colistin Resistance Gene mcr-1 in Klebsiella pneumoniae from Hospital Sewage. Antimicrob. Agents Chemother. 2017, 61, e02229-16. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.S.; Feng, Y.; Lv, X.Y.; Duan, J.H.; Chen, J.; Fang, L.X.; Xia, J.; Liao, X.-P.; Sun, J.; Liu, Y.-H. Emergence of NDM-5- and MCR-1-Producing Escherichia coli Clones ST648 and ST156 from a Single Muscovy Duck (Cairina moschata). Antimicrob. Agents Chemother. 2016, 60, 6899–6902. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Hu, Y.; Li, Z.; Sun, J.; Wang, Q.; Lin, J.; Ye, H.; Liu, F.; Srinivas, S.; Li, D.; et al. Dissemination and Mechanism for the MCR-1 Colistin Resistance. PLoS Pathog. 2016, 12, e1005957. [Google Scholar] [CrossRef]
- Matamoros, S.; van Hattem, J.M.; Arcilla, M.S.; Willemse, N.; Melles, D.C.; Penders, J.; Vinh, T.N.; Thi Hoa, N.; Bootsma, M.C.J.; van Genderen, P.J.; et al. Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci Rep. 2017, 7, 15364. [Google Scholar] [CrossRef]
- Wang, Q.; Sun, J.; Li, J.; Ding, Y.; Li, X.P.; Lin, J.; Hassan, B.; Feng, Y. Expanding landscapes of the diversified mcr-1-bearing plasmid reservoirs. Microbiome 2017, 5, 70. [Google Scholar] [CrossRef]
- Rebelo, A.R.; Bortolaia, V.; Kjeldgaard, J.S.; Pedersen, S.K.; Leekitcharoenphon, P.; Hansen, I.M.; Guerra, B.; Malorny, B.; Borowiak, M.; Hammerl, J.A.; et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 2018, 23, 17-00672. [Google Scholar] [CrossRef]
- Eltai, N.O.; Yassine, H.M.; El-Obeid, T.; Al-Hadidi, S.H.; Al Thani, A.A.; Alali, W.Q. Prevalence of Antibiotic-Resistant Escherichia coli Isolates from Local and Imported Retail Chicken Carcasses. J Food Prot. 2020, 83, 2200–2208. [Google Scholar] [CrossRef]
- Johar, A.; Al-Thani, N.; Al-Hadidi, S.; Dlissi, E.; Mahmoud, M.; Eltai, N. Antibiotic Resistance and Virulence Gene Patterns Associated with Avian Pathogenic Escherichia coli (APEC) from Broiler Chickens in Qatar. Antibiotics 2021, 10, 564. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Khalmeter, G.; Olsson-Lilijequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Arredondo-Alonso, S.; Rogers, M.R.C.; Braat, J.C.; Verschuuren, T.D.; Top, J.; Corander, J.; Willems, R.J.L.; Schürch, A.C.Y. mlplasmids: A user-friendly tool to predict plasmid- and chromosome-derived sequences for single species. Microb. Genom. 2018, 4, e000224. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.H.K.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2020, 76, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Elbediwi, M.; Li, Y.; Paudyal, N.; Pan, H.; Li, X.; Xie, S.; Rajkovic, A.; Feng, Y.; Fang, W.; Rankin, S.C.; et al. Global Burden of Colistin-Resistant Bacteria: Mobilized Colistin Resistance Genes Study (1980–2018). Microorganisms 2019, 7, 461. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, R.; Fujiya, Y.; Yamaguchi, T.; Khong, D.T.; Nguyen, T.N.; Tran, H.T.; Yamamoto, Y. Most Domestic Livestock Possess Colistin-Resistant Commensal Escherichia coli Harboring mcr in a Rural Community in Vietnam. Antimicrob. Agents Chemother. 2019, 63, e00594-19. [Google Scholar] [CrossRef]
- Hmede, Z.; Kassem, I.I. The Colistin Resistance Gene mcr-1 Is Prevalent in Commensal Escherichia coli Isolated from Preharvest Poultry in Lebanon. Antimicrob. Agents Chemother. 2018, 62, e01304-18. [Google Scholar] [CrossRef]
- Rumi, M.V.; Mas, J.; Elena, A.; Cerdeira, L.; Muñoz, M.E.; Lincopan, N.; Gentilini, É.R.; Di Conza, J.; Gutkind, G. Co-occurrence of clinically relevant β-lactamases and MCR-1 encoding genes in Escherichia coli from companion animals in Argentina. Vet Microbiol. 2019, 230, 228–234. [Google Scholar] [CrossRef]
- Merida-Vieyra, J.; Ranero, A.D.C.; Arzate-Barbosa, P.; Garza EA de la Méndez-Tenorio, A.; Murcia-Garzón, J.; Aquino-Andrade, A. First clinical isolate of Escherichia coli harboring mcr-1 gene in Mexico. PLoS ONE 2019, 14, e0214648. [Google Scholar] [CrossRef]
- Eltai, N.O.; Abdfarag, E.A.; Al-Romaihi, H.; Wehedy, E.; Mahmoud, M.H.; Alawad, O.K.; Al-Hajri, M.M.; Al Thani, A.A.; Yassine, H.M. Antibiotic Resistance Profile of Commensal Escherichia coli Isolated from Broiler Chickens in Qatar. J. Food Prot. 2018, 81, 302–307. [Google Scholar] [CrossRef]
- McEwen, S.A.; Collignon, P.J. Antimicrobial Resistance: A One Health Perspective. Microbiol. Spectr. 2018, 6, 255–260. [Google Scholar] [CrossRef]
- Gordon, D.M.; Cowling, A. The distribution and genetic structure of Escherichia coli in Australian vertebrates: Host and geographic effects. Microbiology 2003, 149, 3575–3586. [Google Scholar] [CrossRef]
- Carlos, C.; Pires, M.M.; Stoppe, N.C.; Hachich, E.M.; Sato, M.I.; Gomes, T.A.; Amaral, L.A.; Ottoboni, L.M. Escherichia coli phylogenetic group determination and its application in the identification of the major animal source of fecal contamination. BMC Microbiol. 2010, 10, 161. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Monk, J.M.; Mih, N.; Du, B.; Sastry, A.V.; Kavvas, E.; Seif, Y.; Smarr, L.; Palsson, B.O. Escherichia coli B2 strains prevalent in inflammatory bowel disease patients have distinct metabolic capabilities that enable colonization of intestinal mucosa. BMC Syst. Biol. 2018, 12, 66. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Castillo, D.; Esposito, F.; Cardoso, B.; Dalazen, G.; Moura, Q.; Fuga, B.; Fontana, H.; Cerdeira, L.; Dropa, M.; Rottmann, J.; et al. Genomic data reveal international lineages of critical priority Escherichia coli harbouring wide resistome in Andean condors (Vultur gryphus Linnaeus, 1758). Mol. Ecol. 2020, 29, 1919–1935. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Wang, Y.; Xiao, Y. Prevalence and transmission of mobilized colistin resistance (mcr) gene in bacteria common to animals and humans. Biosaf. Health 2020, 2, 71–78. [Google Scholar] [CrossRef]
- Kassem, I.I.; Mann, D.; Li, S.; Deng, X. Draft genome sequences and resistome analysis of multidrug-resistant mcr-1-harbouring Escherichia coli isolated from pre-harvest poultry in Lebanon. J. Glob. Antimicrob. Resist. 2021, 25, 114–116. [Google Scholar] [CrossRef] [PubMed]
- Hassan, J.; Eddine, R.Z.; Mann, D.; Li, S.; Deng, X.; Saoud, I.P.; Issmat, K.I. The Mobile Colistin Resistance Gene, mcr-1.1, Is Carried on IncX4 Plasmids in Multidrug Resistant E. coli Isolated from Rainbow Trout Aquaculture. Microorganisms 2020, 8, 1636. [Google Scholar] [CrossRef]
- Li, R.; Yu, H.; Xie, M.; Chen, K.; Dong, N.; Lin, D.; Chan, E.W.-C.; Chen, S. Genetic basis of chromosomally-encoded mcr-1 gene. Int. J. Antimicrob. Agents 2017, 51, 578–585. [Google Scholar] [CrossRef]
- Baur, B.; Hanselmann, K.; Schlimme, W.; Jenni, B. Genetic transformation in freshwater: Escherichia coli is able to develop natural competence. Appl. Environ. Microbiol. 1996, 62, 3673–3678. [Google Scholar] [CrossRef]
- Sinha, S.; Cameron, A.D.S.; Redfield, R.J. Sxy Induces a CRP-S Regulon in Escherichia coli. J. Bacteriol. 2009, 191, 5180–5195. [Google Scholar] [CrossRef]
- Sinha, S.; Redfield, R.J. Natural DNA Uptake by Escherichia coli. PLoS ONE 2012, 7, e35620. [Google Scholar] [CrossRef]
- Anjum, M.F.; Duggett, N.A.; AbuOun, M.; Randall, L.; Nunez-Garcia, J.; Ellis, R.J.; Rogers, J.; Horton, R.; Brena, C.; Williamson, S.; et al. Colistin resistance in Salmonella and Escherichia coli isolates from a pig farm in Great Britain. J. Antimicrob. Chemother. 2016, 71, 2306–2313. [Google Scholar] [CrossRef] [PubMed]
- Bowler, P.; Murphy, C.; Wolcott, R. Biofilm exacerbates antibiotic resistance: Is this a current oversight in antimicrobial stewardship? Antimicrob. Resist. Infect. Control. 2020, 9, 162. [Google Scholar] [CrossRef] [PubMed]
- Nang, S.C.; Morris, F.C.; McDonald, M.J.; Han, M.L.; Wang, J.; Strugnell, R.A.; Velkov, T.; Li, J. Fitness cost of mcr-1-mediated polymyxin resistance in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2018, 73, 1604–1610. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing (M100), 31st ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2021. [Google Scholar]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2016, 13, e1005595. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Beghain, J.; Bridier-Nahmias, A.; Le Nagard, H.; Denamur, E.; Clermont, O. ClermonTyping: An easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 2018, 4, e000192. [Google Scholar] [CrossRef]
- Inouye, M.; Dashnow, H.; Raven, L.; Schultz, M.B.; Pope, B.J.; Tomita, T.; Zobel, J.; Holt, K.E. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. bioRxiv 2014, 6, 90. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef] [PubMed]
- Roer, L.; Tchesnokova, V.; Allesøe, R.; Muradova, M.; Chattopadhyay, S.; Ahrenfeldt, J.; Thomsen, M.C.F.; Lund, O.; Hansen, F.; Hammerum, A.M.; et al. Development of a Web Tool for Escherichia coli Subtyping Based on fimH Alleles. J. Clin. Microbiol. 2017, 55, 2538–2543. [Google Scholar] [CrossRef]
- Wong, S.C.Y.; Tse, H.; Chen, J.H.K.; Cheng, V.C.C.; Ho, P.L.; Yuen, K.Y. Colistin-Resistant Enterobacteriaceae Carrying the mcr-1 Gene among Patients in Hong Kong. Emerg. Infect. Dis. 2016, 22, 1667–1669. [Google Scholar] [CrossRef] [PubMed]
- Froger, A.; Hall, J.E. Transformation of Plasmid DNA into E. coli Using the Heat Shock Method. J. Vis. Exp. 2007, 6, e253. [Google Scholar] [CrossRef]
- Hmede, Z.; Sulaiman, A.A.A.; Jaafar, H.; Kassem, I.I. Emergence of plasmid-borne colistin resistance gene mcr-1 in multidrug-resistant Escherichia coli isolated from irrigation water in Lebanon. Int. J. Antimicrob. Agents 2019, 54, 102–104. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2011; Available online: http://www.r-project.org/ (accessed on 24 February 2022).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016; Available online: https://ggplot2.tidyverse.org (accessed on 24 February 2022).
- Yu, G. Using ggtree to Visualize Data on Tree-Like Structures. Curr. Protoc. Bioinform. 2020, 69, e96. [Google Scholar] [CrossRef]
- Yu, G. Aplot: Decorate a “ggplot” with Associated Information. 2021. Available online: https://CRAN.R-project.org/package=aplot (accessed on 24 February 2022).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).