Metabolic Profile, Biotransformation, Docking Studies and Molecular Dynamics Simulations of Bioactive Compounds Secreted by CG3 Strain
Abstract
:1. Introduction
2. Results and Discussion
2.1. Antimicrobial Activity of Strain CG3
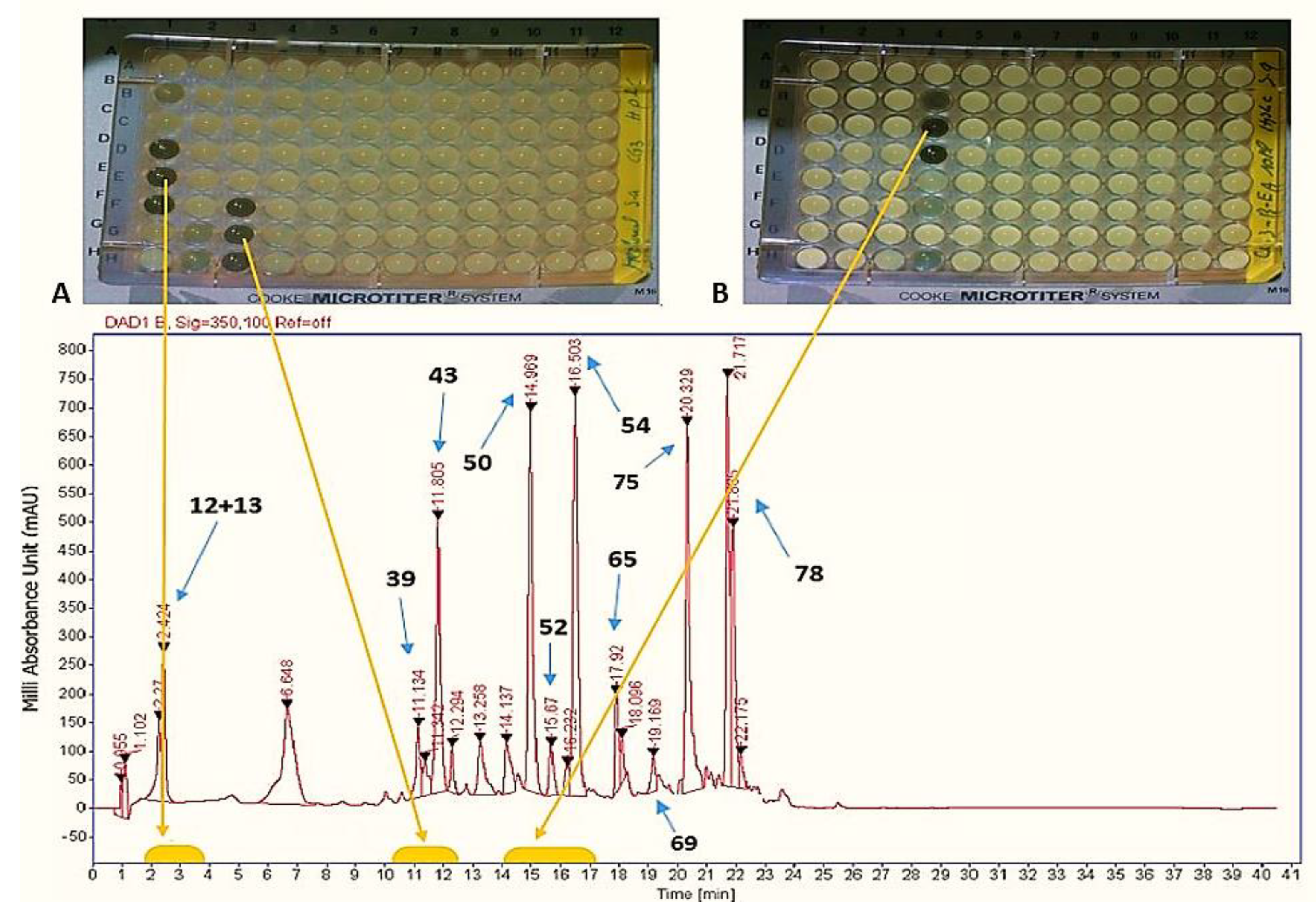
2.2. Fractionation of Crude Extract
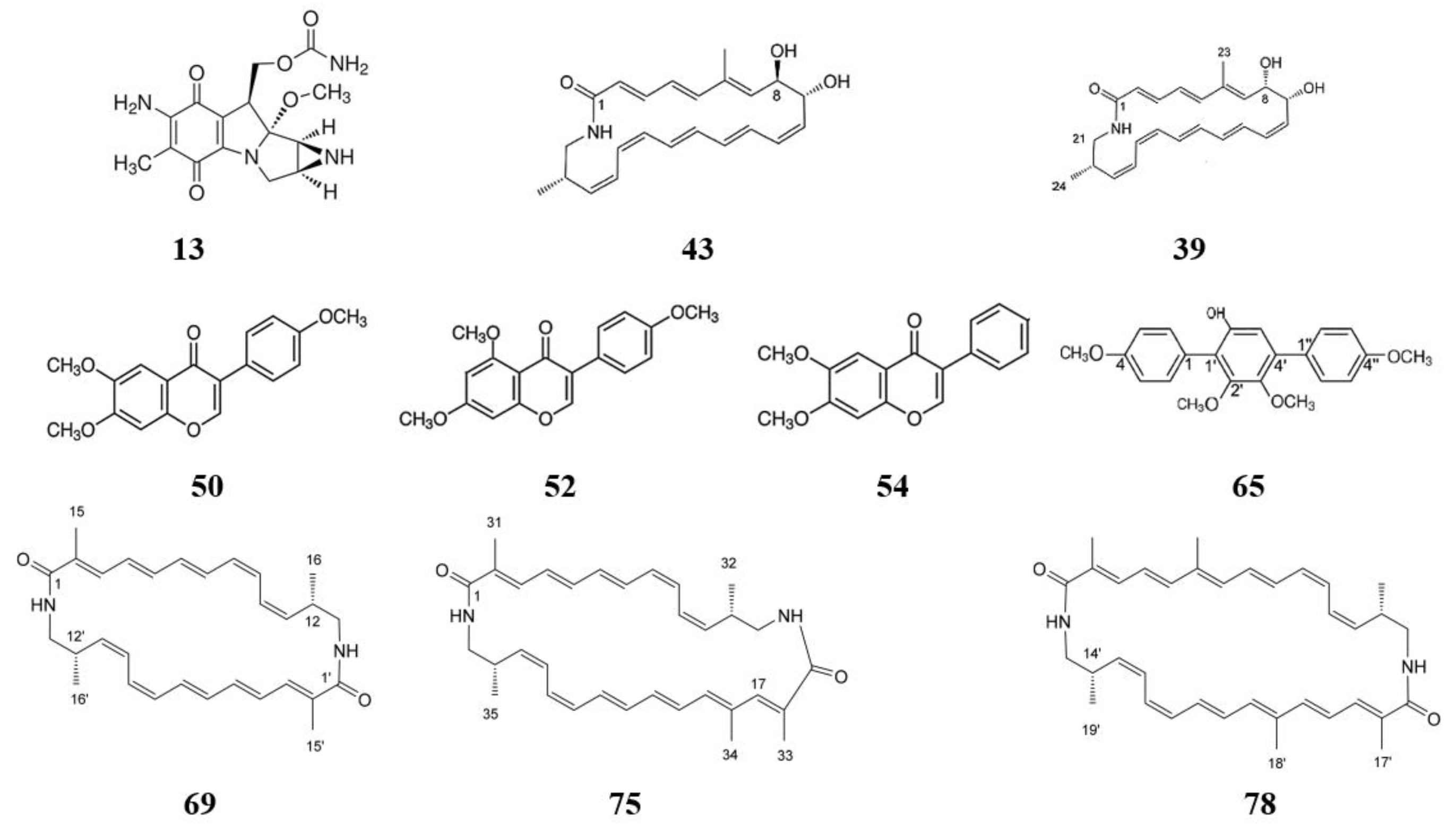
2.3. HPLC-UV-HRESIMS Analysis of Crude Extracts
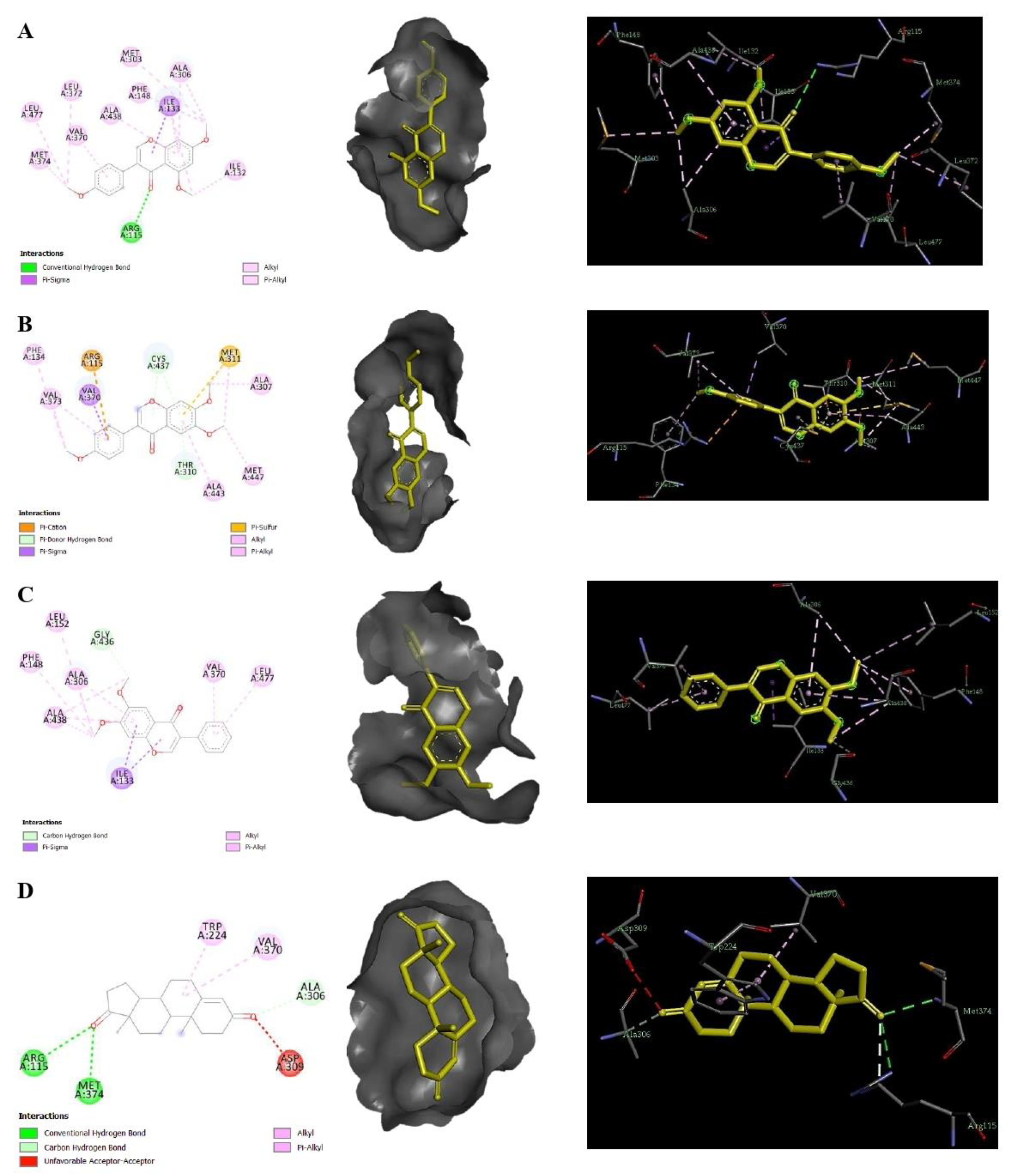
2.4. Molecular Docking Using AutoDock
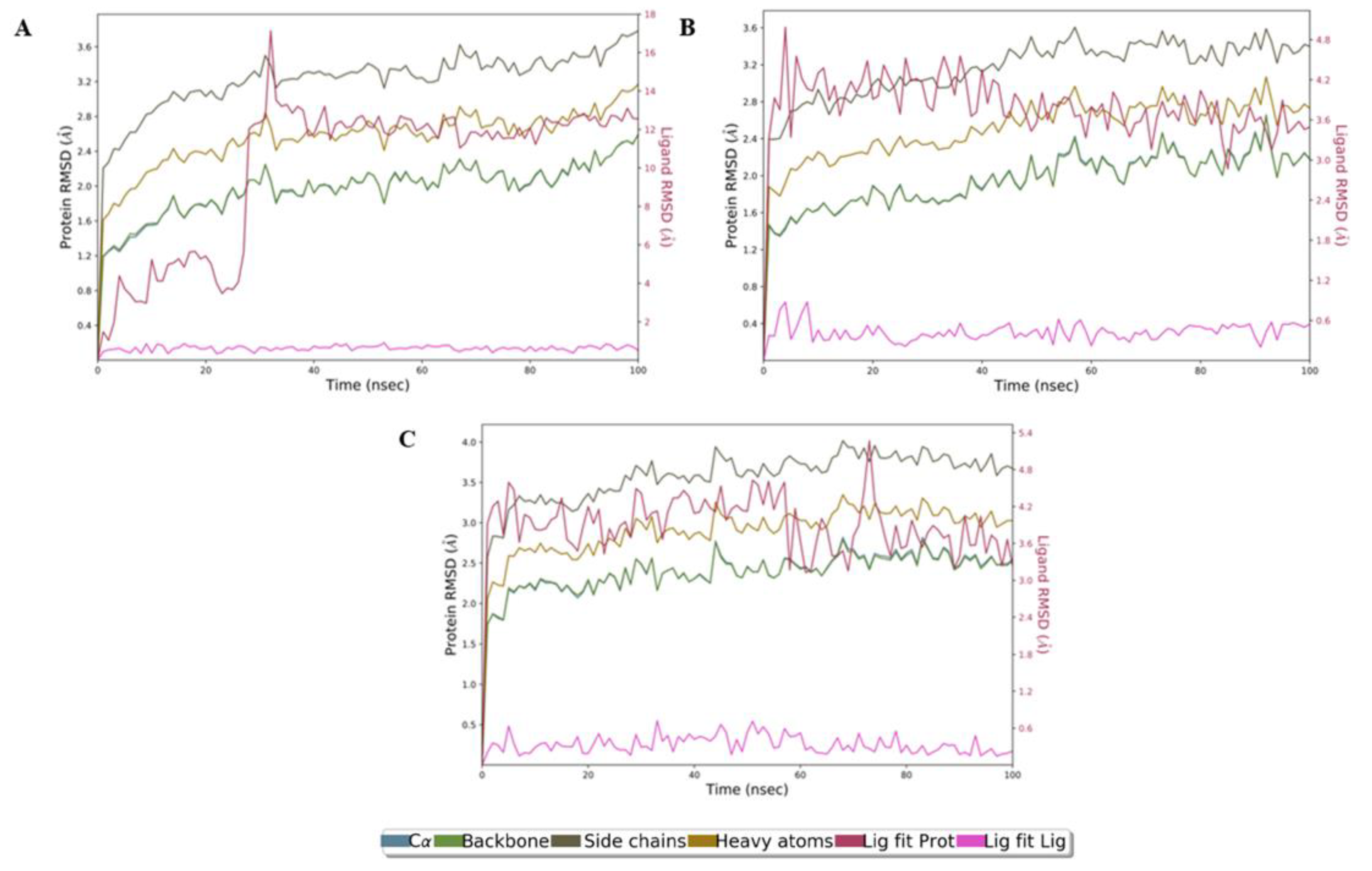
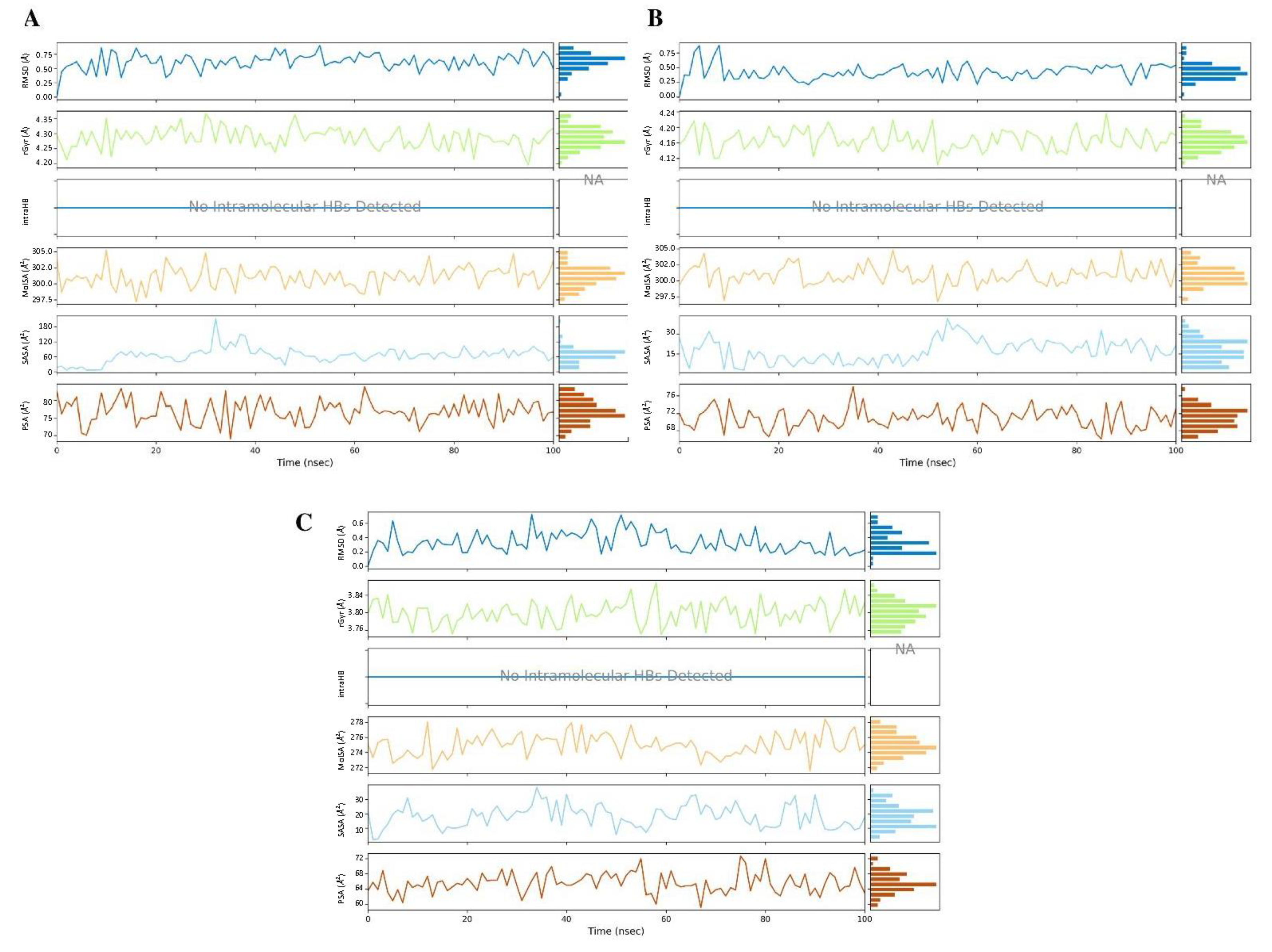
2.5. Molecular Dynamics Simulation
3. Materials and Methods
3.1. Selective Isolation of CG3 and Identification
3.2. Evaluation of Antimicrobial Activity
3.2.1. Preparation of Suspension
3.2.2. Preparation of Crude Extract
3.2.3. Serial Dilution Method for Antimicrobial Activity
3.3. Analytical HPLC and Fractionation of Crude Extract
3.4. Metabolic Profile
3.5. Docking Studies and Molecular Dynamics Simulations
3.5.1. Protein and Ligand Preparation
3.5.2. Molecular Docking Analysis
3.5.3. Molecular Dynamics Simulation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arocha-Garza, H.F.; Castillo RCDEguiarte, L.E.; Souza, V.; De la Torre-Zavala, S. High diversity and suggested endemicity of culturable Actinobacteria in an extremely oligotrophic desert oasis. PeerJ 2017, 5, e3247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Messaoudi, O.; Bendahou, M.; Benamur, I.; Abdelwouhid, D. Identification and preliminary characterization of non-polyene antibiotics secreted by new strain of actinomycete isolated from Sebkha of Kenadsa, Algeria. Asian Pac. J. Trop. Biomed. 2015, 5, 438–445. [Google Scholar] [CrossRef] [Green Version]
- Horikoshi, K. Alkaliphiles: Some applications of their products for biotechnology. Microbiol. Mol. Biol. Rev. 1999, 63, 735–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meklat, A.; Sabaou, N.; Zitouni, A.; Mathieu, F.; Lebrihi, A. Isolation, taxonomy, and antagonistic properties of halophilic actinomycetes in Saharan soils of Algeria. Appl. Environ. Microbiol. 2011, 77, 6710–6714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohamed, H.; Miloud, B.; Zohra, F.; García-Arenzana, J.M.; Veloso, A.; Rodríguez-Couto, S. Isolation and characterization of actinobacteria from Algerian Sahara soils with antimicrobial activities. Int. J. Mol. Cell Med. 2017, 6, 109–120. [Google Scholar]
- Wang, J.; Zhang, R.; Chen, X.; Sun, X.; Yan, Y.; Shen, X.; Yuan, Q. Biosynthesis of aromatic polyketides in microorganisms using type II polyketide synthases. Microb. Cell Factories 2020, 19, 110. [Google Scholar] [CrossRef] [PubMed]
- Yun, C.S.; Nishimoto, K.; Motoyama, T.; Shimizu, T.; Hino, T.; Dohmae, N.; Nagano, S.; Osada, H. Unique features of the ketosynthase domain in a nonribosomal peptide synthetase-polyketide synthase hybrid enzyme, tenuazonic acid synthetase 1. J. Biol. Chem. 2020, 295, 11602–11612. [Google Scholar] [CrossRef]
- Tang, S.; Zhang, W.; Li, Z.; Li, H.; Geng, C.; Huang, X.; Lu, X. Discovery and characterization of a PKS-NRPS hybrid in aspergillus terreus by genome mining. J. Nat. Prod. 2020, 83, 473–480. [Google Scholar] [CrossRef]
- Wei, M.; Wang, S.; Shang, G. Biosynthetic pathways and engineering for bioactive natural products. Curr. Org. Chem. 2010, 14, 1433–1446. [Google Scholar] [CrossRef]
- Flissi, A.; Ricart, E.; Campart, C.; Chevalier, M.; Dufresne, Y.; Michalik, J.; Jacques, P.; Flahaut, C.; Lisacek, F.; Leclère, V.; et al. Norine: Update of the nonribosomal peptide resource. Nucleic Acids Res. 2020, 48, D465–D469. [Google Scholar]
- Seeger, M.; González, M.; Cámara, B.; Muñoz, L.; Ponce, E.; Mejías, L.; Mascayano, C.; Vásquez, Y.; Sepúlveda-Boza, S. Biotransformation of natural and synthetic isoflavonoids by two recombinant microbial enzymes. Appl. Environ. Microbiol. 2003, 69, 5045–5050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Matos, I.L.; Nitschke, M.; Da Fonseca, L.J.P.; Porto, A.L.M. Biotransformation of flavonoids by terrestrial and marine microorganisms. Encycl. Mar. Biotechnol. 2020, 3, 1979–2000. [Google Scholar]
- Coccia, A.; Mosca, L.; Puca, R.; Mangino, G.; Rossi, A.; Lendaro, E. Extra-virgin olive oil phenols block cell cycle progression and modulate chemotherapeutic toxicity in bladder cancer cells. Oncol. Rep. 2016, 36, 3095–3104. [Google Scholar] [CrossRef] [Green Version]
- Soo, V.W.C.; Kwan, B.W.; Quezada, H.; Castillo-Juárez, I.; Pérez-Eretza, B.; García-Contreras, S.J.; Martínez-Vázquez, M.; Wood, T.K.; García-Contreras, R. Repurposing of anticancer drugs for the treatment of bacterial infections. Curr. Top. Med. Chem. 2017, 17, 1157–1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Messaoudi, O.; Sudarman, E.; Bendahou, M.; Jansen, R.; Stadler, M.; Wink, J. Kenalactams A–E, polyene macrolactams isolated from Nocardiopsis CG3. J. Nat. Prod. 2019, 82, 1081–1088. [Google Scholar] [CrossRef] [PubMed]
- Saah, E.F.; Sielinou, V.T.; Kuete, V.; Lacmata, S.T.; Nkengfack, A.E. Antimicrobial and antioxidant isoflavonoid derivatives from the roots of Amphimas pterocarpoides. Z. Naturforsch. B J. Chem. Sci. 2013, 68, 931–938. [Google Scholar] [CrossRef]
- Popa, D.S.; Rusu, M.E. Isoflavones: Vegetable sources, biological activity, and analytical methods for their assessment. In Superfood and Functional Food—The Development of Superfoods and Their Roles as Medicine; Shiomi, N., Waisundara, V., Eds.; InTech: London, UK, 2017; ISBN 978-953-51-2942-4. [Google Scholar]
- Tsao, R. Chemistry and biochemistry of dietary polyphenols. Nutrients 2010, 2, 1231–1246. [Google Scholar] [CrossRef]
- Jung, Y.S.; Rha, C.S.; Baik, M.Y.; Baek, N.I.; Kim, D.O. A brief history and spectroscopic analysis of soy isoflavones. Food Sci. Biotechnol. 2020, 29, 1605–1617. [Google Scholar] [CrossRef]
- Liao, L.; Zhou, Y.; Peng, T.; Guo, Y.; Zhao, Y.; Zeng, Z. Crystal structure of a S-adenosyl-L-methionine-dependent O-methyltransferase-like enzyme from Aspergillus flavus. Proteins 2021, 89, 185–192. [Google Scholar] [CrossRef]
- Setchell, K.D.R.; Brown, N.M.; Desai, P.; Zimmer-Nechemias, L.; Wolfe, B.E.; Brashear, W.T.; Kirschner, A.S.; Cassidy, A.; Heubi, J.E. Bioavailability of pure isoflavones in healthy humans and analysis of commercial soy isoflavone supplements. J. Nutr. 2001, 131, 1362S–1375S. [Google Scholar] [CrossRef] [Green Version]
- Goto, H.; Terao, Y.; Akai, S. Synthesis of various kinds of isoflavones, isoflavanes, and biphenyl-ketones and their 1,1-diphenyl-2-picrylhydrazyl radical-scavenging activities. Chem. Pharm. Bull. 2009, 57, 346–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, G.Y.; Lim, Y.; Koh, D.; Huh, J.S.; Hyun, C.; Kim, Y.M.; Cho, M. TMF and glycitin act synergistically on keratinocytes and fibroblasts to promote wound healing and anti-scarring activity. Exp. Mol. Med. 2017, 49, e302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, S.Z.; Pu, X.; Luo, G.; Zhao, L.X.; Xu, L.H.; Li, W.J.; Luo, Y. Isolation and characterization of new p-Terphenyls with antifungal, antibacterial, and antioxidant activities from halophilic actinomycete Nocardiopsis gilva YIM 90087. J. Agric Food Chem. 2013, 61, 3006–3012. [Google Scholar] [CrossRef]
- Fañanas-Baquero, S.; Orman, I.; Becerra Aparicio, F.; Bermudez de Miguel, S.; Garcia Merino, J.; Yañez, R.; Fernandez Sainz, Y.; Sánchez, R.; Dessy-Rodríguez, M.; Alberquilla, O.; et al. Natural estrogens enhance the engraftment of human hematopoietic stem and progenitor cells in immunodeficient mice. Haematologica 2020, 105, 1–60. [Google Scholar] [CrossRef] [PubMed]
- Hatono, M.; Ikeda, H.; Suzuki, Y.; Kajiwara, Y.; Kawada, K.; Tsukioki, T.; Kochi, M.; Suzawa, K.; Iwamoto, T.; Yamamoto, H.; et al. Effect of isoflavones on breast cancer cell development and their impact on breast cancer treatments. Breast Cancer Res. Treat. 2021, 185, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Vegunta, S.; Kling, J.M.; Kapoor, E. Androgen therapy in women. J. Womens Health 2020, 29, 57–64, Erratum in: J. Womens Health 2020, 29, 1487. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Cao, S.; Zhang, Z.; Li, L.; Chen, F.; Wu, Q. Multiple omics analysis of the protective effects of SFN on estrogen-dependent breast cancer cells. Mol. Biol. Rep. 2020, 47, 3331–3346. [Google Scholar] [CrossRef] [PubMed]
- Khosrow-Khavar, F.; Filion, K.B.; Bouganim, N.; Suissa, S.; Azoulay, L. Aromatase inhibitors and the risk of cardiovascular outcomes in women with breast cancer: A population-based cohort study. Circulation 2020, 141, 549–559. [Google Scholar] [CrossRef]
- Balam, F.H.; Ahmadi, Z.S.; Ghorbani, A. Inhibitory effect of chrysin on estrogen biosynthesis by suppression of enzyme aromatase (CYP19): A systematic review. Heliyon 2020, 6, e03557. [Google Scholar] [CrossRef]
- Kharb, R.; Haider, K.; Neha, K.; Yar, M.S. Aromatase inhibitors: Role in postmenopausal breast cancer. Arch. Pharm. 2020, 353, e2000081. [Google Scholar] [CrossRef]
- Brueggemeier, R.W.; Hackett, J.C.; Diaz-Cruz, E.S. Aromatase inhibitors in the treatment of breast cancer. Endocr. Rev. 2005, 26, 331–345. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, D.; Griswold, J.; Erman, M.; Pangborn, W. Structural basis for androgen specificity and oestrogen synthesis in human aromatase. Nature 2009, 457, 219–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suvannang, N.; Nantasenamat, C.; Isarankura-Na-Ayudhya, C.; Prachayasittikul, V. Molecular docking of aromatase inhibitors. Molecules 2011, 16, 3597–3617. [Google Scholar] [CrossRef] [Green Version]
- Chayawan, C.; Toma, C.; Benfenati, E.; Caballero Alfonso, A.Y. Towards an understanding of the mode of action of human aromatase activity for azoles through quantum chemical descriptors-based regression and structure activity relationship modeling analysis. Molecules 2020, 25, 739. [Google Scholar] [CrossRef] [Green Version]
- Messaoudi, O.; Wink, J.; Bendahou, M. Diversity of Actinobacteria isolated from date palms rhizosphere and saline environments: Isolation, identification and biological activity evaluation. Microorganisms 2020, 8, 1853. [Google Scholar] [CrossRef] [PubMed]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Thakur, D. Antimicrobial biosynthetic potential and diversity of culturable soil actinobacteria from forest ecosystems of Northeast India. Sci. Rep. 2020, 10, 4104. [Google Scholar] [CrossRef]
- Leite, R.F.; Gonçalves, J.L.; Peti, A.P.F.; Figueiró, F.S.; Moraes, L.A.B.; Santos, M.V. Antimicrobial activity of crude extracts from actinomycetes against mastitis pathogens. J. Dairy Sci. 2018, 101, 10116–10125. [Google Scholar] [CrossRef]
- Ohikhena, F.U.; Wintola, O.A.; Afolayan, A.J. Evaluation of the antibacterial and antifungal properties of Phragmanthera capitata (Sprengel) Balle (Loranthaceae), a mistletoe growing on rubber tree, using the dilution techniques. Sci. World J. 2017, 2017, 9658598. [Google Scholar] [CrossRef] [Green Version]
- Messaoudi, O. Isolement et Caractérisation de Nouvelles Molécules Bioactives à Partir D’Actinomycètes Isolées du sol Algérien. Ph.D. Thesis, Université de Tlemcen, Tlemcen, Algeria, 2020. [Google Scholar]
- Jordan, K.W.; Nordenstam, J.; Lauwers, G.Y.; Rothenberger, D.A.; Alavi, K.; Garwood, M.; Cheng, L.L. Metabolomic characterization of human rectal adenocarcinoma with intact tissue magnetic resonance spectroscopy. Dis. Colon Rectum. 2009, 52, 520–525. [Google Scholar] [CrossRef] [Green Version]
- Messaoudi, O.; Gouzi, H.; El-Hoshoudy, A.; Benaceur, F.; Patel, C.; Goswami, D.; Boukerouis, D.; Bendahou, M. Berries anthocyanins as potential SARS-CoV–2 inhibitors targeting the viral attachment and replication; molecular docking simulation. Egypt J. Pet. 2021, 30, 33–43. [Google Scholar] [CrossRef]
- Schrödinger Release 2021-4: Desmond Molecular Dynamics System; D. E. Shaw Research: New York, NY, USA, 2021. Maestro-Desmond Interoperability Tools; Schrödinger: New York, NY, USA, 2021. Available online: https://www.schrodinger.com/products/desmond (accessed on 11 April 2022).




| MIC (µg mL−1) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| SM | 2.08 | 66.66 | - | 1.04 | 0.52 | 2.08 | 0.52 | 66.66 | 66.66 | 16.66 |
| ISP2 | 2.08 | 66.66 | - | 1.04 | 0.52 | 2.08 | 0.52 | 66.66 | - | - |
| NC | - | - | - | - | - | - | - | - | - | - |
| PC | 1.66 | 1.66 | 1.66 | 0.52 | 2.08 | 0.05 | 0.52 | 1.66 | 8.8 | 4.16 |
| Compounds | Target Proteins (3EQM); Binding Energy (kcal mol−1). |
|---|---|
| 5,7-Dimethoxy-3-(4-methoxyphenyl)chromen-4-one (52) | −7.5 |
| 6,7-Dimethoxy-3-(4-methoxyphenyl)chromen-4-one (50) | −7.1 |
| 6,7-Dimethoxy-3-phenylchromen-4-one (54) | −7.3 |
| Androstenedione | −9.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Messaoudi, O.; Sudarman, E.; Patel, C.; Bendahou, M.; Wink, J. Metabolic Profile, Biotransformation, Docking Studies and Molecular Dynamics Simulations of Bioactive Compounds Secreted by CG3 Strain. Antibiotics 2022, 11, 657. https://doi.org/10.3390/antibiotics11050657
Messaoudi O, Sudarman E, Patel C, Bendahou M, Wink J. Metabolic Profile, Biotransformation, Docking Studies and Molecular Dynamics Simulations of Bioactive Compounds Secreted by CG3 Strain. Antibiotics. 2022; 11(5):657. https://doi.org/10.3390/antibiotics11050657
Chicago/Turabian StyleMessaoudi, Omar, Enge Sudarman, Chirag Patel, Mourad Bendahou, and Joachim Wink. 2022. "Metabolic Profile, Biotransformation, Docking Studies and Molecular Dynamics Simulations of Bioactive Compounds Secreted by CG3 Strain" Antibiotics 11, no. 5: 657. https://doi.org/10.3390/antibiotics11050657
APA StyleMessaoudi, O., Sudarman, E., Patel, C., Bendahou, M., & Wink, J. (2022). Metabolic Profile, Biotransformation, Docking Studies and Molecular Dynamics Simulations of Bioactive Compounds Secreted by CG3 Strain. Antibiotics, 11(5), 657. https://doi.org/10.3390/antibiotics11050657








