Derivatives of Esculentin-1 Peptides as Promising Candidates for Fighting Infections from Escherichia coli O157:H7
Abstract
:1. Introduction
2. Results
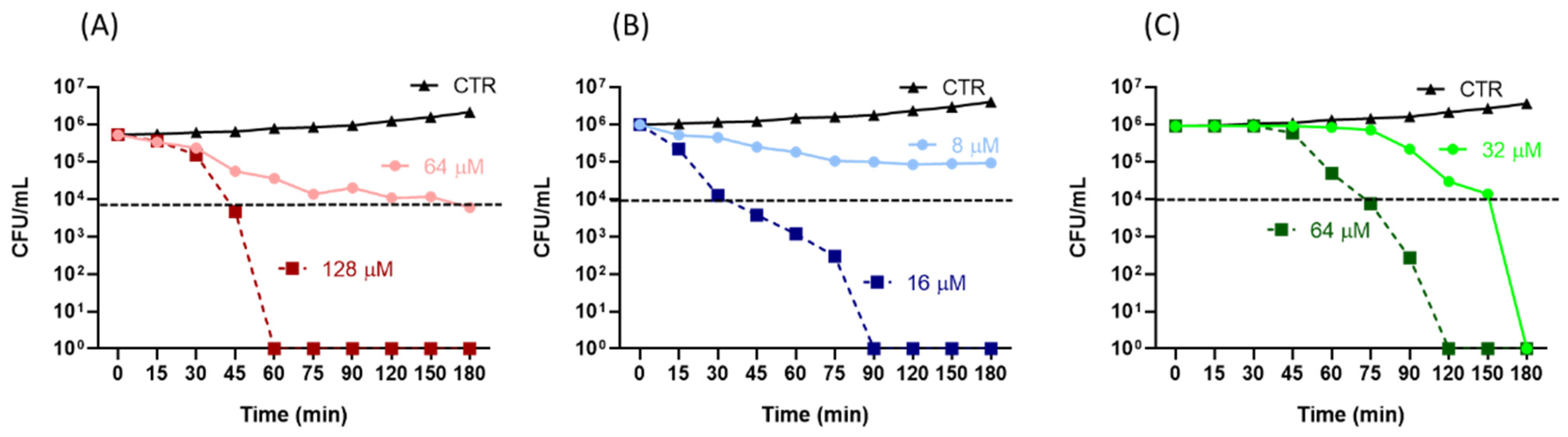
2.1. Antibacterial Activity of the Peptides
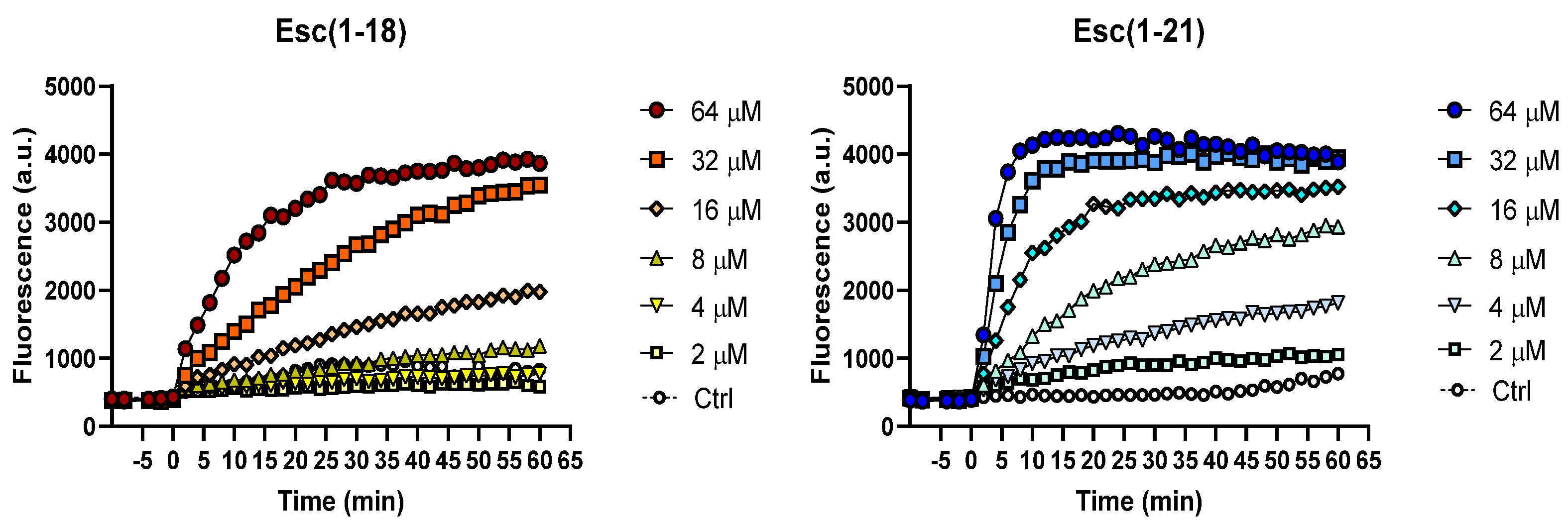
2.2. Inner Membrane Permeation
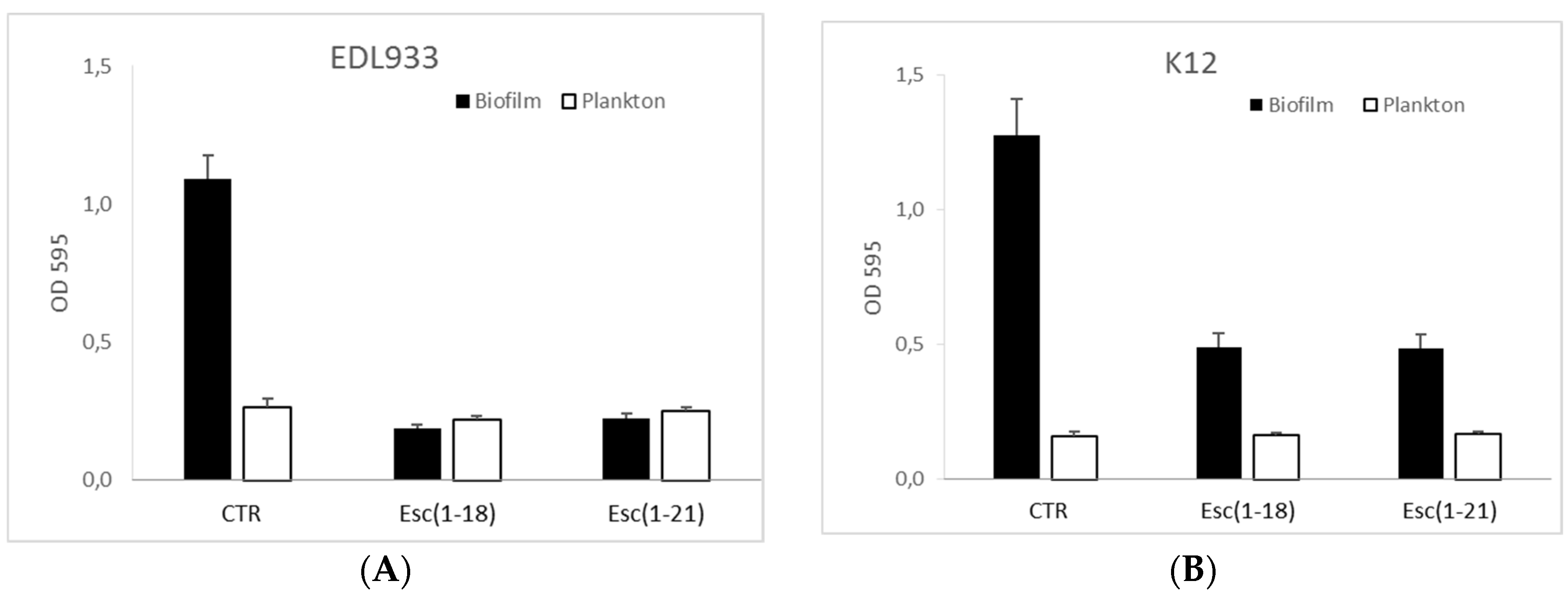
2.3. Antibiofilm Activity
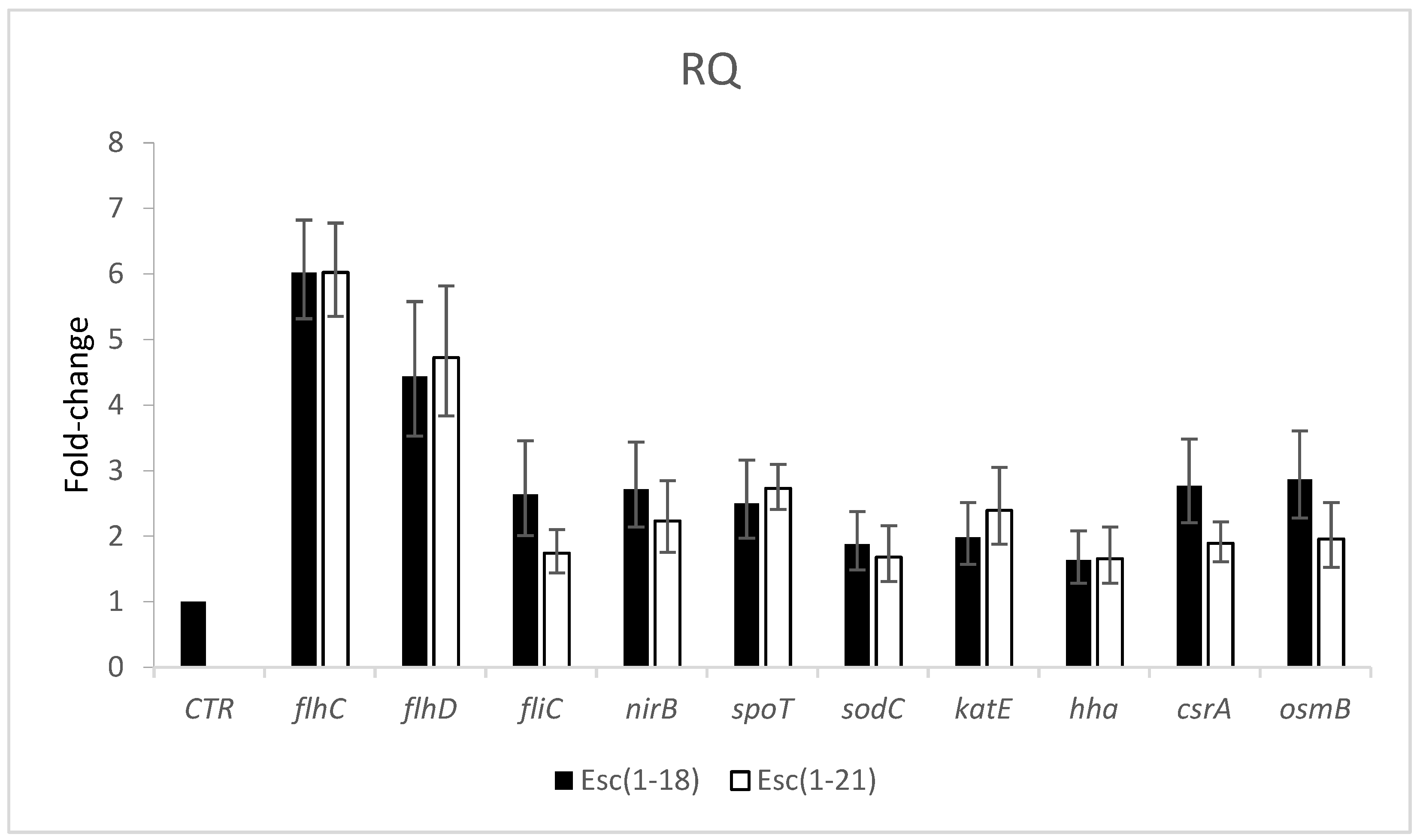
2.4. Effects of Peptides on Gene Expression
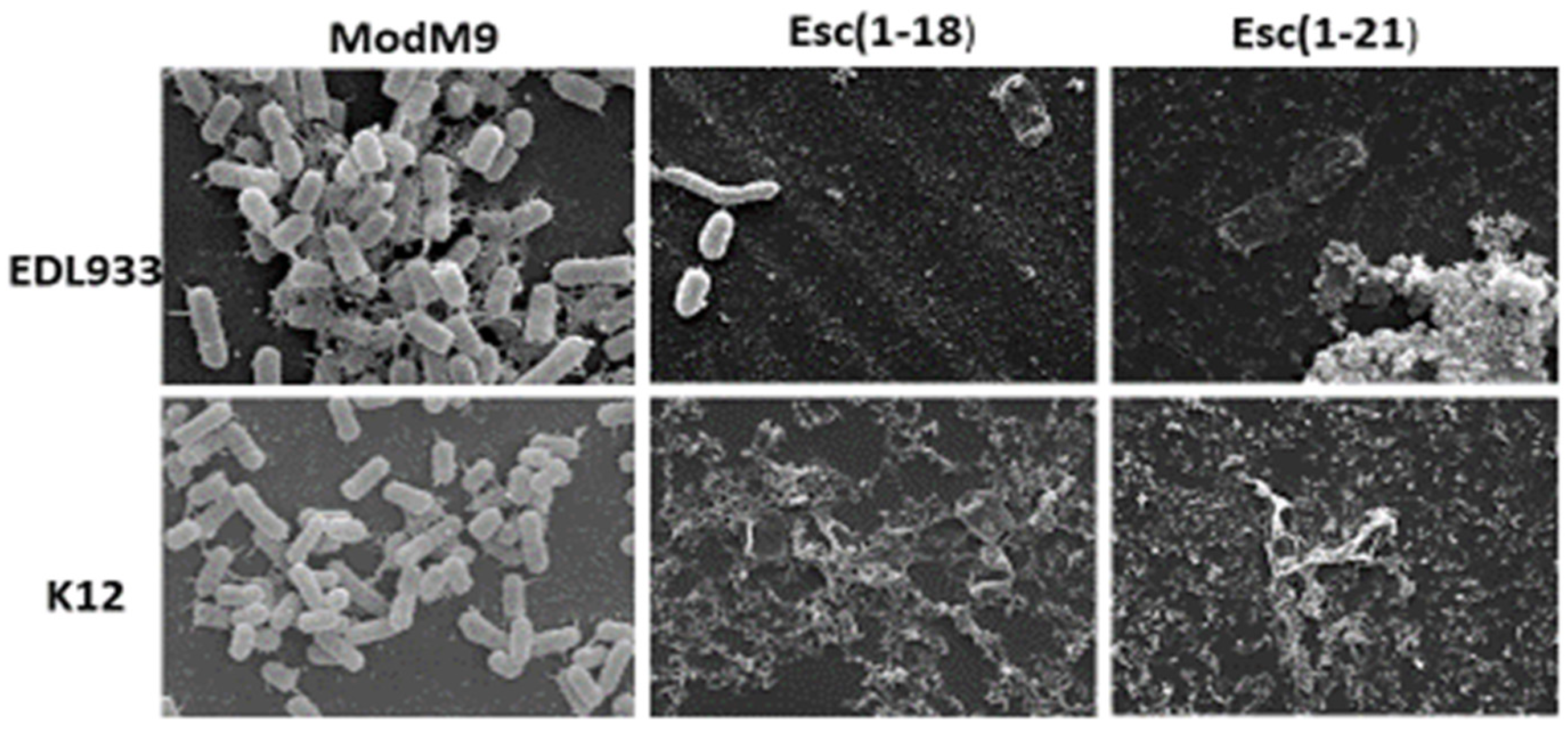
2.5. Scanning Electron Microscopy (SEM)
3. Discussion
4. Materials and Methods
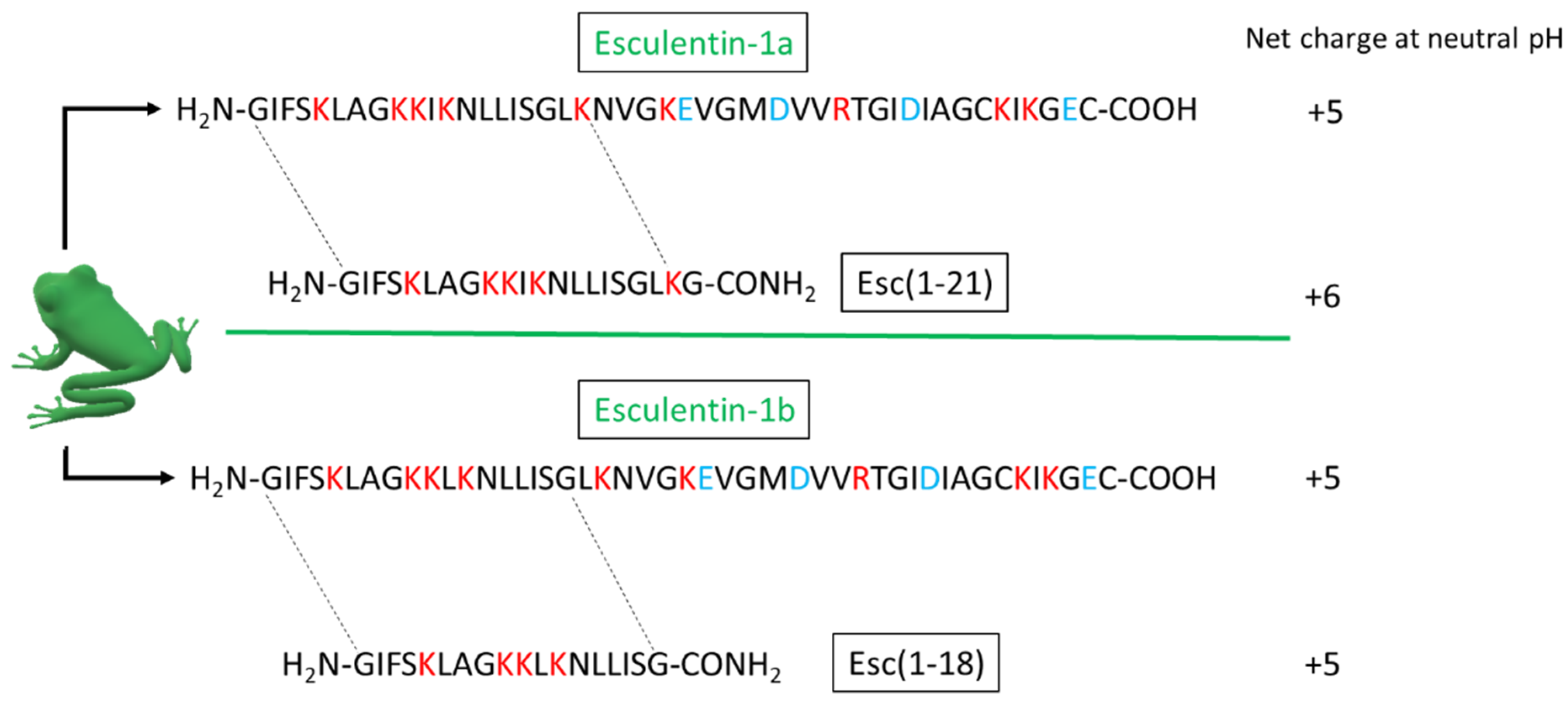
4.1. Peptides
4.2. Bacterial Strains
4.3. MIC and MBC Assays
4.4. Biofilm Formation Assay
4.5. Time-Kill Assay
4.6. Permeation of the Bacterial IM
4.7. RNA Isolation and Quantitative Real-Time RT-PCR
4.8. Scanning Electron Microscopy (SEM)
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hancock, R.E.W.; Alford, M.A.; Haney, E.F. Antibiofilm activity of host defense peptides: Complexity provides opportunities. Nat. Rev. Microbiol. 2021, 19, 786–797. [Google Scholar] [CrossRef] [PubMed]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Römling, U.; Balsalobre, C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 2012, 272, 541–561. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente-Núñez, C.; Reffuveille, F.; Fernández, L.; Hancock, R.E. Bacterial biofilm development as a multicellular adaptation: Antibiotic resistance and new therapeutic strategies. Curr. Opin. Microbiol. 2013, 16, 580–589. [Google Scholar] [CrossRef]
- Paton, J.C.; Paton, A.W. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli Infections. Clin. Microbiol. Rev. 1998, 11, 450–479. [Google Scholar] [CrossRef] [Green Version]
- Lazzaro, B.P.; Zasloff, M.; Rolff, J. Antimicrobial peptides: Application informed by evolution. Science 2020, 368, eaau5480. [Google Scholar] [CrossRef]
- Hancock, R.E.; Sahl, H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef]
- de la Fuente-Núñez, C.; Korolik, V.; Bains, M.; Nguyen, U.; Breidenstein, E.B.; Horsman, S.; Lewenza, S.; Burrows, L.; Hancock, R.E. Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob. Agents Chemother. 2012, 56, 2696–2704. [Google Scholar] [CrossRef] [Green Version]
- Reffuveille, F.; de la Fuente-Núñez, C.; Mansour, S.; Hancock, R.E. A broad-spectrum antibiofilm peptide enhances antibiotic action against bacterial biofilms. Antimicrob. Agents Chemother. 2014, 58, 5363–5371. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.W.; Chen, F.Y.; Lee, M.T. Molecular mechanism of Peptide-induced pores in membranes. Phys. Rev. Lett. 2004, 92, 198304. [Google Scholar] [CrossRef] [Green Version]
- Nicolas, P. Multifunctional host defense peptides: Intracellular-targeting antimicrobial peptides. FEBS J. 2009, 276, 6483–6496. [Google Scholar] [CrossRef]
- Ladram, A.; Nicolas, P. Antimicrobial peptides from frog skin: Biodiversity and therapeutic promises. Front. Biosci. 2016, 21, 1341–1371. [Google Scholar] [CrossRef] [Green Version]
- Simmaco, M.; Mignogna, G.; Barra, D. Antimicrobial peptides from amphibian skin: What do they tell us? Biopolymers 1998, 47, 435–450. [Google Scholar] [CrossRef]
- Mangoni, M.L.; Luca, V.; McDermott, A.M. Fighting microbial infections: A lesson from amphibian skin-derived esculentin-1 peptides. Peptides 2015, 71, 286–295. [Google Scholar] [CrossRef]
- Simmaco, M.; Mignogna, G.; Barra, D.; Bossa, F. Antimicrobial peptides from skin secretions of Rana esculenta. Molecular cloning of cDNAs encoding esculentin and brevinins and isolation of new active peptides. J. Biol. Chem. 1994, 269, 11956–11961. [Google Scholar] [CrossRef]
- Islas-Rodrìguez, A.E.; Marcellini, L.; Orioni, B.; Barra, D.; Stella, L.; Mangoni, M.L. Esculentin 1–21: A linear antimicrobial peptide from frog skin with inhibitory effect on bovine mastitis-causing bacteria. J. Pept. Sci. 2009, 15, 607–614. [Google Scholar] [CrossRef]
- Marcellini, L.; Borro, M.; Gentile, G.; Rinaldi, A.C.; Stella, L.; Aimola, P.; Barra, D.; Mangoni, M.L. Esculentin-1b(1-18)-a membrane-active antimicrobial peptide that synergizes with antibiotics and modifies the expression level of a limited number of proteins in Escherichia coli. FEBS J. 2009, 276, 5647–5664. [Google Scholar] [CrossRef] [Green Version]
- Luca, V.; Stringaro, A.; Colone, M.; Pini, A.; Mangoni, M.L. Esculentin(1-21), an amphibian skin membrane-active peptide with potent activity on both planktonic and biofilm cells of the bacterial pathogen Pseudomonas aeruginosa. Cell Mol. Life Sci. 2013, 70, 2773–2786. [Google Scholar] [CrossRef]
- Casciaro, B.; Lin, Q.; Afonin, S.; Loffredo, M.R.; de Turris, V.; Middel, V.; Ulrich, A.S.; Di, Y.P.; Mangoni, M.L. Inhibition of Pseudomonas aeruginosa biofilm formation and expression of virulence genes by selective epimerization in the peptide Esculentin-1a(1-21)NH2. FEBS J. 2019, 286, 3874–3891. [Google Scholar] [CrossRef]
- Luca, V.; Olivi, M.; Di Grazia, A.; Palleschi, C.; Uccelletti, D.; Mangoni, M.L. Anti-Candida activity of 1-18 fragment of the frog skin peptide esculentin-1b: In vitro and in vivo studies in a Caenorhabditis elegans infection model. Cell Mol. Life Sci. 2014, 71, 2535–2546. [Google Scholar] [CrossRef]
- Casciaro, B.; Loffredo, M.R.; Cappiello, F.; Verrusio, W.; Corleto, V.D.; Mangoni, M.L. Frog Skin-Derived Peptides Against Corynebacterium jeikeium: Correlation between Antibacterial and Cytotoxic Activities. Antibiotics 2020, 9, 448. [Google Scholar] [CrossRef] [PubMed]
- Mangoni, M.L.; Fiocco, D.; Mignogna, G.; Barra, D.; Simmaco, M. Functional characterisation of the 1-18 fragment of esculentin-1b, an antimicrobial peptide from Rana esculenta. Peptides 2003, 24, 1771–1777. [Google Scholar] [CrossRef] [PubMed]
- Scotti, R.; Stringaro, A.; Nicolini, L.; Zanellato, M.; Boccia, P.; Maggi, F.; Gabbianelli, R. Effects of Essential Oils from Cymbopogon spp. and Cinnamomum verum on Biofilm and Virulence Properties of Escherichia coli O157:H7. Antibiotics 2021, 10, 113. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.L.; Brun-Zinkernagel, A.M.; Simecka, J.W.; Prüss, B.M.; Babitzke, P.; Romeo, T. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 2001, 40, 245–256. [Google Scholar] [CrossRef]
- Liu, X.; Matsumura, P. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J. Bacteriol. 1994, 176, 7345–7351. [Google Scholar] [CrossRef] [Green Version]
- Timmermans, J.; Van Melderen, L. Post-transcriptional global regulation by CsrA in bacteria. Cell Mol. Life Sci. 2010, 67, 2897–2908. [Google Scholar] [CrossRef]
- Sharma, V.K.; Bearson, B.L. Hha Controls Escherichia coli O157:H7 Biofilm Formation by Differential Regulation of Global Transcriptional Regulators FlhDC and CsgD. Appl. Environ. Microbiol. 2013, 79, 2384–2396. [Google Scholar] [CrossRef] [Green Version]
- Durfee, T.; Hansen, A.M.; Zhi, H.; Blattner, F.R.; Jin, D.J. Transcription profiling of the stringent response in Escherichia coli. J. Bacteriol. 2008, 190, 1084–1096. [Google Scholar] [CrossRef] [Green Version]
- Sanyal, R.; Vimala, A.; Harinarayanan, R. Studies on the Regulation of (p)ppGpp Metabolism and Its Perturbation Through the Over-Expression of Nudix Hydrolases in Escherichia coli. Front. Microbiol. 2020, 11, 562804, e Collection 2020. [Google Scholar] [CrossRef]
- Pacios, O.; Blasco, L.; Bleriot, I.; Fernandez-Garcia, L.; Ambroa, A.; López, M.; Bou, G.; Cantón, R.; Garcia-Contreras, R.; Wood, T.K.; et al. (p)ppGpp and Its Role in Bacterial Persistence: New Challenges. Antimicrob. Agents Chemother. 2020, 64, e01283-20. [Google Scholar] [CrossRef]
- de la Fuente-Núñez, C.; Reffuveille, F.; Haney, E.F.; Straus, S.K.; Hancock, R.E. Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog. 2014, 10, e1004152. [Google Scholar] [CrossRef] [Green Version]
- Pratt, L.A.; Kolter, R. Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 1998, 30, 285–293. [Google Scholar] [CrossRef]
- Prigent-Combaret, C.; Prensier, G.; Le Thi, T.T.; Vidal, O.; Lejeune, P.; Dorel, C. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: Role of flagella, curli and colanic acid. Environ. Microbiol. 2000, 2, 450–464. [Google Scholar] [CrossRef]
- Pesavento, C.; Becker, G.; Sommerfeldt, N.; Possling, A.; Tschowri, N.; Mehlis, A.; Hengge, R. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 2008, 22, 2434–2446. [Google Scholar] [CrossRef] [Green Version]
- Khlebodarova, T.M.; Ree, N.A.; Likhoshvai, V.A. On the control mechanisms of the nitrite level in Escherichia coli cells: The mathematical model. BMC Microbiol. 2016, 16 (Suppl. 1), 7. [Google Scholar] [CrossRef] [Green Version]
- McDougald, D.; Rice, S.A.; Barraud, N.; Steinberg, P.D.; Kjelleberg, S. Should we stay or should we go: Mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 2011, 10, 39–50. [Google Scholar] [CrossRef]
- Arora, D.P.; Hossain, S.; Xu, Y.; Boon, E.M. Nitric Oxide Regulation of Bacterial Biofilms. Biochemistry 2015, 54, 3717–3728. [Google Scholar] [CrossRef]
- Park, J.-S.; Choi, H.-Y.; Kim, W.-G. The Nitrite Transporter Facilitates Biofilm Formation via Suppression of Nitrite Reductase and is a New Antibiofilm Target in Pseudomonas aeruginosa. mBIO 2020, 11, e00878-20. [Google Scholar] [CrossRef]
- Barraud, N.; Storey, M.V.; Moore, Z.P.; Webb, J.S.; Rice, S.A.; Kjelleberg, S. Nitric oxide-mediated dispersal in single- and multi-species biofilms of clinically and industrially relevant microorganisms. Microb. Biotechnol. 2009, 2, 370–378. [Google Scholar] [CrossRef] [Green Version]
- Barraud, N.; Kelso, M.J.; Rice, S.A.; Kjelleberg, S. Nitric oxide: A key mediator of biofilm dispersal with applications in infectious diseases. Curr. Pharm. Des. 2015, 21, 31–42. [Google Scholar] [CrossRef] [Green Version]
- Zhu, C.; Chen, J.; Wang, Y.; Wang, J.; Wang, L.; Guo, X.; Chen, N.; Zheng, P.; Sun, J.; Ma, Y. Enhancing 5-aminolevulinic acid tolerance and production by engineering the antioxidant defense system of Escherichia coli. Biotechnol. Bioeng. 2019, 116, 2018–2028. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A Common Mechanism of Cellular Death Induced by Bactericidal Antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.; Yang, Z.; Weisshaar, J.C. Single-cell, real-time detection of oxidative stress induced in Escherichia coli by the antimicrobial peptide CM15. Proc. Natl. Acad. Sci. USA 2015, 112, E303–E310. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Dong, S.L.; Xu, F.; Wang, X.Q.; Withers, T.R.; Yu, H.D.; Wang, X. Effect of intracellular expression of antimicrobial peptide LL-37 on growth of Escherichia coli strain TOP10 under aerobic and anaerobic conditions. Antimicrob. Agents Chemother. 2013, 57, 4707–4716. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.; Hwang, J.S.; Lee, D.G. Antibacterial action of lactoferricin B like peptide against Escherichia coli: Reactive oxygen species-induced apoptosis-like death. J. Appl. Microbiol. 2020, 129, 287–295. [Google Scholar] [CrossRef]
- Charoenwong, D.; Andrews, S.; Mackey, B. Role of rpoS in the development of cell envelope resilience and pressure resistance in stationary-phase Escherichia coli. Appl. Environ. Microbiol. 2011, 77, 5220–5229. [Google Scholar] [CrossRef] [Green Version]
- Gabbianelli, R.; Scotti, R.; Ammendola, S.; Petrarca, P.; Nicolini, L.; Battistoni, A. Role of ZnuABC and ZinT in Escherichia coli O157:H7 zinc acquisition and interaction with epithelial cells. BMC Microbiol. 2011, 11, 36. [Google Scholar] [CrossRef]
- M07-A9; Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. 9th ed. Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2012; Volume 32.
- Scotti, R.; Nicolini, L.; Stringaro, A.; Gabbianelli, R. A study on prophagic and chromosomal sodC genes involvement in Escherichia coli O157:H7 biofilm formation and biofilm resistance to H2O2. Ann. Ist. Super Sanità 2015, 51, 62–66. [Google Scholar] [CrossRef]
- Jackson, D.W.; Suzuki, K.; Oakford, L.; Simecka, J.W.; Mark E Hart, M.E.; Romeo, R. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 2002, 184, 290–301. [Google Scholar] [CrossRef] [Green Version]
- González, M.G.; Robino, L.; Iribarnegaray, V.; Zunino, P.; Scavone, P. Effect of different antibiotics on biofilm produced by uropathogenic Escherichia coli isolated from children with urinary tract infection. Pathog. Dis. 2017, 75, ftx053. [Google Scholar] [CrossRef] [Green Version]
- M26-A; Methods for Determining Bactericidal Activity of Antimicrobial Agents. Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 1999; Volume 19.
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
| Strain | Esc(1-18) | Esc(1-21) | Kanamycin | |||
|---|---|---|---|---|---|---|
| EDL933 | MIC | 32 | MIC | 4 | MIC | 16 |
| MBC | 64 | MBC | 8 | MBC | 32 | |
| K12 | MIC | 16 | MIC | 2 | MIC | 16 |
| MBC | 32 | MBC | 4 | MBC | 32 | |
| Oligo Name | Sequence 5′-3′ | |
|---|---|---|
| 16s | For | Rev |
| CATCCACAGAACTTTCCAGAG | CCAACATTTCACAACACGAG | |
| csrA | For | Rev |
| TTAGTAACTGGACTGCTGGG | GTTGGTGAGACCCTCATGAT | |
| flhC | For | Rev |
| CGACTGGTTTATGACTTGGG | CTGGTGAGCGTGGGTAATAA | |
| flhD | For | Rev |
| CCGAGTTGCTGAAACACATTTA | ATTTATGCCGAGACGAAACA | |
| fliC | For | Rev |
| TTGCCGACTATACAGTCTCTTAC | TTGGTAGTGGTGTTGTTCAG | |
| Hha | For | Rev |
| AGGAAGGGATCTTGTCGTACAG | TCGTTGCCAGACAATTGACAC | |
| katE | For | Rev |
| GTATTCATACCTTCCGCCTG | GTGCTTCATCCCAAACGAG | |
| nirB | For | Rev |
| TTACCTCGACGAAAGCAAAG | GCAGTTTATCAACACCGATAGA | |
| osmB | For | Rev |
| GTTCTAACTGGTCTAAACGGG | CCTAATGTACCCAACGTACTG | |
| relA | For | Rev |
| GATTACTGCTTCCGTTATCTCC | CGACCATACACTTCAGCTTTA | |
| sodC | For | Rev |
| GTCGAGATGAACCTCGTCAG | TCCAGACCTTTATCGGTTTCA | |
| spoT | For | Rev |
| ATGGCTGTGGAATGGGATAA | TCTTTCTCTTCCGTATTCAAACT | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scotti, R.; Casciaro, B.; Stringaro, A.; Morgia, F.; Mangoni, M.L.; Gabbianelli, R. Derivatives of Esculentin-1 Peptides as Promising Candidates for Fighting Infections from Escherichia coli O157:H7. Antibiotics 2022, 11, 656. https://doi.org/10.3390/antibiotics11050656
Scotti R, Casciaro B, Stringaro A, Morgia F, Mangoni ML, Gabbianelli R. Derivatives of Esculentin-1 Peptides as Promising Candidates for Fighting Infections from Escherichia coli O157:H7. Antibiotics. 2022; 11(5):656. https://doi.org/10.3390/antibiotics11050656
Chicago/Turabian StyleScotti, Raffaella, Bruno Casciaro, Annarita Stringaro, Fabrizio Morgia, Maria Luisa Mangoni, and Roberta Gabbianelli. 2022. "Derivatives of Esculentin-1 Peptides as Promising Candidates for Fighting Infections from Escherichia coli O157:H7" Antibiotics 11, no. 5: 656. https://doi.org/10.3390/antibiotics11050656
APA StyleScotti, R., Casciaro, B., Stringaro, A., Morgia, F., Mangoni, M. L., & Gabbianelli, R. (2022). Derivatives of Esculentin-1 Peptides as Promising Candidates for Fighting Infections from Escherichia coli O157:H7. Antibiotics, 11(5), 656. https://doi.org/10.3390/antibiotics11050656









