The Use of Bacteriophages in Biotechnology and Recent Insights into Proteomics
Abstract
:1. Introduction
2. Exploiting Bacteriophage Proteomes
3. Phage Based Methods
3.1. Phage Display
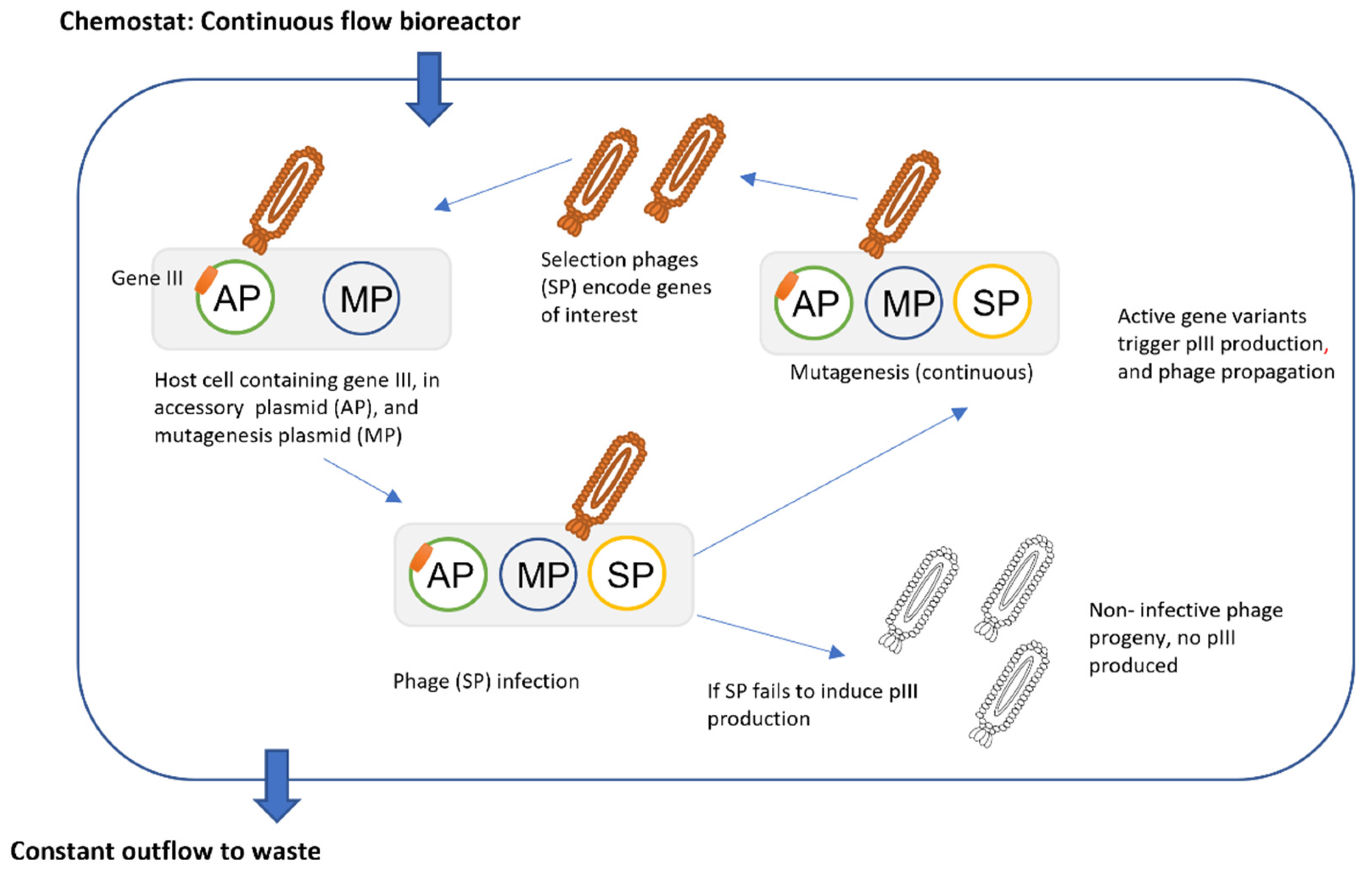
3.2. Phage Assisted Evolution
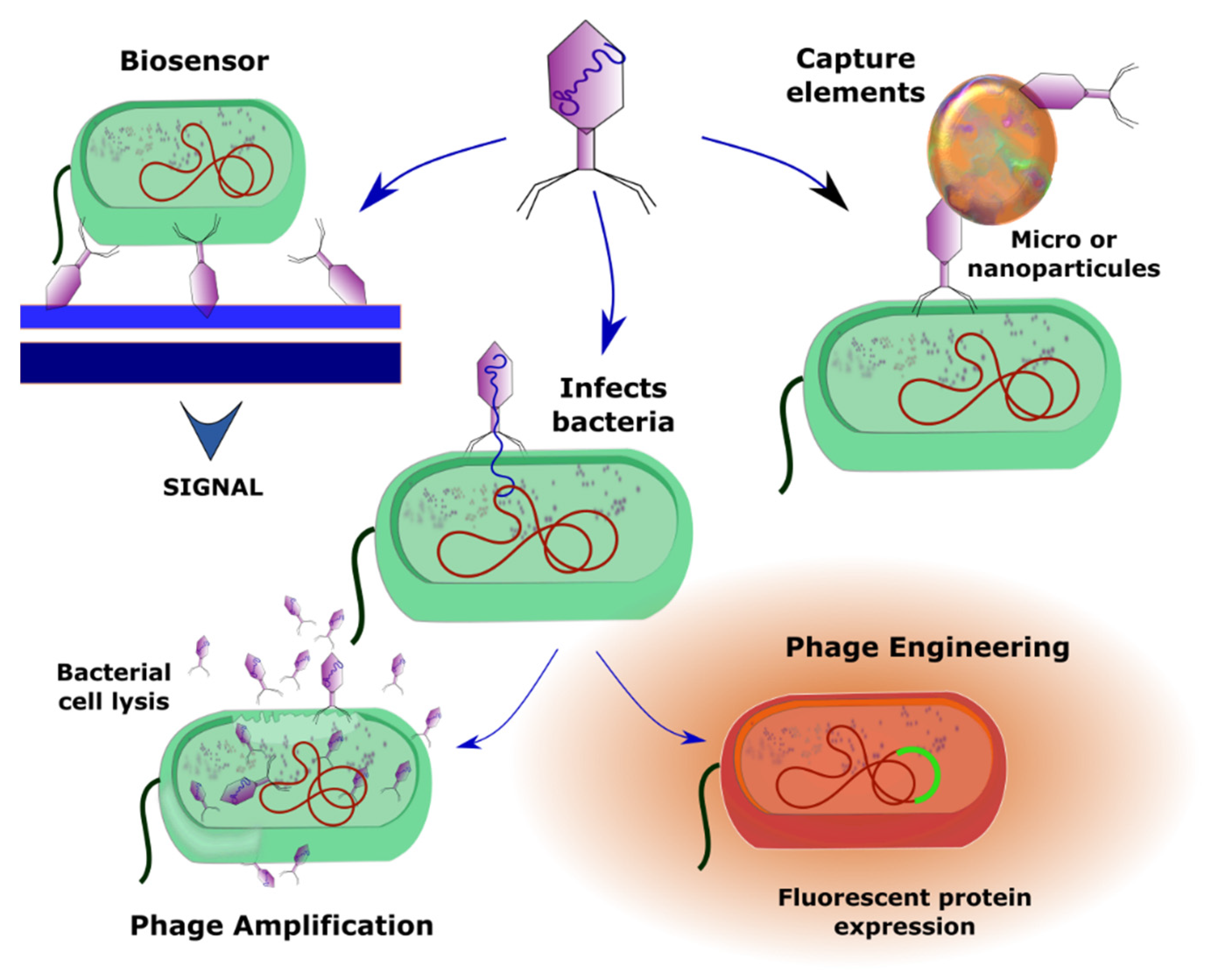
3.3. Phage Amplification-Based Detection
3.4. Phage Engineering
3.5. Biosensors
4. Mass Spectrometry (MS)-Based Proteomics
4.1. Discovery Proteomics
4.2. Targeted Proteomics
4.3. Identification of Bacteriophage-Derived Proteins for Bacteria Detection by MS-Based Phage Proteomics
| Bacteriophages | Sample Source | Analytical Method | Reference |
|---|---|---|---|
| Triaviruses, Phietaviruses, Biseptimaviruses, Kayviruses, Twortvirus, P68virus | reference and isolates | MALDI-TOF MS | [193] |
| Kayvirus K1/420 | medical isolate | CZE, MALDI-TOF MS | [197] |
| Staphylococcal phages (K1/420, 11, P68) | physiological saline solution, human serum | MALDI-TOF MS | [199] |
| Staphylococcal phages (K1/420, 11, P68, 3A, 77) | blood, serum | MALDI-TOF MS | [196] |
| Yersinia pestis phage ϕA1122 and E. coli phage MS2 | MALDI-TOF MS | [191] | |
| Methicillin-resistant Staphylococcus aureus phages | MALDI-TOF MS | [192] | |
| Streptococcus spp. bacteriophages | Dairy products from mastitis | LC-ESI-MS/MS | [194] |
| Staphylococcus spp. bacteriophages | Dairy products from mastitis | LC-ESI-MS/MS | [195] |
| E. coli lamda phage | LC-ESI-MS/MS | [23] |
5. Bacteriophage as Antimicrobials
6. Bacteriophage as Vaccines
7. Concluding Remarks and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Hess, K.L.; Jewell, C.M. Phage display as a tool for vaccine and immunotherapy development. Bioeng. Transl. Med. 2020, 5, e10142. [Google Scholar] [CrossRef] [PubMed]
- Koskella, B.; Brockhurst, M.A. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol. Rev. 2014, 38, 916–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fortier, L.C.; Sekulovic, O. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence 2013, 4, 354–365. [Google Scholar] [CrossRef]
- Menouni, R.; Hutinet, G.; Agn‘, M.-A.; Petit, A.; Ansaldi, M. Bacterial genome remodeling through bacteriophage recombination. FEMS Microbiol. Lett. 2015, 362, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Canchaya, C.; Fournous, G.; Brüssow, H. The impact of prophages on bacterial chromosomes. Mol. Microbiol. 2004, 53, 9–18. [Google Scholar] [CrossRef]
- Feiner, R.; Argov, T.; Rabinovich, L.; Sigal, N.; Borovok, I.; Herskovits, A.A. A new perspective on lysogeny: Prophages as active regulatory switches of bacteria. Nat. Rev. Microbiol. 2015, 13, 641–650. [Google Scholar] [CrossRef]
- Argov, T.; Azulay, G.; Pasechnek, A.; Stadnyuk, O.; Ran-Sapir, S.; Borovok, I.; Sigal, N.; Herskovits, A.A. Temperate bacteriophages as regulators of host behavior. Curr. Opin. Microbiol. 2017, 38, 81–87. [Google Scholar] [CrossRef]
- Paczesny, J.; Richter, Ł.; Hołyst, R. Recent Progress in the Detection of Bacteria Using Bacteriophages: A Review. Viruses 2020, 12, 845. [Google Scholar] [CrossRef]
- Santos, S.B.; Costa, A.R.; Carvalho, C.; Nóbrega, F.L.; Azeredo, J. Exploiting Bacteriophage Proteomes: The Hidden Biotechnological Potential. Trends Biotechnol. 2018, 36, 966–984. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, A.; Rabsch, W.; Broeker, N.K.; Barbirz, S. Bacteriophage tailspike protein based assay to monitor phase variable glucosylations in Salmonella O-antigens. BMC Microbiol. 2016, 16, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Zampara, A.; Sørensen, M.C.H.; Grimon, D.; Antenucci, F.; Vitt, A.R.; Bortolaia, V.; Briers, Y.; Brøndsted, L. Exploiting phage receptor binding proteins to enable endolysins to kill Gram-negative bacteria. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Merino, S.; Tomás, J.M. Bacterial Capsules and Evasion of Immune Responses. eLS 2015, 1–10. [Google Scholar] [CrossRef]
- Hsieh, P.F.; Lin, H.H.; Lin, T.L.; Chen, Y.Y.; Wang, J.T. Two T7-like Bacteriophages, K5-2 and K5-4, Each Encodes Two Capsule Depolymerases: Isolation and Functional Characterization. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Topka-Bielecka, G.; Dydecka, A.; Necel, A.; Bloch, S.; Nejman-Faleńczyk, B.; Węgrzyn, G.; Węgrzyn, A. Bacteriophage-Derived Depolymerases against Bacterial Biofilm. Antibiotics 2021, 10, 175. [Google Scholar] [CrossRef]
- Lammens, E.M.; Nikel, P.I.; Lavigne, R. Exploring the synthetic biology potential of bacteriophages for engineering non-model bacteria. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef]
- Murphy, K.C. Phage Recombinases and Their Applications. Adv. Virus Res. 2012, 83, 367–414. [Google Scholar]
- Wannier, T.M.; Ciaccia, P.N.; Ellington, A.D.; Filsinger, G.T.; Isaacs, F.J.; Javanmardi, K.; Jones, M.A.; Kunjapur, A.M.; Nyerges, A.; Pal, C.; et al. Recombineering and MAGE. Nat. Rev. Methods Prim. 2021, 1, 1–24. [Google Scholar] [CrossRef]
- Lemire, S.; Yehl, K.M.; Lu, T.K. Phage-Based Applications in Synthetic Biology. Annu. Rev. Virol. 2018, 5, 453–476. [Google Scholar] [CrossRef]
- Inniss, M.C.; Bandara, K.; Jusiak, B.; Lu, T.K.; Weiss, R.; Wroblewska, L.; Zhang, L. A novel Bxb1 integrase RMCE system for high fidelity site-specific integration of mAb expression cassette in CHO Cells. Biotechnol. Bioeng. 2017, 114, 1837–1846. [Google Scholar] [CrossRef]
- Hsiao, V.; Hori, Y.; Rothemund, P.W.; Murray, R.M. A population-based temporal logic gate for timing and recording chemical events. Mol. Syst. Biol. 2016, 12, 869. [Google Scholar] [CrossRef]
- Young, R. Phage lysis: Three steps, three choices, one outcome. J. Microbiol. 2014, 52, 243. [Google Scholar] [CrossRef]
- Young, R. Bacteriophage lysis: Mechanism and regulation. Microbiol. Rev. 1992, 56, 430–481. [Google Scholar] [CrossRef]
- Serafim, V.; Ring, C.J.; Pantoja, L.; Shah, H.; Shah, A.J. Rapid Identification of E. coli Bacteriophages using Mass Spectrometry. J. Proteom. Enzymol. 2017, 6, 1–5. [Google Scholar]
- Chang, Y.; Kim, M.; Ryu, S. Characterization of a novel endolysin LysSA11 and its utility as a potent biocontrol agent against Staphylococcus aureus on food and utensils. Food Microbiol. 2017, 68, 112–120. [Google Scholar] [CrossRef]
- Kong, M.; Shin, J.H.; Heu, S.; Park, J.K.; Ryu, S. Lateral flow assay-based bacterial detection using engineered cell wall binding domains of a phage endolysin. Biosens. Bioelectron. 2017, 96, 173–177. [Google Scholar] [CrossRef]
- Yu, J.; Zhang, Y.; Zhang, Y.; Li, H.; Yang, H.; Wei, H. Sensitive and rapid detection of Staphylococcus aureus in milk via cell binding domain of lysin. Biosens. Bioelectron. 2016, 77, 366–371. [Google Scholar] [CrossRef]
- Dewey, J.S.; Savva, C.G.; White, R.L.; Vitha, S.; Holzenburg, A.; Young, R. Micron-scale holes terminate the phage infection cycle. Proc. Natl. Acad. Sci. USA 2010, 107, 2219–2223. [Google Scholar] [CrossRef] [Green Version]
- Horii, T.; Suzuki, Y.; Kobayashi, M. Characterization of a holin (HolNU3-1) in methicillin-resistant Staphylococcus aureus host. FEMS Immunol. Med. Microbiol. 2002, 34, 307–310. [Google Scholar] [CrossRef] [Green Version]
- Agu, C.A.; Klein, R.; Schwab, S.; König-Schuster, M.; Kodajova, P.; Ausserlechner, M.; Binishofer, B.; Bläsi, U.; Salmons, B.; Günzburg, W.H.; et al. The cytotoxic activity of the bacteriophage λ-holin protein reduces tumour growth rates in mammary cancer cell xenograft models. J. Gene Med. 2006, 8, 229–241. [Google Scholar] [CrossRef]
- Rodríguez-Rubio, L.; Martínez, B.; Rodríguez, A.; Donovan, D.M.; García, P. Enhanced staphylolytic activity of the Staphylococcus aureus bacteriophage vB_SauS-phiIPLA88 HydH5 virion-associated peptidoglycan hydrolase: Fusions, deletions, and synergy with LysH5. Appl. Environ. Microbiol. 2012, 78, 2241–2248. [Google Scholar] [CrossRef] [Green Version]
- Vipra, A.A.; Desai, S.N.; Roy, P.; Patil, R.; Raj, J.M.; Narasimhaswamy, N.; Paul, V.D.; Chikkamadaiah, R.; Sriram, B. Antistaphylococcal activity of bacteriophage derived chimeric protein P128. BMC Microbiol. 2012, 12, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutiérrez, D.; Fernández, L.; Rodríguez, A.; García, P. Are phage lytic proteins the secret weapon to kill Staphylococcus aureus? MBio 2018, 9, e01923-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-Rubio, L.; Quiles-Puchalt, N.; Martínez, B.; Rodríguez, A.; Penadés, J.R.; García, P. The peptidoglycan hydrolase of Staphylococcus aureus bacteriophage Ψ11 plays a structural role in the viral particle. Appl. Environ. Microbiol. 2013, 79, 6187–6190. [Google Scholar] [CrossRef] [Green Version]
- Burmistrz, M.; Krakowski, K.; Krawczyk-Balska, A. RNA-Targeting CRISPR–Cas Systems and Their Applications. Int. J. Mol. Sci. 2020, 21, 1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanley, S.Y.; Maxwell, K.L. Phage-Encoded Anti-CRISPR Defenses. Annu. Rev. Genet. 2018, 52, 445–464. [Google Scholar] [CrossRef]
- Payaslian, F.; Gradaschi, V.; Piuri, M. Genetic manipulation of phages for therapy using BRED. Curr. Opin. Biotechnol. 2021, 68, 8–14. [Google Scholar] [CrossRef]
- Hatoum-Aslan, A. Phage Genetic Engineering Using CRISPR–Cas Systems. Viruses 2018, 10, 335. [Google Scholar] [CrossRef] [Green Version]
- Li, H.F.; Wang, X.F.; Tang, H. Predicting Bacteriophage Enzymes and Hydrolases by Using Combined Features. Front. Bioeng. Biotechnol. 2020, 8, 183. [Google Scholar] [CrossRef]
- Roux, S.; Páez-Espino, D.; Chen, I.M.A.; Palaniappan, K.; Ratner, A.; Chu, K.; Reddy, T.; Nayfach, S.; Schulz, F.; Call, L.; et al. IMG/VR v3: An integrated ecological and evolutionary framework for interrogating genomes of uncultivated viruses. Nucleic Acids Res. 2021, 49, D764–D775. [Google Scholar] [CrossRef]
- Zayed, A.A.; Lü Cking, D.; Mohssen, M.; Cronin, D.; Bolduc, B.; Gregory, A.C.; Hargreaves, K.R.; Piehowski, P.D.; White Iii, R.A.; Huang, E.L.; et al. efam: An expanded, metaproteome-supported HMM profile database of viral protein families. Bioinformatics 2021, 37, 4202–4208. [Google Scholar] [CrossRef]
- Richter, Ł.; Janczuk-richter, M.; Niedzió, J.; Paczesny, J.; Ho, R. Recent advances in bacteriophage- based methods for bacteria detection. Drug Discov. Today 2018, 23, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Zwietering, M.H.; Jacxsens, L.; Membré, J.M.; Nauta, M.; Peterz, M. Relevance of microbial finished product testing in food safety management. Food Control. 2016, 60, 31–43. [Google Scholar] [CrossRef] [Green Version]
- Imdad, A.; Retzer, F.; Thomas, L.S.; McMillian, M.; Garman, K.; Rebeiro, P.F.; Deppen, S.A.; Dunn, J.R.; Woron, A.M. Impact of Culture-Independent Diagnostic Testing on Recovery of Enteric Bacterial Infections. Clin. Infect. Dis. 2018, 66, 1892–1898. [Google Scholar] [CrossRef] [Green Version]
- Gordillo Altamirano, F.L.; Barr, J.J. Phage therapy in the postantibiotic era. Clin. Microbiol. Rev. 2019, 32, e00066-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ISO. ISO 6579-1:2017-Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 1: Detection of Salmonella spp. Available online: https://www.iso.org/standard/56712.html (accessed on 2 May 2022).
- Feucherolles, M.; Cauchie, H.M.; Penny, C. MALDI-TOF Mass Spectrometry and Specific Biomarkers: Potential New Key for Swift Identification of Antimicrobial Resistance in Foodborne Pathogens. Microorganisms 2019, 7, 593. [Google Scholar] [CrossRef] [Green Version]
- Meile, S.; Kilcher, S.; Loessner, M.J.; Dunne, M. Reporter Phage-Based Detection of Bacterial Pathogens: Design Guidelines and Recent Developments. Viruses 2020, 12, 944. [Google Scholar] [CrossRef]
- Farooq, U.; Yang, Q.; Ullah, M.W.; Wang, S. Bacterial biosensing: Recent advances in phage-based bioassays and biosensors. Biosens. Bioelectron. 2018, 118, 204–216. [Google Scholar] [CrossRef]
- Machera, S.J.; Niedziółka-Jönsson, J.; Szot-Karpińska, K. Phage-Based Sensors in Medicine: A Review. Chemosensors 2020, 8, 61. [Google Scholar] [CrossRef]
- Craigie, J.; Yen, C.H. The Demonstration of Types of B. Typhosus by Means of Preparations of Type II Vi Phage: I. Principles and Technique on JSTOR. Can. Public Health J. 1938, 29, 484–496. [Google Scholar]
- Kretzer, J.W.; Schmelcher, M.; Loessner, M.J. Ultrasensitive and Fast Diagnostics of Viable Listeria Cells by CBD Magnetic Separation Combined with A511::luxAB Detection. Viruses 2018, 10, 626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, M.; Sim, J.; Kang, T.; Nguyen, H.H.; Park, H.K.; Chung, B.H.; Ryu, S. A novel and highly specific phage endolysin cell wall binding domain for detection of Bacillus cereus. Eur. Biophys. J. 2015, 44, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Torres, N.; Ávila, M.; Narbad, A.; Mayer, M.J.; Garde, S. Use of fluorescent CTP1L endolysin cell wall-binding domain to study the evolution of Clostridium tyrobutyricum during cheese ripening. Food Microbiol. 2019, 78, 11–17. [Google Scholar] [CrossRef]
- Sumrall, E.T.; Keller, A.P.; Shen, Y.; Loessner, M.J. Structure and function of Listeria teichoic acids and their implications. Mol. Microbiol. 2020, 113, 627–637. [Google Scholar] [CrossRef] [Green Version]
- Buth, S.A.; Shneider, M.M.; Scholl, D.; Leiman, P.G. Structure and Analysis of R1 and R2 Pyocin Receptor-Binding Fibers. Viruses 2018, 10, 427. [Google Scholar] [CrossRef] [Green Version]
- Kunstmann, S.; Scheidt, T.; Buchwald, S.; Helm, A.; Mulard, L.A.; Fruth, A.; Barbirz, S. Bacteriophage Sf6 Tailspike Protein for Detection of Shigella flexneri Pathogens. Viruses 2018, 10, 431. [Google Scholar] [CrossRef] [Green Version]
- Uchiyama, J.; Matsui, H.; Murakami, H.; Kato, S.I.; Watanabe, N.; Nasukawa, T.; Mizukami, K.; Ogata, M.; Sakaguchi, M.; Matsuzaki, S.; et al. Potential Application of Bacteriophages in Enrichment Culture for Improved Prenatal Streptococcus agalactiae Screening. Viruses 2018, 10, 552. [Google Scholar] [CrossRef] [Green Version]
- Nagano, K.; Tsutsumi, Y. Phage Display Technology as a Powerful Platform for Antibody Drug Discovery. Viruses 2021, 13, 178. [Google Scholar] [CrossRef]
- Bao, Q.; Li, X.; Han, G.; Zhu, Y.; Mao, C.; Yang, M. Phage-based vaccines. Adv. Drug Deliv. Rev. 2019, 145, 40–56. [Google Scholar] [CrossRef]
- Nam, K.T.; Kim, D.W.; Yoo, P.J.; Chiang, C.Y.; Meethong, N.; Hammond, P.T.; Chiang, Y.M.; Belcher, A.M. Virus-enabled synthesis and assembly of nanowires for lithium ion battery electrodes. Science 2006, 312, 885–888. [Google Scholar] [CrossRef] [Green Version]
- Karimi, M.; Mirshekari, H.; Moosavi Basri, S.M.; Bahrami, S.; Moghoofei, M.; Hamblin, M.R. Bacteriophages and phage-inspired nanocarriers for targeted delivery of therapeutic cargos. Adv. Drug Deliv. Rev. 2016, 106, 45–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.J.; Yi, H.; Kim, W.J.; Kang, K.; Yun, D.S.; Strano, M.S.; Ceder, G.; Belcher, A.M. Fabricating genetically engineered high-power lithium-ion batteries using multiple virus genes. Science 2009, 324, 1051–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larman, H.B.; Zhao, Z.; Laserson, U.; Li, M.Z.; Ciccia, A.; Gakidis, M.A.M.; Church, G.M.; Kesari, S.; Leproust, E.M.; Solimini, N.L.; et al. Autoantigen discovery with a synthetic human peptidome. Nat. Biotechnol. 2011, 29, 535–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Straus, S.K.; Scott, W.R.P.; Symmons, M.F.; Marvin, D.A. On the structures of filamentous bacteriophage Ff (fd, f1, M13). Eur. Biophys. J. 2008, 37, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Hay, I.D.; Lithgow, T. Filamentous phages: Masters of a microbial sharing economy. EMBO Rep. 2019, 20, e47427. [Google Scholar] [CrossRef]
- Talwar, H.; Hanoudi, S.N.; Draghici, S.; Samavati, L. Novel T7 Phage Display Library Detects Classifiers for Active Mycobacterium tuberculosis Infection. Viruses 2018, 10, 375. [Google Scholar] [CrossRef] [Green Version]
- Petrenko, V.A. Landscape Phage: Evolution from Phage Display to Nanobiotechnology. Viruses 2018, 10, 311. [Google Scholar] [CrossRef] [Green Version]
- Santos, S.B.; Azeredo, J. Bacteriophage-Based Biotechnological Applications. Viruses 2019, 11, 737. [Google Scholar] [CrossRef] [Green Version]
- Caberoy, N.B.; Zhou, Y.; Jiang, X.; Alvarado, G.; Li, W. Efficient identification of tubby-binding proteins by an improved system of T7 phage display. J. Mol. Recognit. 2010, 23, 74–83. [Google Scholar] [CrossRef] [Green Version]
- Caberoy, N.B.; Zhou, Y.; Alvarado, G.; Fan, X.; Li, W. Efficient identification of phosphatidylserine-binding proteins by ORF phage display. Biochem. Biophys. Res. Commun. 2009, 386, 197–201. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Caberoy, N.B. New perspective for phage display as an efficient and versatile technology of functional proteomics. Appl. Microbiol. Biotechnol. 2010, 85, 909–919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, K.B.; Jensen, O.N.; Ravn, P.; Clark, B.F.C.; Kristensen, P. Identification of Keratinocyte-specific Markers Using Phage Display and Mass Spectrometry *. Mol. Cell. Proteomics 2003, 2, 61–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Wang, Y.; Li, Z.; Li, J.; Dong, Z. Humanization of a mouse monoclonal antibody neutralizing TNF-α by guided selection. J. Immunol. Methods 2000, 241, 171–184. [Google Scholar] [CrossRef]
- Chen, Y.; Wiesmann, C.; Fuh, G.; Li, B.; Christinger, H.W.; McKay, P.; De Vos, A.M.; Lowman, H.B. Selection and analysis of an optimized anti-VEGF antibody: Crystal structure of an affinity-matured fab in complex with antigen. J. Mol. Biol. 1999, 293, 865–881. [Google Scholar] [CrossRef] [Green Version]
- Baker, K.P.; Edwards, B.M.; Main, S.H.; Choi, G.H.; Wager, R.E.; Halpern, W.G.; Lappin, P.B.; Riccobene, T.; Abramian, D.; Sekut, L.; et al. Generation and characterization of LymphoStat-B, a human monoclonal antibody that antagonizes the bioactivities of B lymphocyte stimulator. Arthritis Rheum. 2003, 48, 3253–3265. [Google Scholar] [CrossRef]
- Mazumdar, S. Raxibacumab. J. Mol. Biol. 2009, 1, 531–538. [Google Scholar] [CrossRef]
- Lu, D.; Shen, J.; Vil, M.D.; Zhang, H.; Jimenez, X.; Bohlen, P.; Witte, L.; Zhut, Z. Tailoring in Vitro Selection for a Picomolar Affinity Human Antibody Directed against Vascular Endothelial Growth Factor Receptor 2 for Enhanced Neutralizing Activity. J. Biol. Chem. 2003, 278, 43496–43507. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Kussie, P.; Ferguson, K.M. Structural Basis for EGF Receptor Inhibition by the Therapeutic Antibody IMC-11F8. Structure 2008, 16, 216–227. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Lu, J.; Allan, B.W.; Tang, Y.; Tetreault, J.; Chow, C.K.; Barmettler, B.; Nelson, J.; Bina, H.; Huang, L.; et al. Generation and characterization of ixekizumab, a humanized monoclonal antibody that neutralizes interleukin-17A. J. Inflamm. Res. 2016, 9, 39. [Google Scholar] [CrossRef] [Green Version]
- Herbst, R.S.; Soria, J.C.; Kowanetz, M.; Fine, G.D.; Hamid, O.; Gordon, M.S.; Sosman, J.A.; McDermott, D.F.; Powderly, J.D.; Gettinger, S.N.; et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014, 515, 563–567. [Google Scholar] [CrossRef] [Green Version]
- Markham, A. Atezolizumab: First Global Approval. Drugs 2016, 76, 1227–1232. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.S. Avelumab: First Global Approval. Drugs 2017, 77, 929–937. [Google Scholar] [CrossRef] [PubMed]
- Boehncke, W.H.; Brembilla, N.C.; Nissen, M.J. Guselkumab: The First Selective IL-23 Inhibitor for Active Psoriatic Arthritis in Adults. Expert Rev. Clin. Immunol. 2020, 17, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Duggan, S. Correction to: Caplacizumab: First Global Approval. Drugs 2018, 78, 1955. [Google Scholar] [CrossRef] [Green Version]
- Locatelli, F.; Jordan, M.B.; Allen, C.; Cesaro, S.; Rizzari, C.; Rao, A.; Degar, B.; Garrington, T.P.; Sevilla, J.; Putti, M.-C.; et al. Emapalumab in Children with Primary Hemophagocytic Lymphohistiocytosis. N. Engl. J. Med. 2020, 382, 1811–1822. [Google Scholar] [CrossRef]
- Dhillon, S. Moxetumomab Pasudotox: First Global Approval. Drugs 2018, 78, 1763–1767. [Google Scholar] [CrossRef] [Green Version]
- Syed, Y.Y. Lanadelumab: First Global Approval. Drugs 2018, 78, 1633–1637. [Google Scholar] [CrossRef]
- Esvelt, K.M.; Carlson, J.C.; Liu, D.R. A system for the continuous directed evolution of biomolecules. Nature 2011, 472, 499–503. [Google Scholar] [CrossRef] [Green Version]
- Miller, S.M.; Wang, T.; Liu, D.R. Phage-assisted continuous and non-continuous evolution. Nat. Protoc. 2020, 15, 4101–4127. [Google Scholar] [CrossRef]
- DeBenedictis, E.A.; Chory, E.J.; Gretton, D.W.; Wang, B.; Golas, S.; Esvelt, K.M. Systematic molecular evolution enables robust biomolecule discovery. Nat. Methods 2021, 19, 55–64. [Google Scholar] [CrossRef]
- Vaidya, A.; Ravindranath, S.; Annapure, U.S. Detection and differential identification of typhoidal Salmonella using bacteriophages and resazurin. 3 Biotech. 2020, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Jiang, M.; Xiong, J.; Li, J.; Wei, H.; Yu, J. Rapid ultrasensitive diagnosis of pneumonia caused by Acinetobacter baumannii using a combination of enrichment and phage-based qPCR assay. Res. Sq. 2020. [Google Scholar] [CrossRef] [Green Version]
- Garrido-Maestu, A.; Fuciños, P.; Azinheiro, S.; Carvalho, C.; Carvalho, J.; Prado, M. Specific detection of viable Salmonella Enteritidis by phage amplification combined with qPCR (PAA-qPCR) in spiked chicken meat samples. Food Control. 2019, 99, 79–83. [Google Scholar] [CrossRef] [Green Version]
- Almutairi, M.; Imam, M.; Alammari, N.; Hafiz, R.; Patel, F.; Alajel, S. Using Phages to Reduce Salmonella Prevalence in Chicken Meat: A Systematic Review. HAGE 2022, 3, 15–27. [Google Scholar] [CrossRef]
- Mido, T.; Schaffer, E.M.; Dorsey, R.W.; Sozhamannan, S.; Hofmann, E.R. Sensitive detection of live Escherichia coli by bacteriophage amplification-coupled immunoassay on the Luminex® MAGPIX instrument. J. Microbiol. Methods 2018, 152, 143–147. [Google Scholar] [CrossRef]
- Anany, H.; Brovko, L.; El Dougdoug, N.K.; Sohar, J.; Fenn, H.; Alasiri, N.; Jabrane, T.; Mangin, P.; Monsur Ali, M.; Kannan, B.; et al. Print to detect: A rapid and ultrasensitive phage-based dipstick assay for foodborne pathogens. Anal. Bioanal. Chem. 2018, 410, 1217–1230. [Google Scholar] [CrossRef]
- Schofield, D.; Sharp, N.J.; Westwater, C. Phage-based platforms for the clinical detection of human bacterial pathogens. Bacteriophage 2012, 2, 105–121. [Google Scholar] [CrossRef] [Green Version]
- Pizarro-Bauerle, J.; Ando, H. Engineered Bacteriophages for Practical Applications. Biol. Pharm. Bull. 2020, 43, 240–249. [Google Scholar] [CrossRef] [Green Version]
- Alcaine, S.D.; Pacitto, D.; Sela, D.A.; Nugen, S.R. Phage & phosphatase: A novel phage-based probe for rapid, multi-platform detection of bacteria. Analyst 2015, 140, 7629–7636. [Google Scholar]
- Franche, N.; Vinay, M.; Ansaldi, M. Substrate-independent luminescent phage-based biosensor to specifically detect enteric bacteria such as E. coli. Environ. Sci. Pollut. Res. 2017, 24, 42–51. [Google Scholar] [CrossRef] [Green Version]
- Fu, X.; Ding, M.; Zhang, N.; Li, J. Mycobacteriophages: An important tool for the diagnosis of Mycobacterium tuberculosis (Review). Mol. Med. Rep. 2015, 12, 13–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobs, W.R., Jr. Gene Transfer in Mycobacterium tuberculosis: Shuttle Phasmids to Enlightenment. Microbiol. Spectr. 2014, 2, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenneman, B.R.; Fernbach, J.; Loessner, M.J.; Lu, T.K.; Kilcher, S. Enhancing phage therapy through synthetic biology and genome engineering. Curr. Opin. Biotechnol. 2021, 68, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Hupfeld, M.; Trasanidou, D.; Ramazzini, L.; Klumpp, J.; Loessner, M.J.; Kilcher, S. A functional type II-A CRISPR–Cas system from Listeria enables efficient genome editing of large non-integrating bacteriophage. Nucleic Acids Res. 2018, 46, 6920–6933. [Google Scholar] [CrossRef]
- Zhang, D.; Coronel-Aguilera, C.P.; Romero, P.L.; Perry, L.; Minocha, U.; Rosenfield, C.; Gehring, A.G.; Paoli, G.C.; Bhunia, A.K.; Applegate, B. The Use of a Novel NanoLuc -Based Reporter Phage for the Detection of Escherichia coli O157:H7. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Meile, S.; Sarbach, A.; Du, J.; Schuppler, M.; Saez, C.; Loessner, M.J.; Kilcher, S. Engineered reporter phages for rapid bioluminescence-based detection and differentiation of viable Listeria cells. Appl. Environ. Microbiol. 2020, 86, 33235. [Google Scholar] [CrossRef] [Green Version]
- Hinkley, T.C.; Garing, S.; Jain, P.; Williford, J.; Le Ny, A.L.M.; Nichols, K.P.; Peters, J.E.; Talbert, J.N.; Nugen, S.R. A Syringe-Based Biosensor to Rapidly Detect Low Levels of Escherichia coli (ECOR13) in Drinking Water Using Engineered Bacteriophages. Sensors 2020, 20, 1953. [Google Scholar] [CrossRef] [Green Version]
- Kozak, S.; Alcaine, S.D. Phage-based forensic tool for spatial visualization of bacterial contaminants in cheese. J. Dairy Sci. 2020, 103, 5964–5971. [Google Scholar] [CrossRef]
- Wang, D.; Hinkley, T.; Chen, J.; Talbert, J.N.; Nugen, S.R. Phage based electrochemical detection of Escherichia coli in drinking water using affinity reporter probes. Analyst 2019, 144, 1345–1352. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Hinkley, T.; Nugen, S.R.; Talbert, J.N. Colorimetric detection of Escherichia coli using engineered bacteriophage and an affinity reporter system. Anal. Bioanal. Chem. 2019, 411, 7273–7279. [Google Scholar] [CrossRef]
- Alcaine, S.D.; Law, K.; Ho, S.; Kinchla, A.J.; Sela, D.A.; Nugen, S.R. Bioengineering bacteriophages to enhance the sensitivity of phage amplification-based paper fluidic detection of bacteria. Biosens. Bioelectron. 2016, 82, 14–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinkley, T.C.; Singh, S.; Garing, S.; Le Ny, A.L.M.; Nichols, K.P.; Peters, J.E.; Talbert, J.N.; Nugen, S.R. A phage-based assay for the rapid, quantitative, and single CFU visualization of E. coli (ECOR #13) in drinking water. Sci. Rep. 2018, 8, 1–8. [Google Scholar]
- Wisuthiphaet, N.; Yang, X.; Young, G.M.; Nitin, N. Rapid detection of Escherichia coli in beverages using genetically engineered bacteriophage T7. AMB Express 2019, 9, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rondón, L.; Urdániz, E.; Latini, C.; Payaslian, F.; Matteo, M.; Sosa, E.J.; Do Porto, D.F.; Turjanski, A.G.; Nemirovsky, S.; Hatfull, G.F.; et al. Fluoromycobacteriophages can detect viable Mycobacterium tuberculosis and determine phenotypic rifampicin resistance in 3–5 days from sputum collection. Front. Microbiol. 2018, 9, 1471. [Google Scholar] [CrossRef]
- Dunne, M.; Prokhorov, N.S.; Loessner, M.J.; Leiman, P.G. Reprogramming bacteriophage host range: Design principles and strategies for engineering receptor binding proteins. Curr. Opin. Biotechnol. 2021, 68, 272–281. [Google Scholar] [CrossRef]
- Yehl, K.; Lemire, S.; Yang, A.C.; Ando, H.; Mimee, M.; Torres, M.D.T.; de la Fuente-Nunez, C.; Lu, T.K. Engineering Phage Host-Range and Suppressing Bacterial Resistance through Phage Tail Fiber Mutagenesis. Cell 2019, 179, 459–469.e9. [Google Scholar] [CrossRef]
- Hwang, H.J.; Ryu, M.Y.; Park, J.P. Identification of high affinity peptides for capturing norovirus capsid proteins. RSC Adv. 2015, 5, 55300–55302. [Google Scholar] [CrossRef]
- Wu, J.; Park, J.P.; Dooley, K.; Cropek, D.M.; West, A.C.; Banta, S. Rapid Development of New Protein Biosensors Utilizing Peptides Obtained via Phage Display. PLoS ONE 2011, 6, e24948. [Google Scholar] [CrossRef] [Green Version]
- Padmanaban, G.; Park, H.; Choi, J.S.; Cho, Y.W.; Kang, W.C.; Moon, C.I.; Kim, I.S.; Lee, B.H. Identification of peptides that selectively bind to myoglobin by biopanning of phage displayed-peptide library. J. Biotechnol. 2014, 187, 43–50. [Google Scholar] [CrossRef]
- Wu, J.; Cropek, D.M.; West, A.C.; Banta, S. Development of a troponin i biosensor using a peptide obtained through phage display. Anal. Chem. 2010, 82, 8235–8243. [Google Scholar] [CrossRef]
- Shin, H.J.; Lim, W.K. Rapid label-free detection of E. coli using a novel SPR biosensor containing a fragment of tail protein from phage lambda. Prep. Biochem. Biotechnol. 2018, 48, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Poshtiban, S.; Evoy, S. Recent advances in bacteriophage based biosensors for food-borne pathogen detection. Sensors 2013, 13, 1763–1786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denyes, J.M.; Dunne, M.; Steiner, S.; Mittelviefhaus, M.; Weiss, A.; Schmidt, H.; Klumpp, J.; Loessner, M.J. Modified bacteriophage S16 long tail fiber proteins for rapid and specific immobilization and detection of Salmonella cells. Appl. Environ. Microbiol. 2017, 83, 277–294. [Google Scholar] [CrossRef] [Green Version]
- Sumrall, E.T.; Röhrig, C.; Hupfeld, M.; Selvakumar, L.; Du, J.; Dunne, M.; Schmelcher, M.; Shen, Y.; Loessner, M.J. Glycotyping and specific separation of Listeria monocytogenes with a novel bacteriophage protein tool kit. Appl. Environ. Microbiol. 2020, 86, e00612-20. [Google Scholar] [CrossRef]
- Park, C.; Kong, M.; Lee, J.H.; Ryu, S.; Park, S. Detection of Bacillus Cereus Using Bioluminescence Assay with Cell Wall-binding Domain Conjugated Magnetic Nanoparticles. BioChip J. 2018, 12, 287–293. [Google Scholar] [CrossRef]
- Ferapontova, E.E. Electrochemical assays for microbial analysis: How far they are from solving microbiota and microbiome challenges. Curr. Opin. Electrochem. 2020, 19, 153–161. [Google Scholar] [CrossRef]
- Janczuk-Richter, M.; Marinović, I.; Niedziółka-Jönsson, J.; Szot-Karpińska, K. Recent applications of bacteriophage-based electrodes: A mini-review. Electrochem. Commun. 2019, 99, 11–15. [Google Scholar] [CrossRef]
- Xu, J.; Chau, Y.; Lee, Y. kuen Phage-based Electrochemical Sensors: A Review. Micromachines 2019, 10, 855. [Google Scholar] [CrossRef] [Green Version]
- Ogata, A.F.; Edgar, J.M.; Majumdar, S.; Briggs, J.S.; Patterson, S.V.; Tan, M.X.; Kudlacek, S.T.; Schneider, C.A.; Weiss, G.A.; Penner, R.M. Virus-enabled biosensor for human serum albumin. Anal. Chem. 2017, 89, 1373–1381. [Google Scholar] [CrossRef] [Green Version]
- Yue, H.; He, Y.; Fan, E.; Wang, L.; Lu, S.; Fu, Z. Label-free electrochemiluminescent biosensor for rapid and sensitive detection of pseudomonas aeruginosa using phage as highly specific recognition agent. Biosens. Bioelectron. 2017, 94, 429–432. [Google Scholar] [CrossRef]
- Hiremath, N.; Guntupalli, R.; Vodyanoy, V.; Chin, B.A.; Park, M.K. Detection of methicillin-resistant Staphylococcus aureus using novel lytic phage-based magnetoelastic biosensors. Sensors Actuators B Chem. 2015, 210, 129–136. [Google Scholar] [CrossRef]
- Yang, L.M.C.; Tam, P.Y.; Murray, B.J.; McIntire, T.M.; Overstreet, C.M.; Weiss, G.A.; Penner, R.M. Virus Electrodes for Universal Biodetection. Anal. Chem. 2006, 78, 3265–3270. [Google Scholar] [CrossRef] [PubMed]
- Donavan, K.C.; Arter, J.A.; Pilolli, R.; Cioffi, N.; Weiss, G.A.; Penner, R.M. Virus-poly(3,4-ethylenedioxythiophene) composite films for impedance-based biosensing. Anal. Chem. 2011, 83, 2420–2424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhasin, A.; Sanders, E.C.; Ziegler, J.M.; Briggs, J.S.; Drago, N.P.; Attar, A.M.; Santos, A.M.; True, M.Y.; Ogata, A.F.; Yoon, D.V.; et al. Virus Bioresistor (VBR) for Detection of Bladder Cancer Marker DJ-1 in Urine at 10 pM in One Minute. Anal. Chem. 2020, 92, 6654–6666. [Google Scholar] [CrossRef]
- Jia, Y.; Qin, M.; Zhang, H.; Niu, W.; Li, X.; Wang, L.; Li, X.; Bai, Y.; Cao, Y.; Feng, X. Label-free biosensor: A novel phage-modified Light Addressable Potentiometric Sensor system for cancer cell monitoring. Biosens. Bioelectron. 2007, 22, 3261–3266. [Google Scholar] [CrossRef]
- Donavan, K.C.; Arter, J.A.; Weiss, G.A.; Penner, R.M. Virus-Poly(3,4-ethylenedioxythiophene) biocomposite films. Langmuir 2012, 28, 12581–12587. [Google Scholar] [CrossRef]
- O’Donnell, M.R.; Larsen, M.H.; Brown, T.S.; Jain, P.; Munsamy, V.; Wolf, A.; Uccellini, L.; Karim, F.; de Oliveira, T.; Mathema, B.; et al. Early detection of emergent extensively drug-resistant tuberculosis by flow cytometry-based phenotyping and whole-genome sequencing. Antimicrob. Agents Chemother. 2019, 63, e01834-18. [Google Scholar] [CrossRef] [Green Version]
- Urdániz, E.; Rondón, L.; Martí, M.A.; Hatfull, G.F.; Piuri, M. Rapid whole-cell assay of antitubercular drugs using second-generation Fluoromycobacteriophages. Antimicrob. Agents Chemother. 2016, 60, 3253–3256. [Google Scholar] [CrossRef] [Green Version]
- Timme, T.L.; Brennan, P.J. Induction of bacteriophage from members of the Mycobacterium avium, Mycobacterium intracellulare, Mycobacterium scrofulaceum serocomplex. J. Gen. Microbiol. 1984, 130, 2059–2066. [Google Scholar] [CrossRef] [Green Version]
- Ford, M.E.; Stenstrom, C.; Hendrix, R.W.; Hatfull, G.F. Mycobacteriophage TM4: Genome structure and gene expression. Tuber. Lung Dis. 1998, 79, 63–73. [Google Scholar] [CrossRef]
- Messing, J. [2] New M13 vectors for cloning. Methods Enzymol. 1983, 101, 20–78. [Google Scholar] [PubMed]
- Chanishvili, N. Nanotechnology to Aid Chemical and Biological Defense; Springer: Berlin/Heidelberg, Germany, 2015; pp. 17–33. [Google Scholar]
- Lavigne, R.; Ceyssens, P.J.; Robben, J. Phage Proteomics: Applications of Mass Spectrometry. Methods Mol. Biol. 2009, 502, 239–251. [Google Scholar] [PubMed]
- Kim, J.; Vu, B.; Kourentzi, K.; Willson, R.C.; Conrad, J.C. Increasing Binding Efficiency via Reporter Shape and Flux in a Viral Nanoparticle Lateral-Flow Assay. ACS Appl. Mater. Interfaces 2017, 9, 6878–6884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Ramasamy, R.P. Isolation and separation of Listeria monocytogenes using bacteriophage P100-modified magnetic particles. Colloids Surfaces B Biointerfaces 2019, 175, 421–427. [Google Scholar] [CrossRef]
- Franco, D.; De Plano, L.M.; Rizzo, M.G.; Scibilia, S.; Lentini, G.; Fazio, E.; Neri, F.; Guglielmino, S.P.P.; Mezzasalma, A.M. Bio-hybrid gold nanoparticles as SERS probe for rapid bacteria cell identification. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 224, 117394. [Google Scholar] [CrossRef]
- Peng, H.; Chen, I.A. Rapid Colorimetric Detection of Bacterial Species through the Capture of Gold Nanoparticles by Chimeric Phages. ACS Nano 2019, 13, 1244–1252. [Google Scholar] [CrossRef]
- Imai, M.; Mine, K.; Tomonari, H.; Uchiyama, J.; Matuzaki, S.; Niko, Y.; Hadano, S.; Watanabe, S. Dark-Field Microscopic Detection of Bacteria using Bacteriophage-Immobilized SiO2@AuNP Core-Shell Nanoparticles. Anal. Chem. 2019, 91, 12352–12357. [Google Scholar] [CrossRef]
- Liana, A.E.; Marquis, C.P.; Gunawan, C.; Gooding, J.J.; Amal, R. T4 bacteriophage conjugated magnetic particles for E. coli capturing: Influence of bacteriophage loading, temperature and tryptone. Colloids Surfaces B Biointerfaces 2017, 151, 47–57. [Google Scholar] [CrossRef]
- Bhardwaj, N.; Bhardwaj, S.K.; Mehta, J.; Nayak, M.K.; Deep, A. Bacteriophage conjugated IRMOF-3 as a novel opto-sensor for S. arlettae. New J. Chem. 2016, 40, 8068–8073. [Google Scholar] [CrossRef]
- Bhardwaj, N.; Bhardwaj, S.K.; Mehta, J.; Kim, K.H.; Deep, A. MOF-bacteriophage biosensor for highly sensitive and specific detection of Staphylococcus aureus. ACS Appl. Mater. Interfaces 2017, 9, 33589–33598. [Google Scholar] [CrossRef]
- Abril, A.G.; Carrera, M.; Böhme, K.; Barros-Velázquez, J.; Rama, J.L.R.; Calo-Mata, P.; Sánchez-Pérez, A.; Villa, T.G. Proteomic Characterization of Antibiotic Resistance, and Production of Antimicrobial and Virulence Factors in Streptococcus Species Associated with Bovine Mastitis. Could Enzybiotics Represent Novel Therapeutic Agents Against These Pathogens? Antibiotics 2020, 9, 302. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Mann, M. Proteomics to study genes and genomes. Nature 2000, 405, 837–846. [Google Scholar] [CrossRef] [PubMed]
- Rabilloud, T.; Lelong, C. Two-dimensional gel electrophoresis in proteomics: A tutorial. J. Proteomics 2011, 74, 1829–1841. [Google Scholar] [CrossRef] [PubMed]
- Carrera, M.; Cañas, B.; Piñeiro, C.; Vázquez, J.; Gallardo, J.M. De novo mass spectrometry sequencing and characterization of species-specific peptides from nucleoside diphosphate kinase B for the classification of commercial fish species belonging to the family merlucciidae. J. Proteome Res. 2007, 6, 3070–3080. [Google Scholar] [CrossRef] [PubMed]
- Geng, F.; Huang, X.; Majumder, K.; Zhu, Z.; Cai, Z.; Ma, M. Mass Spectrometry and Two-Dimensional Electrophoresis to Characterize the Glycosylation of Hen Egg White Ovomacroglobulin. J. Agric. Food Chem. 2015, 63, 8209–8215. [Google Scholar] [CrossRef]
- Mayer, K.; Albrecht, S.; Schaller, A. Targeted Analysis of Protein Phosphorylation by 2D Electrophoresis. Methods Mol. Biol. 2015, 1306, 167–176. [Google Scholar]
- Martinotti, S.; Ranzato, E. 2-DE Gel Analysis: The Spot Detection. Methods Mol. Biol. 2016, 1384, 155–164. [Google Scholar]
- Abril, A.G.; Ortea, I.; Barros-Velázquez, J.; Villa, T.G.; Calo-Mata, P. Shotgun Proteomics for FoodMicroorganism Detection. Methods Mol. Biol. 2021, 2259, 205–213. [Google Scholar]
- Zhang, Y.; Fonslow, B.R.; Shan, B.; Baek, M.C.; Yates, J.R. Protein analysis by shotgun/bottom-up proteomics. Chem. Rev. 2013, 113, 2343–2394. [Google Scholar] [CrossRef] [Green Version]
- Carrera, M.; Cañas, B.; Lopez-Ferrer, D. Fast Global Phosphoproteome Profiling of Jurkat T Cells by HIFU-TiO2-SCX-LC-MS/MS. Anal. Chem. 2017, 89, 8853–8862. [Google Scholar] [CrossRef]
- Wolters, D.A.; Washburn, M.P.; Yates, J.R. An Automated Multidimensional Protein Identification Technology for Shotgun Proteomics. Anal. Chem. 2001, 73, 5683–5690. [Google Scholar] [CrossRef] [PubMed]
- Carrera, M.; Ezquerra-Brauer, J.M.; Aubourg, S.P. Characterization of the Jumbo Squid (Dosidicus gigas) Skin By-Product by Shotgun Proteomics and Protein-Based Bioinformatics. Mar. Drugs 2019, 18, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eng, J.K.; McCormack, A.L.; Yates, J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994, 5, 976–989. [Google Scholar] [CrossRef] [Green Version]
- Perkins, D.N.; Pappin, D.J.C.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Keller, A.; Nesvizhskii, A.I.; Kolker, E.; Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002, 74, 5383–5392. [Google Scholar] [CrossRef]
- Käll, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 2007, 4, 923–925. [Google Scholar] [CrossRef]
- Shevchenko, A.; Wilm, M.; Mann, M. Peptide Sequencing by Mass Spectrometry for Homology Searches and Cloning of Genes. J. Protein Chem. 1997, 16, 481–490. [Google Scholar] [CrossRef]
- Bern, M.; Kil, Y.J.; Becker, C. Byonic: Advanced Peptide and Protein Identification Software. Curr. Protoc. Bioinforma. 2012, 40, 13.20.1–13.20.14. [Google Scholar] [CrossRef] [Green Version]
- Ma, B.; Zhang, K.; Hendrie, C.; Liang, C.; Li, M.; Doherty-Kirby, A.; Lajoie, G. PEAKS: Powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 2337–2342. [Google Scholar] [CrossRef]
- Carrera, M.; Cañas, B.; Vázquez, J.; Gallardo, J.M. Extensive de Novo sequencing of new parvalbumin isoforms using a novel combination of bottom-up proteomics, accurate molecular mass measurement by FTICR-MS, and selected MS/MS ion monitoring. J. Proteome Res. 2010, 9, 4393–4406. [Google Scholar] [CrossRef]
- Mateos, J.; Landeira-Abia, A.; Fafián-Labora, J.A.; Fernández-Pernas, P.; Lesende-Rodríguez, I.; Fernández-Puente, P.; Fernández-Moreno, M.; Delmiro, A.; Martín, M.A.; Blanco, F.J.; et al. iTRAQ-based analysis of progerin expression reveals mitochondrial dysfunction, reactive oxygen species accumulation and altered proteostasis. Stem Cell Res. Ther. 2015, 6, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robotti, E.; Marengo, E. 2D-DIGE and Fluorescence Image Analysis. Methods Mol. Biol. 2018, 1664, 25–39. [Google Scholar] [PubMed]
- Stryiński, R.; Mateos, J.; Pascual, S.; González, Á.F.; Gallardo, J.M.; Łopieńska-Biernat, E.; Medina, I.; Carrera, M. Proteome profiling of L3 and L4 Anisakis simplex development stages by TMT-based quantitative proteomics. J. Proteomics 2019, 201, 1–11. [Google Scholar] [CrossRef] [PubMed]
- López-Ferrer, D.; Ramos-Fernández, A.; Martínez-Bartolomé, S.; García-Ruiz, P.; Vázquez, J. Quantitative proteomics using 16O/18O labeling and linear ion trap mass spectrometry. Proteomics 2006, 6, S4–S11. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.E.; Blagoev, B.; Kratchmarova, I.; Kristensen, D.B.; Steen, H.; Pandey, A.; Mann, M. Stable Isotope Labeling by Amino Acids in Cell Culture, SILAC, as a Simple and Accurate Approach to Expression Proteomics *. Mol. Cell. Proteom. 2002, 1, 376–386. [Google Scholar] [CrossRef] [Green Version]
- Mueller, L.N.; Rinner, O.; Schmidt, A.; Letarte, S.; Bodenmiller, B.; Brusniak, M.Y.; Vitek, O.; Aebersold, R.; Müller, M. SuperHirn–A novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics 2007, 7, 3470–3480. [Google Scholar] [CrossRef]
- Fornelli, L.; Toby, T.K.; Schachner, L.F.; Doubleday, P.F.; Srzentić, K.; DeHart, C.J.; Kelleher, N.L. Top-down proteomics: Where we are, where we are going? J. Proteom. 2018, 175, 3. [Google Scholar] [CrossRef]
- Carrera, M.; Weisbrod, C.; Lopez-Ferrer, D.; Huguet, R.; Manuel Gallardo, J.; Schwartz, J.; Huhmer, A.; Fisher Scientifi, T.; Jose, S.; Manu, J.; et al. Top-Down, High-Throughput of Thermo-Stable Allergens Using Complementary MS/MS Fragmentation Strategies Top-Down, High-Throughput of Thermo-Stable Allergens Using Compleme. Foods 2015, 9, 1134. [Google Scholar] [CrossRef]
- Borràs, E.; Sabidó, E. What is targeted proteomics? A concise revision of targeted acquisition and targeted data analysis in mass spectrometry. Proteomics 2017, 17, 1700180. [Google Scholar] [CrossRef]
- Aebersold, R.; Bensimon, A.; Collins, B.; Ludwig, C.; Sabido, L. Applications and developments in targeted proteomics: From SRM to DIA/SWATH. Wiley Online Libr. 2016, 16, 2065–2067. [Google Scholar] [CrossRef] [Green Version]
- Lange, V.; Picotti, P.; Domon, B.; Aebersold, R. Selected reaction monitoring for quantitative proteomics: A tutorial. Mol. Syst. Biol. 2008, 4, 222. [Google Scholar] [CrossRef] [PubMed]
- Jorge, I.; Casas, E.M.; Villar, M.; Ortega-Pérez, I.; López-Ferrer, D.; Martínez-Ruiz, A.; Carrera, M.; Marina, A.; Martínez, P.; Serrano, H.; et al. High-sensitivity analysis of specific peptides in complex samples by selected MS/MS ion monitoring and linear ion trap mass spectrometry: Application to biological studies. J. Mass Spectrom. 2007, 42, 1391–1403. [Google Scholar] [CrossRef] [PubMed]
- Carrera, M.; Cañas, B.; López-Ferrer, D.; Piñeiro, C.; Vázquez, J.; Gallardo, J.M. Fast monitoring of species-specific peptide biomarkers using high-intensity-focused-ultrasound-assisted tryptic digestion and selected MS/MS ion monitoring. Anal. Chem. 2011, 83, 5688–5695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrera, M.; Gallardo, J.M.; Pascual, S.; González, Á.F.; Medina, I. Protein biomarker discovery and fast monitoring for the identification and detection of Anisakids by parallel reaction monitoring (PRM) mass spectrometry. J. Proteom. 2016, 142, 130–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillet, L.C.; Navarro, P.; Tate, S.; Röst, H.; Selevsek, N.; Reiter, L.; Bonner, R.; Aebersold, R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: A new concept for consistent and accurate proteome analysis. Mol. Cell. Proteom. 2012, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beynon, R.J.; Doherty, M.K.; Pratt, J.M.; Gaskell, S.J. Multiplexed absolute quantification in proteomics using artificial QCAT proteins of concatenated signature peptides. Nat. Methods 2005, 2, 587–589. [Google Scholar] [CrossRef]
- Bereman, M.S.; Maclean, B.; Tomazela, D.M.; Liebler, D.C.; Maccoss, M.J. The development of selected reaction monitoring methods for targeted proteomics via empirical refinement. Proteomics 2012, 12, 1134–1141. [Google Scholar] [CrossRef] [Green Version]
- Röst, H.; Malmström, L.; Aebersold, R. A computational tool to detect and avoid redundancy in selected reaction monitoring. Mol. Cell. Proteom. 2012, 11, 540–549. [Google Scholar] [CrossRef] [Green Version]
- Cox, C.R.; Rees, J.C.; Voorhees, K.J. Modeling bacteriophage amplification as a predictive tool for optimized MALDI-TOF MS-based bacterial detection. J. Mass Spectrom. 2012, 47, 1435–1441. [Google Scholar] [CrossRef]
- Rees, J.C.; Barr, J.R. Detection of methicillin-resistant Staphylococcus aureus using phage amplification combined with matrix-assisted laser desorption / ionization mass spectrometry. Anal. Bioanal. Chem. 2017, 409, 1379–1386. [Google Scholar] [CrossRef] [Green Version]
- Štveráková, D.; Šedo, O.; Benešík, M.; Zdráhal, Z.; Doškař, J.; Pantůček, R. Rapid Identification of Intact Staphylococcal Bacteriophages Using Matrix-Assisted Laser Desorption Ionization-Time-of-Flight Mass Spectrometry. Viruses 2018, 10, 176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abril, A.G.; Carrera, M.; Böhme, K.; Barros-Velázquez, J.; Cañas, B.; Rama, J.L.R.; Villa, T.G.; Calo-Mata, P. Characterization of Bacteriophage Peptides of Pathogenic Streptococcus by LC-ESI-MS/MS: Bacteriophage Phylogenomics and Their Relationship to Their Host. Front. Microbiol. 2020, 11, 1241. [Google Scholar] [CrossRef] [PubMed]
- Abril, A.G.; Carrera, M.; Böhme, K.; Barros-Velázquez, J.; Cañas, B.; Rama, J.L.R.; Villa, T.G.; Calo-Mata, P. Proteomic Characterization of Bacteriophage Peptides from the Mastitis Producer Staphylococcus aureus by LC-ESI-MS/MS and the Bacteriophage Phylogenomic Analysis. Foods 2021, 10, 799. [Google Scholar] [CrossRef] [PubMed]
- Horká, M.; Karásek, P.; Šalplachta, J.; Růžička, F.; Štveráková, D.; Pantůček, R.; Roth, M. Nano-etched fused-silica capillary used for on-line preconcentration and electrophoretic separation of bacteriophages from large blood sample volumes with off-line MALDI-TOF mass spectrometry identification. Microchim. Acta 2020, 187, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Horká, M.; Štveráková, D.; Šalplachta, J.; Šlais, K.; Šiborová, M.; Růžička, F.; Pantůček, R. Electrophoretic techniques for purification, separation and detection of Kayvirus with subsequent control by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and microbiological methods. J. Chromatogr. A 2018, 1570, 155–163. [Google Scholar]
- Buszewski, B.; Maślak, E.; Złoch, M.; Railean-Plugaru, V.; Kłodzińska, E.; Pomastowski, P. A new approach to identifying pathogens, with particular regard to viruses, based on capillary electrophoresis and other analytical techniques. TrAC Trends Anal. Chem. 2021, 139, 116250. [Google Scholar] [CrossRef] [PubMed]
- Horká, M.; Šalplachta, J.; Karásek, P.; Růžička, F.; Štveráková, D.; Pantůček, R.; Roth, M. Rapid Isolation, Propagation, and Online Analysis of a Small Number of Therapeutic Staphylococcal Bacteriophages from a Complex Matrix. ACS Infect. Dis. 2020, 6, 2745–2755. [Google Scholar] [CrossRef]
- Wilson, V.G. Beyond Antibiotics–Are Phages Our Allies? Viruses Intim. Invaders 2022, 279–302. [Google Scholar] [CrossRef]
- Schooley, R.T.; Biswas, B.; Gill, J.J.; Hernandez-Morales, A.; Lancaster, J.; Lessor, L.; Barr, J.J.; Reed, S.L.; Rohwer, F.; Benler, S.; et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob. Agents Chemother. 2017, 61, e00954-17. [Google Scholar] [CrossRef] [Green Version]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Burrowes, B.H.; Molineux, I.J.; Fralick, J.A. Directed in Vitro Evolution of Therapeutic Bacteriophages: The Appelmans Protocol. Viruses 2019, 11, 241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Menéndez, E.; Fernández, L.; Gutiérrez, D.; Pando, D.; Martínez, B.; Rodríguez, A.; García, P. Strategies to Encapsulate the Staphylococcus aureus Bacteriophage phiIPLA-RODI. Viruses 2018, 10, 495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ssekatawa, K.; Byarugaba, D.K.; Kato, C.D.; Wampande, E.M.; Ejobi, F.; Tweyongyere, R.; Nakavuma, J.L. A review of phage mediated antibacterial applications. Alex. J. Med. 2020, 57, 1–20. [Google Scholar] [CrossRef]
- Hudson, J.A.; Billington, C.; Carey-Smith, G.; Greening, G. Bacteriophages as Biocontrol Agents in Food. J. Food Prot. 2005, 68, 426–437. [Google Scholar] [CrossRef]
- Smith, G.P.; Petrenko, V.A. Phage display. Chem. Rev. 1997, 97, 391–410. [Google Scholar] [CrossRef]
- Aghebati-Maleki, L.; Bakhshinejad, B.; Baradaran, B.; Motallebnezhad, M.; Aghebati-Maleki, A.; Nickho, H.; Yousefi, M.; Majidi, J. Phage display as a promising approach for vaccine development. J. Biomed. Sci. 2016, 23, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Wang, Y.; Liu, Z.; Wang, Z. Phage display and its application in vaccine design. Ann. Microbiol. 2010, 60, 13–19. [Google Scholar] [CrossRef]
- Xu, H.; Bao, X.; Lu, Y.; Liu, Y.; Deng, B.; Wang, Y.; Xu, Y.; Hou, J. Immunogenicity of T7 bacteriophage nanoparticles displaying G-H loop of foot-and-mouth disease virus (FMDV). Vet. Microbiol. 2017, 205, 46–52. [Google Scholar] [CrossRef]
- Bahadir, A.O.; Balcioglu, B.K.; Uzyol, K.S.; Hatipoglu, I.; Sogut, I.; Basalp, A.; Erdag, B. Phage displayed HBV core antigen with immunogenic activity. Appl. Biochem. Biotechnol. 2011, 165, 1437–1447. [Google Scholar] [CrossRef]
- Gao, J.; Liu, Z.; Huang, M.; Li, X.; Wang, Z. T7 phage displaying latent membrane protein 1 of Epstein-Barr virus elicits humoral and cellular immune responses in rats. Acta Virol. 2011, 55, 117–121. [Google Scholar] [CrossRef] [Green Version]
- Vázquez, R.; Díez-Martínez, R.; Domingo-Calap, P.; García, P.; Gutiérrez, D.; Muniesa, M.; Ruiz-Ruigómez, M.; Sanjuán, R.; Tomás, M.; Tormo-Mas, M.Á.; et al. Essential Topics for the Regulatory Consideration of Phages as Clinically Valuable Therapeutic Agents: A Perspective from Spain. Microorganisms 2022, 10, 717. [Google Scholar] [CrossRef] [PubMed]



| Product Name | Nonproprietary Name | Target Antigen | First Application | Approved Year | Special Note on Phage Display Technology |
|---|---|---|---|---|---|
| Humira® | Adalimumab | TNFα | RA | 2002 | Humanization using guided selection method [74] |
| Lucentis® | Ranibizumab | VEGFA | nAMD | 2006 | In vitro affinity maturation [75] |
| Benlysta® | Belimumab | BLyS | SLE | 2011 | Isolation from CAT’s library (human naïve scFv library) [76] |
| ABthrax® | Raxibacumab | Bacillus anthracis PA | Inhaled anthrax | 2012 | Isolation from CAT’s library (human naïve scFv library) [77] |
| Cyramza® | Ramucirumab | VEGFR2 | GC NSCLC | 2014 | Isolation from Dyax’s library (human naïve Fab library) [78] |
| Portrazza® | Necitumumab | EGFR | NSCLC | 2015 | Isolation from Dyax’s library (human naïve Fab library) [79] |
| Taltz® | Ixekizumab | IL-17A | Psoriasis | 2016 | Isolation from mouse immune Fab library [80] |
| Tecentriq® | Atezolizumab | PD-L1 | UC NSCLC | 2016 | Isolation from Genentech’s library (human naïve library) [81,82] |
| Bavencio® | Avelumab | PD-L1 | MCC | 2017 | Isolation from Dyax’s library (human naïve Fab library) [83] |
| Tremfya® | Guselkumab | IL-23 | Psoriasis | 2017 | Isolation from HuCAL GOLD® library (Synthetic Fab library) [84] |
| Cablivi® | Caplacizumab | vWF | aTTP | 2018 | Isolation from Camelidae-derived nanobody library [85] |
| Gamifant® | Emapalumab | IFNγ | HLH | 2018 | Isolation from CAT’s library (human naïve scFv library) [86] |
| Lumoxiti® | Moxetumomab pasudotox | CD22 | HCL | 2018 | In vitro affinity maturation [87] |
| Takhzyro® | Lanadelumab | pKal | HAE | 2018 | Isolation from Dyax’s library (human naïve Fab library) [88] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abril, A.G.; Carrera, M.; Notario, V.; Sánchez-Pérez, Á.; Villa, T.G. The Use of Bacteriophages in Biotechnology and Recent Insights into Proteomics. Antibiotics 2022, 11, 653. https://doi.org/10.3390/antibiotics11050653
Abril AG, Carrera M, Notario V, Sánchez-Pérez Á, Villa TG. The Use of Bacteriophages in Biotechnology and Recent Insights into Proteomics. Antibiotics. 2022; 11(5):653. https://doi.org/10.3390/antibiotics11050653
Chicago/Turabian StyleAbril, Ana G., Mónica Carrera, Vicente Notario, Ángeles Sánchez-Pérez, and Tomás G. Villa. 2022. "The Use of Bacteriophages in Biotechnology and Recent Insights into Proteomics" Antibiotics 11, no. 5: 653. https://doi.org/10.3390/antibiotics11050653
APA StyleAbril, A. G., Carrera, M., Notario, V., Sánchez-Pérez, Á., & Villa, T. G. (2022). The Use of Bacteriophages in Biotechnology and Recent Insights into Proteomics. Antibiotics, 11(5), 653. https://doi.org/10.3390/antibiotics11050653







