Does Chlorination Promote Antimicrobial Resistance in Waterborne Pathogens? Mechanistic Insight into Co-Resistance and Its Implication for Public Health
Abstract
1. Introduction
2. Water Quality and Human Health
3. Fundamentals of Chlorine Disinfection
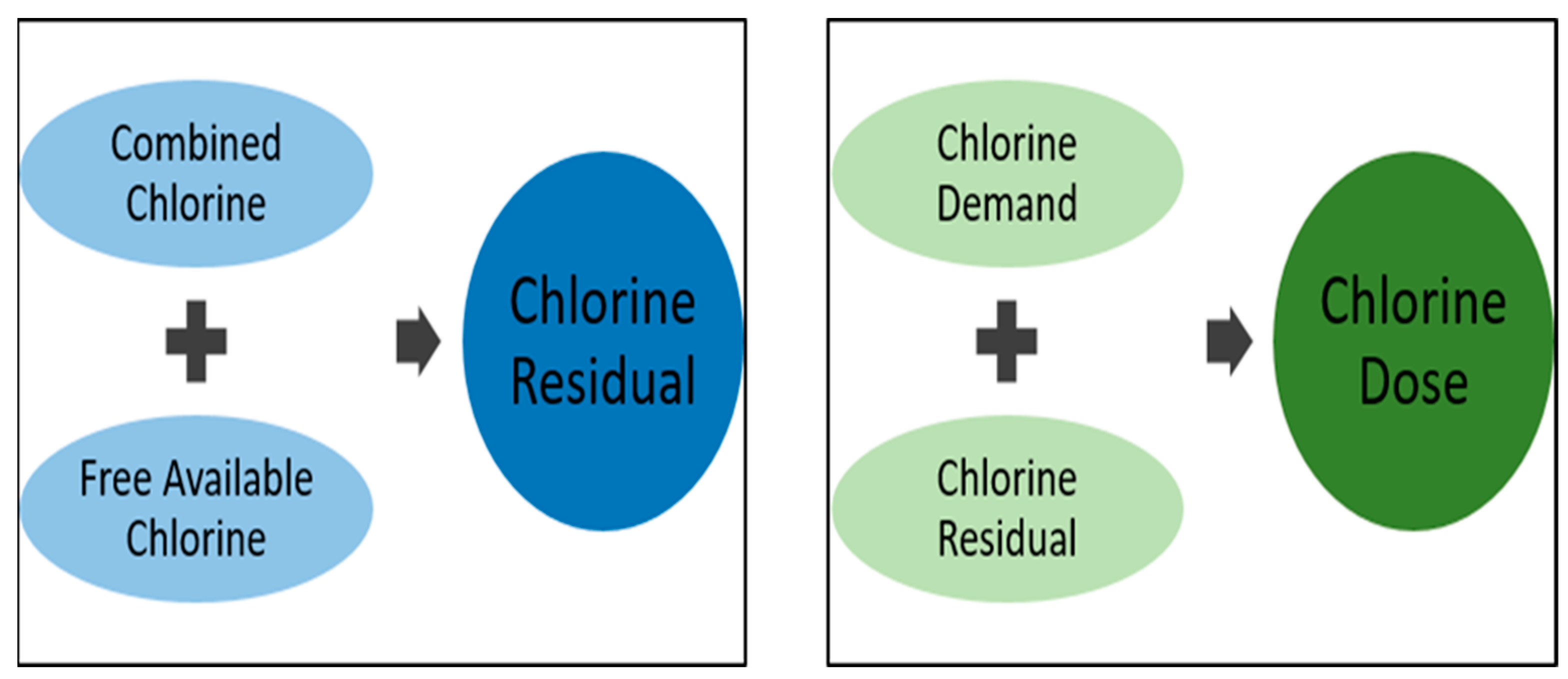
3.1. Breakpoint Chlorination and Factors Influencing Disinfection
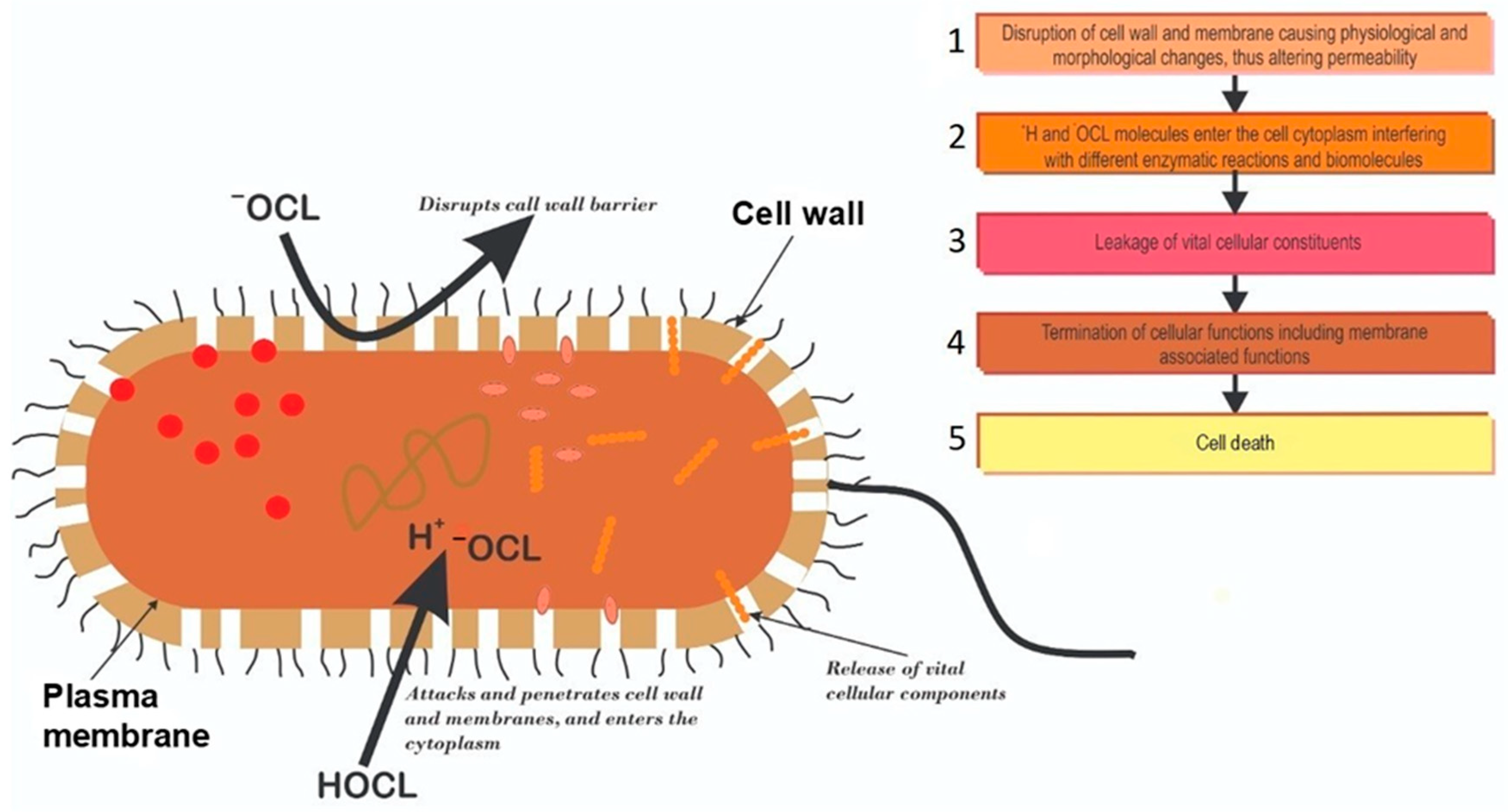
3.2. Mechanisms of Chlorine Disinfection
3.3. Incidences of Chlorine Tolerant Microorganisms from Treated Water Sources
4. Major Drivers of Antimicrobial Resistance (AMR) in Aquatic Environments
4.1. Stepwise Accumulation of Drug Resistance Mutations
4.2. Contribution of Chlorination to AMR Expansion via Horizontal Gene Transfer
5. Association between AMR and Disinfectant Resistance in Microorganisms
6. Proteome Mediated Chlorine Tolerance
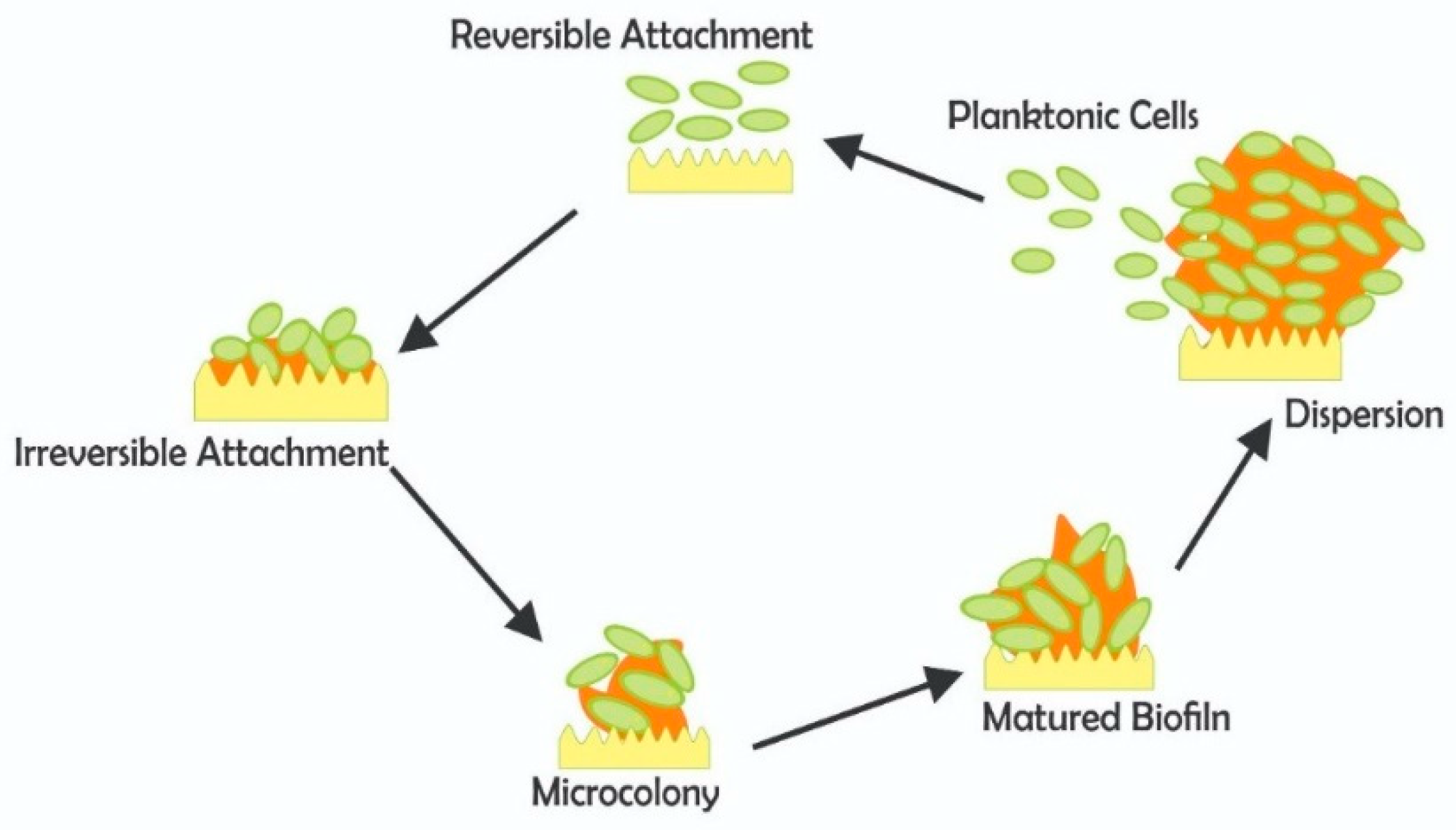
7. Biofilm and Pathogen Survival in Water Treatment
Resistance and Pathogen Protection in Biofilms
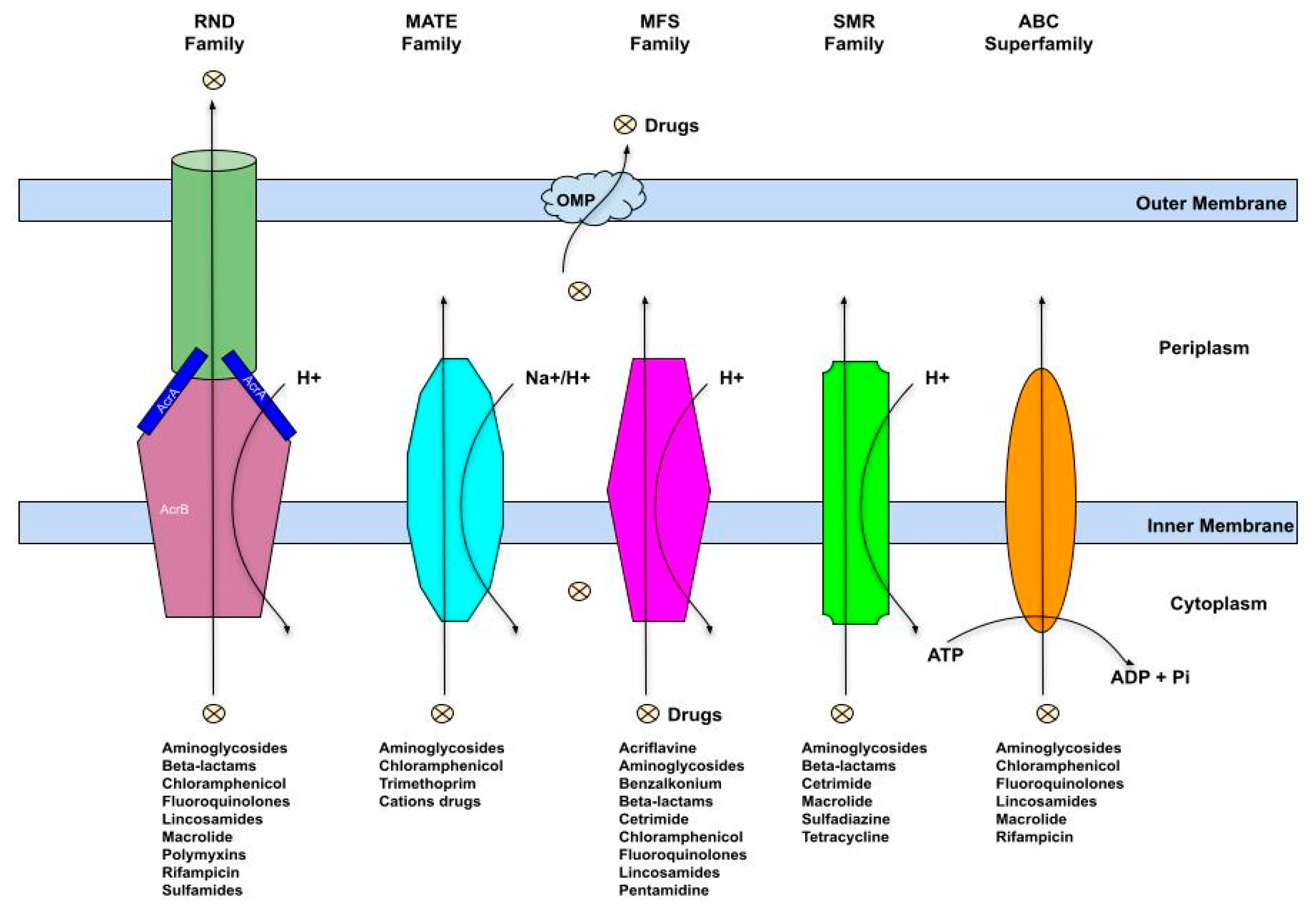
8. Multidrug Efflux Pumps Induction and Biocide Resistance
9. Conclusions and Future Directives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- DeNegre, A.A.; Mbah, M.L.N.; Myers, K.; Fefferman, N.H. Emergence of antibiotic resistance in immunocompromised host populations: A case study of emerging antibiotic resistant tuberculosis in AIDS patients. PLoS ONE 2019, 14, e0212969. [Google Scholar] [CrossRef] [PubMed]
- León-Buitimea, A.; Garza-Cárdenas, C.R.; Garza-Cervantes, J.A.; Lerma-Escalera, J.A.; Morones-Ramírez, J.R. The demand for new antibiotics: Antimicrobial peptides, nanoparticles, and combinatorial therapies as future strategies in antibacterial agent design. Front. Microbiol. 2020, 2020, 1669. [Google Scholar] [CrossRef] [PubMed]
- Sumi, C.D.; Heffernan, A.J.; Lipman, J.; Roberts, J.A.; Sime, F.B. What antibiotic exposures are required to suppress the emergence of resistance for Gram-negative bacteria? A systematic review. Clin. Pharmacokinet. 2019, 58, 1407–1443. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Drlica, K. Restricting the selection of antibiotic-resistant mutants: A general strategy derived from fluoroquinolone studies. Clin. Infect. Dis. 2001, 33 (Suppl. S3), S147–S156. [Google Scholar] [CrossRef]
- Baquero, F. Resistance to quinolones in gram-negative microorganisms: Mechanisms and prevention. Eur. Urol. 1990, 17, 3–12. [Google Scholar] [CrossRef]
- Zhang, N.; Ye, X.; Wu, Y.; Huang, Z.; Gu, X.; Cai, Q.; Shen, X.; Jiang, H.; Ding, H. Determination of the mutant selection window and evaluation of the killing of Mycoplasma gallisepticum by danofloxacin, doxycycline, tilmicosin, tylvalosin and valnemulin. PLoS ONE 2017, 12, e0169134. [Google Scholar] [CrossRef]
- Huang, Z.; Mao, C.; Wei, Y.; Gu, X.; Cai, Q.; Shen, X.; Ding, H. Analysis of the mutant selection window and killing of Mycoplasma hyopneumoniae for doxycycline, tylosin, danofloxacin, tiamulin, and valnemulin. PLoS ONE 2020, 15, e0220350. [Google Scholar] [CrossRef]
- García-León, G.; Sánchez, M.B.; Martínez, J.L. The inactivation of intrinsic antibiotic resistance determinants widens the mutant selection window for quinolones in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2012, 56, 6397–6399. [Google Scholar] [CrossRef]
- Khan, S.; Beattie, T.K.; Knapp, C.W. Relationship between antibiotic-and disinfectant-resistance profiles in bacteria harvested from tap water. Chemosphere 2016, 152, 132–141. [Google Scholar] [CrossRef]
- Yuan, Q.B.; Guo, M.T.; Yang, J. Fate of antibiotic resistant bacteria and genes during wastewater chlorination: Implication for antibiotic resistance control. PLoS ONE 2015, 10, e0119403. [Google Scholar] [CrossRef]
- Liu, S.S.; Qu, H.M.; Yang, D.; Hu, H.; Liu, W.L.; Qiu, Z.G.; Hou, A.M.; Guo, J.; Li, J.W.; Shen, Z.Q.; et al. Chlorine disinfection increases both intracellular and extracellular antibiotic resistance genes in a full-scale wastewater treatment plant. Water Res. 2018, 136, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Ouyang, W.; Qian, Y.; Su, C.; Su, J.; Chen, H. High-throughput profiling of antibiotic resistance genes in drinking water treatment plants and distribution systems. Environ. Pollut. 2016, 213, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Liu, L.; Wang, D.N.; Yang, D.; Liu, W.L.; Yin, J.; Yang, Z.W.; Wang, H.R.; Qiu, Z.G.; Shen, Z.Q.; et al. Chlorine disinfection promotes the exchange of antibiotic resistance genes across bacterial genera by natural transformation. ISME J. 2020, 114, 1847–1856. [Google Scholar] [CrossRef]
- World Health Organisation. Drinking-Water. 2019. Available online: https://www.who.int/news-room/fact-sheets/detail/drinking-water (accessed on 12 July 2021).
- World Health Organisation. Water. 2020. Available online: https://www.afro.who.int/health-topics/water (accessed on 11 July 2021).
- Sabri, N.A.; Schmitt, H.; Van der Zaan, B.; Gerritsen, H.W.; Zuidema, T.; Rijnaarts, H.H.M.; Langenhoff, A.A.M. Prevalence of antibiotics and antibiotic resistance genes in a wastewater effluent-receiving river in the Netherlands. J. Environ. Chem. Eng. 2020, 8, 102245. [Google Scholar] [CrossRef]
- Murray, G.E.; Tobin, R.S.; Junkins, B.; Kushner, D.J. Effect of chlorination on antibiotic resistance profiles of sewage-related bacteria. Appl. Environ. Microbiol. 1984, 48, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Rutala, W.A.; Stiegel, M.M.; Sarubbi, F.A.; Weber, D.J. Susceptibility of antibiotic-susceptible and antibiotic-resistant hospital bacteria to disinfectants. Infect. Control Hosp. Epidemiol. 1997, 18, 417–421. [Google Scholar] [CrossRef]
- Maertens, H.; De Reu, K.; Meyer, E.; Van Coillie, E.; Dewulf, J. Limited association between disinfectant use and either antibiotic or disinfectant susceptibility of Escherichia coli in both poultry and pig husbandry. BMC Vet. Res. 2019, 15, 310. [Google Scholar] [CrossRef]
- Al-Abri, M.; Al-Ghafri, B.; Bora, T.; Dobretsov, S.; Dutta, J.; Castelletto, S.; Rosa, L.; Boretti, A. Chlorination disadvantages and alternative routes for biofouling control in reverse osmosis desalination. NPJ Clean Water 2019, 2, 2. [Google Scholar] [CrossRef]
- Goyal, R.V.; Patel, H.M. Analysis of residual chlorine in simple drinking water distribution system with intermittent water supply. Appl. Water Sci. 2015, 5, 311–319. [Google Scholar] [CrossRef]
- Musz-Pomorska, A.; Widomski, M.K. Variant analysis of the chlorination efficiency of water in a selected water supply network. Proc. ECOpole 2020, 14, 19–28. [Google Scholar]
- Calomiris, J.; Christman, K. How Does Chlorine Added to Drinking Water Kill Bacteria and Other Harmful Organisms? Why Doesn’t It Harm Us? Scientific American. Available online: https://www.scientificamerican.com/article/how-does-chlorine-added-t/ (accessed on 5 August 2021).
- Venkobachar, C.; Iyengar, L.; Rao, A.P. Mechanism of disinfection: Effect of chlorine on cell membrane functions. Water Res. 1977, 11, 727–729. [Google Scholar] [CrossRef]
- Haas, C.N. Mechanisms of Inactivation of New Indicators of Disinfection Efficiency by Free Available Chlorine. Ph.D. Thesis, Department of Civil Engineering, the University of Illinois at Urbana-Champaign, Champaign, IL, USA, 1978; p. 158. [Google Scholar]
- Chang, S.L. Modern concept of disinfection. JSEDA 1971, 97, 689–707. [Google Scholar] [CrossRef]
- Mizozoe, M.; Otaki, M.; Aikawa, K. The mechanism of chlorine damage using enhanced green fluorescent protein-expressing Escherichia coli. Water 2019, 11, 2156. [Google Scholar] [CrossRef]
- Bridges, D.F.; Lacombe, A.; Wu, V.C. Integrity of the Escherichia coli O157: H7 cell wall and membranes after chlorine dioxide treatment. Front. Microbiol. 2020, 11, 888. [Google Scholar] [CrossRef] [PubMed]
- Reed, B.; Shaw, R.; Chatterton, K. Technical Notes on Drinking-Water, Sanitation and Hygiene in Emergencies; WHO, Water, Engineering and Development Centre: Loughborough, UK, 2013. [Google Scholar]
- Adefisoye, M.A.; Okoh, A.I. Ecological and public health implications of the discharge of multidrug-resistant bacteria and physicochemical contaminants from treated wastewater effluents in the Eastern Cape, South Africa. Water 2017, 9, 562. [Google Scholar] [CrossRef]
- National Water Act. Revision of General Authorization in Terms of Section 39 of the National Water Act, 1998 (Act No. 36 of 1998) (The Act); Gazette No. 19182, Notice No. 1091; National Water Act: 1998; South African Government Gazette: Pretoria, South Africa, 1998; Volume 26187.
- Yang, L.; Chen, X.; She, Q.; Cao, G.; Liu, Y.; Chang, V.W.C.; Tang, C.Y. Regulation, formation, exposure, and treatment of disinfection by-products (DBPs) in swimming pool waters: A critical review. Environ. Int. 2018, 121, 1039–1057. [Google Scholar] [CrossRef]
- Al-Berfkani, M.I.; Zubair, A.I.; Bayazed, H. Assessment of chlorine resistant bacteria and their susceptibility to antibiotic from water distribution system in Duhok province. J. Appl. Biol. Biotechnol. 2014, 2, 010–013. [Google Scholar]
- Owoseni, M.C.; Olaniran, A.O.; Okoh, A.I. Chlorine tolerance and inactivation of Escherichia coli recovered from wastewater treatment plants in the Eastern Cape, South Africa. Appl. Sci. 2017, 7, 810. [Google Scholar] [CrossRef]
- Jathar, S.; Shinde, D.; Dakhni, S.; Fernandes, A.; Jha, P.; Desai, N.; Jobby, R. Identification and characterization of chlorine-resistant bacteria from water distribution sites of Mumbai. Arch. Microbiol. 2021, 203, 5241–5248. [Google Scholar] [CrossRef]
- Martin, D.J.; Wesgate, R.L.; Denyer, S.P.; McDonnell, G.; Maillard, J.Y. Bacillus subtilis vegetative isolate surviving chlorine dioxide exposure: An elusive mechanism of resistance. J. Appl. Microbiol. 2015, 119, 1541–1551. [Google Scholar] [CrossRef]
- Momba, M.N.B.; Kfir, R.; Venter, S.N.; Cloete, T.E. An overview of biofilm formation in distribution systems and its impact on the deterioration of water quality. Water SA 2000, 26, 59–66. [Google Scholar]
- Bridier, A.; Briandet, R.; Thomas, V.; Dubois-Brissonnet, F. Resistance of bacterial biofilms to disinfectants: A review. Biofouling 2011, 27, 1017–1032. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Nandakumar, R.; Nickerson, K.W.; Li, X. Proteomic adaptations to starvation prepare Escherichia coli for disinfection tolerance. Water Res. 2015, 69, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Falkinham, J.O.; Pruden, A.; Edwards, M. Opportunistic premise plumbing pathogens: Increasingly important pathogens in drinking water. Pathogens 2015, 4, 373–386. [Google Scholar] [CrossRef]
- Shrivastava, R.; Upreti, R.K.; Jain, S.R.; Prasad, K.N.; Seth, P.K.; Chaturvedi, U.C. Suboptimal chlorine treatment of drinking water leads to selection of multidrug-resistant Pseudomonas aeruginosa. Ecotoxicol. Environ. Saf. 2004, 58, 277–283. [Google Scholar] [CrossRef]
- Karumathil, D.P.; Yin, H.B.; Kollanoor-Johny, A.; Venkitanarayanan, K. Effect of chlorine exposure on the survival and antibiotic gene expression of multidrug-resistant Acinetobacter baumannii in water. Int. J. Environ. Res. Public Health 2014, 11, 1844–1854. [Google Scholar] [CrossRef]
- Martins, S.C.S.; Júnior, G.D.S.F.; de Melo Machado, B.; Martins, C.M. Chlorine and antibiotic-resistant bacilli isolated from an effluent treatment plant. Acta Sci. Technol. 2013, 35, 3–9. [Google Scholar] [CrossRef][Green Version]
- Owoseni, M.; Okoh, A. Assessment of chlorine tolerance profile of Citrobacter species recovered from wastewater treatment plants in Eastern Cape, South Africa. EMA 2017, 189, 201. [Google Scholar] [CrossRef]
- Ridgway, H.F.; Olson, B.H. Chlorine resistance patterns of bacteria from two drinking water distribution systems. Appl. Environ. Microbiol. 1982, 44, 972–987. [Google Scholar] [CrossRef]
- LeChevallier, M.W.; Hassenauer, T.S.; Camper, A.K.; McFETERS, G.A. Disinfection of bacteria attached to granular activated carbon. Appl. Environ. Microbiol. 1984, 48, 918–923. [Google Scholar] [CrossRef]
- King, C.H.; Shotts, E.B.; Wooley, R.E.; Porter, K.G. Survival of coliforms and bacterial pathogens within protozoa during chlorination. Appl. Environ. Microbiol. 1988, 54, 3023–3033. [Google Scholar] [CrossRef] [PubMed]
- Owoseni, M.; Okoh, A. Evidence of emerging challenge of chlorine tolerance of Enterococcus species recovered from wastewater treatment plants. Int. Biodeterior. Biodegrad. 2017, 120, 216–223. [Google Scholar] [CrossRef]
- Dupuy, M.; Mazoua, S.; Berne, F.; Bodet, C.; Garrec, N.; Herbelin, P.; Ménard-Szczebara, F.; Oberti, S.; Rodier, M.H.; Soreau, S.; et al. Efficiency of water disinfectants against Legionella pneumophila and Acanthamoeba. Water Res. 2011, 45, 1087–1094. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Hernández, S.; Vázquez-Rodríguez, G.A.; Beltrán-Hernández, R.I.; Prieto-García, F.; Miranda-López, J.M.; Franco-Abuín, C.M.; Álvarez-Hernández, A.; Iturbe, U.; Coronel-Olivares, C. Resistance and inactivation kinetics of bacterial strains isolated from the non-chlorinated and chlorinated effluents of a WWTP. Int. J. Environ. Res. Public Health 2013, 10, 3363–3383. [Google Scholar] [CrossRef] [PubMed]
- Adefisoye, M.A.; Okoh, A.I. Identification and antimicrobial resistance prevalence of pathogenic Escherichia coli strains from treated wastewater effluents in Eastern Cape, South Africa. Microbiologyopen 2016, 5, 143–151. [Google Scholar] [CrossRef]
- Buelow, E.; Ploy, M.C.; Dagot, C. Role of pollution on the selection of antibiotic resistance and bacterial pathogens in the environment. Curr. Opin. Microbiol. 2021, 64, 117–124. [Google Scholar] [CrossRef]
- Cantón, R.; Morosini, M.I. Emergence and spread of antibiotic resistance following exposure to antibiotics. FEMS Microbiol. Rev. 2011, 35, 977–991. [Google Scholar] [CrossRef]
- Li, X.; Zhao, X.; Drlica, K. Selection of Streptococcus pneumoniae mutants having reduced susceptibility to levofloxacin and moxifloxacin. Antimicrob. Agents Chemother. 2002, 46, 522–524. [Google Scholar] [CrossRef]
- Drlica, K. The mutant selection window and antimicrobial resistance. J. Antimicrob. Chemother. 2003, 52, 11–17. [Google Scholar] [CrossRef]
- Urban, C.; Rahman, N.; Zhao, X. Fluoroquinolone resistant Streptococcus pneumoniae associated with levofloxacin therapy. J. Infect. Dis. 2001, 184, 794–798. [Google Scholar] [CrossRef][Green Version]
- Guo, M.T.; Yuan, Q.B.; Yang, J. Distinguishing effects of ultraviolet exposure and chlorination on the horizontal transfer of antibiotic resistance genes in municipal wastewater. Environ. Sci. Technol. 2015, 49, 5771–5778. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Yang, H.; Guan, L.; Liu, X.; Zhang, T. Risks of antibiotic resistance genes and antimicrobial resistance under chlorination disinfection with public health concerns. Environ. Int. 2022, 158, 106978. [Google Scholar] [CrossRef] [PubMed]
- Ojemaye, M.O.; Adefisoye, M.A.; Okoh, A.I. Nanotechnology as a viable alternative for the removal of antimicrobial resistance determinants from discharged municipal effluents and associated watersheds: A review. J. Environ. Manag. 2020, 275, 111234. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Fernández, E.Z.; Schuldiner, S. Acidification of cytoplasm in Escherichia coli provides a strategy to cope with stress and facilitates development of antibiotic resistance. Sci. Rep. 2020, 10, 9954. [Google Scholar] [CrossRef]
- Bourély, C.; Cazeau, G.; Jarrige, N.; Haenni, M.; Lupo, A.; Madec, J.Y.; Leblond, A.; Gay, E. Co-resistance to amoxicillin and tetracycline as an indicator of multidrug resistance in Escherichia coli isolates from animals. Front. Microbiol. 2019, 10, 2288. [Google Scholar] [CrossRef]
- Landecker, H. Antimicrobials before antibiotics: War, peace, and disinfectants. Palgrave Commun. 2019, 5, 1–11. [Google Scholar] [CrossRef]
- Voth, W.; Jakob, U. Stress-activated chaperones: A first line of defense. Trends Biochem. Sci. 2017, 42, 899–913. [Google Scholar] [CrossRef]
- Goemans, C.V.; Collet, J.F. Stress-induced chaperones: A first line of defense against the powerful oxidant hypochlorous acid. F1000Research 2019, 8, 1678. [Google Scholar] [CrossRef]
- da Cruz Nizer, W.S.; Inkovskiy, V.; Overhage, J. Surviving reactive chlorine stress: Responses of gram-negative bacteria to hypochlorous acid. Microorganisms 2020, 8, 1220. [Google Scholar] [CrossRef]
- Cornelis, P.; Wei, Q.; Andrews, S.C.; Vinckx, T. Iron homeostasis and management of oxidative stress response in bacteria. Metallomics 2011, 3, 540–549. [Google Scholar] [CrossRef]
- Wang, Z.; Fang, Y.; Zhi, S.; Simpson, D.J.; Gill, A.; McMullen, L.M.; Neumann, N.F.; Gänzle, M.G. The locus of heat resistance confers resistance to chlorine and other oxidizing chemicals in Escherichia coli. Appl. Environ. Microbiol. 2020, 86, e02123-19. [Google Scholar] [CrossRef] [PubMed]
- Butler, C.S.; Boltz, J.P. Biofilm processes and control in water and wastewater treatment. In Comprehensive Water Quality and Purification; Ahuja, S., Ed.; Elsevier: Waltham, MA, USA, 2014; pp. 90–107. [Google Scholar]
- Simões, L.C.; Simões, M. Biofilms in drinking water: Problems and solutions. Rsc Adv. 2013, 3, 2520–2533. [Google Scholar] [CrossRef]
- Sehar, S.; Naz, I. Role of the biofilms in wastewater treatment. In Microbial Biofilms-Importance and Applications; Dhanasekaran, D., Thajuddin, N., Eds.; IntechOpen: Rijeka, Croatia, 2016; pp. 121–144. [Google Scholar]
- Muhammad, M.H.; Idris, A.L.; Fan, X.; Guo, Y.; Yu, Y.; Jin, X.; Qiu, J.; Guan, X.; Huang, T. Beyond risk: Bacterial biofilms and their regulating approaches. Front. Microbiol. 2020, 11, 928. [Google Scholar] [CrossRef] [PubMed]
- Cepas, V.; López, Y.; Muñoz, E.; Rolo, D.; Ardanuy, C.; Martí, S.; Xercavins, M.; Horcajada, J.P.; Bosch, J.; Soto, S.M. Relationship between biofilm formation and antimicrobial resistance in gram-negative Bacteria. Microb. Drug Resist. 2019, 25, 72–79. [Google Scholar] [CrossRef]
- Hall, C.W.; Mah, T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Singh, S.; Singh, S.K.; Chowdhury, I.; Singh, R. Understanding the mechanism of bacterial biofilms resistance to antimicrobial agents. Open Microbiol. J. 2017, 11, 53–62. [Google Scholar] [CrossRef]
- Bas, S.; Kramer, M.; Stopar, D. Biofilm surface density determines biocide effectiveness. Front. Microbiol. 2017, 8, 2443. [Google Scholar] [CrossRef]
- Cai, Y. Molecular Mechanism of Nitric Oxide-Mediated Regulation of Intracellular Cyclic-di-GMP in Pseudomonas Aeruginosa Biofilms. Ph.D. Dissertation, University of Southampton, England, UK, 2018. [Google Scholar]
- Mah, T.F. Biofilm-specific antibiotic resistance. Future Microbiol. 2012, 7, 1061–1072. [Google Scholar] [CrossRef]
- Hall, C.W.; Hinz, A.J.; Gagnon, L.B.P.; Zhang, L.; Nadeau, J.P.; Copeland, S.; Saha, B.; Mah, T.F. Pseudomonas aeruginosa biofilm antibiotic resistance gene ndvB expression requires the RpoS stationary-phase sigma factor. Appl. Environ. Microbiol. 2018, 84, e02762-17. [Google Scholar] [CrossRef]
- Alcalde-Rico, M.; Hernando-Amado, S.; Blanco, P.; Martínez, J.L. Multidrug efflux pumps at the crossroad between antibiotic resistance and bacterial virulence. Front. Microbiol. 2016, 7, 1483. [Google Scholar] [CrossRef]
- Slipski, C.J.; Zhanel, G.G.; Bay, D.C. Biocide selective TolC-independent efflux pumps in Enterobacteriaceae. J. Membr. Biol. 2018, 251, 15–33. [Google Scholar] [CrossRef] [PubMed]
- Abdi, S.N.; Ghotaslou, R.; Ganbarov, K.; Mobed, A.; Tanomand, A.; Yousefi, M.; Asgharzadeh, M.; Kafil, H.S. Acinetobacter baumannii efflux pumps and antibiotic resistance. Infect. Drug. Resist. 2020, 13, 423. [Google Scholar] [CrossRef] [PubMed]
- Ughachukwu, P.O.; Unekwe, P.C. Efflux pump-mediated resistance in chemotherapy. Ann. Med. Health Sci. Res. 2012, 2, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Delmar, J.A.; Su, C.C.; Yu, E.W. Bacterial multidrug efflux transporters. Annu. Rev. Biophys. 2014, 43, 93–117. [Google Scholar] [CrossRef] [PubMed]
- Rahman, T.; Yarnall, B.; Doyle, D.A. Efflux drug transporters at the forefront of antimicrobial resistance. Eur. Biophys. J. 2017, 46, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Perland, E.; Fredriksson, R. Classification systems of secondary active transporters. TIPS 2017, 38, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Cacciotto, P.; Ramaswamy, V.K.; Malloci, G.; Ruggerone, P.; Vargiu, A.V. Molecular modeling of multidrug properties of resistance nodulation division (RND) transporters. In Bacterial Multidrug Exporters; Yamaguchi, A., Nishino, K., Eds.; Humana Press: New York, NY, USA, 2018; pp. 179–219. [Google Scholar]
- Morita, Y.; Tomida, J.; Kawamura, Y. Responses of Pseudomonas aeruginosa to antimicrobials. Front Microbiol. 2014, 4, 422. [Google Scholar] [CrossRef]
- Van der Deen, M.; De Vries, E.G.; Timens, W.; Scheper, R.J.; Timmer-Bosscha, H.; Postma, D.S. ATP-binding cassette (ABC) transporters in normal and pathological lung. Respir. Res. 2005, 6, 59. [Google Scholar] [CrossRef]
- Wilkens, S. Structure and mechanism of ABC transporters. F1000prime Rep. 2015, 7, 14. [Google Scholar] [CrossRef]
- Blanco, P.; Hernando-Amado, S.; Reales-Calderon, J.A.; Corona, F.; Lira, F.; Alcalde-Rico, M.; Bernardini, A.; Sanchez, M.B.; Martinez, J.L. Bacterial multidrug efflux pumps: Much more than antibiotic resistance determinants. Microorganisms 2016, 4, 14. [Google Scholar] [CrossRef]
- Gould, V.C.; Okazaki, A.; Avison, M.B. Coordinate hyperproduction of SmeZ and SmeJK efflux pumps extends drug resistance in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 2013, 57, 655–657. [Google Scholar] [CrossRef] [PubMed]
- Poonsuk, K.; Tribuddharat, C.; Chuanchuen, R. Simultaneous overexpression of multidrug efflux pumps in Pseudomonas aeruginosa non-cystic fibrosis clinical isolates. Can. J. Microbiol. 2014, 60, 437–443. [Google Scholar] [CrossRef] [PubMed]

| Source | Microorganism | Disinfectant Concentration/Time | Mechanism(s) of Resistance | Reference |
|---|---|---|---|---|
| Drinking water | P. aeruginosa | ≤0.5 mg/L Cl− | Natural resistance due to the permeability barrier caused by outer membrane lipopolysaccharides; biofilm formation | [41] |
| Experimental isolates | Acinetobacter baumannii | 0.2–4 mg/L | Increased expression of efflux pumps other antibiotic resistance genes | [42] |
| Drinking water reservoir | Acinetobater species, Serratia species | 2 mg/L | Not determined | [35] |
| Sewage | Bacillus species | 0.1 mg/L NaOCl | Probable spore formation | [43] |
| Secondary effluent | Citrobacter species | 0.5 mg/L Ca(OCl)2 for 30 min | Not determined | [44] |
| Drinking water | Bacillus species, Actinomycete | 10 mg/L NaOCl for 2 min | Cellular aggregation or adhesion to suspended particulate. Production of extracellular slime or capsular material | [45] |
| Drinking water and experimental isolates | Heterotrophic bacteria, faecal coliforms, E. coli, Salmonella typhimurium, Yersinia enterocolitica, Shigella sonnie | 2.0 mg/L free chlorine for 1 h | Bacterial attachment to surface and production of extracellular slime layer | [46] |
| Chlorine-demand–free buffer solution | Coliform isolated from drinking water systems and Enteric bacterial from culture collections cocultured with protozoa (Ciliates and amoebae). | 2–4 mg/L free chlorine for 1–2 h | Shielding of bacteria from chlorine by ingesting protozoans (cysts) and, thus, enhancing resistance | [47] |
| Treated drinking water | S. aureus, Micrococcus varians, Aeromonas hydrophila | 1–100 mg/L Ca(OCl)2 solution for 30 min | Possible synthesis of unique proteins or aggregation of bacteria or encapsulation | [33] |
| Environmental isolates (Wastewater clarifier effluent) suspended in phosphate buffer saline | Enterococcus species | 0.5 mg/L Ca(OCl)2 for 30 min | Not determined | [48] |
| Environmental strains cultured in sterile phosphate buffer solution | Legionella pneumophila from environmental water cocultured with Acanthamoeba species | 2–3 mg/L Cl2 for 1 h with a residual Cl2 of 1 mg/L after 1 h | Possible phenotypic modification of Legionella pneumophila due to intra-cellular growth with Acanthamoeba sp. | [49] |
| Environmental isolates from wastewater treatment plants suspended in saline | Bacillus species, Citrobacter freundii, Enterobacter species, Kluyvera cryocrescens, Kluyvera intermedia | 0.5 mg/L NaOCl for 30 min | Authors suggested the possible expression of certain stress factor genes which may reduce bacterial metabolism or change the permeability of cell membranes | [50] |
| Environmental isolates (Wastewater clarifier effluent) suspended in phosphate buffer saline | E. coli | 0.5 mg/L NaOCl for 30 min | Not determined | [34] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adefisoye, M.A.; Olaniran, A.O. Does Chlorination Promote Antimicrobial Resistance in Waterborne Pathogens? Mechanistic Insight into Co-Resistance and Its Implication for Public Health. Antibiotics 2022, 11, 564. https://doi.org/10.3390/antibiotics11050564
Adefisoye MA, Olaniran AO. Does Chlorination Promote Antimicrobial Resistance in Waterborne Pathogens? Mechanistic Insight into Co-Resistance and Its Implication for Public Health. Antibiotics. 2022; 11(5):564. https://doi.org/10.3390/antibiotics11050564
Chicago/Turabian StyleAdefisoye, Martins A., and Ademola O. Olaniran. 2022. "Does Chlorination Promote Antimicrobial Resistance in Waterborne Pathogens? Mechanistic Insight into Co-Resistance and Its Implication for Public Health" Antibiotics 11, no. 5: 564. https://doi.org/10.3390/antibiotics11050564
APA StyleAdefisoye, M. A., & Olaniran, A. O. (2022). Does Chlorination Promote Antimicrobial Resistance in Waterborne Pathogens? Mechanistic Insight into Co-Resistance and Its Implication for Public Health. Antibiotics, 11(5), 564. https://doi.org/10.3390/antibiotics11050564






