Antimicrobial Resistance in Escherichia coli from the Broiler Farm Environment, with Detection of SHV-12-Producing Isolates
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Collection
2.2. Antimicrobial Susceptibility Test
2.3. Characterization of Antimicrobial Resistance Genes, Integrons, and Plasmids
2.4. Molecular Typing
2.5. Statistical Analysis
3. Results
3.1. Antimicrobial Susceptibility Testing
3.2. Molecular Characterization and Typing of ESBL-Producing E. coli
3.3. Characterization of Antimicrobial Resistance Genes among Non-ESBL-Producing Isolates
3.4. Correlation between Phenotypic and Genotypic Resistance Profile of E. coli Isolates
3.5. Characterization of Integrons among STX-Resistant E. coli Isolates
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- EFSA Panel on Biological Hazards (BIOHAZ); Koutsoumanis, K.; Allende, A.; Álvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; Cesare, A.; de Herman, L.; et al. Role played by the environment in the emergence and spread of antimicrobial resistance (AMR) through the food chain. EFSA J. 2021, 19, 6651. [Google Scholar] [CrossRef]
- World Organization for Animal Health (OIE). Annual Report on Antimicrobial Agents Intended for Use in Animals. Fifth Report. 2021. Available online: https://www.oie.int/en/document/fifth-oie-annual-report-on-antimicrobial-agents-intended-for-use-in-animals/ (accessed on 10 March 2022).
- Swelum, A.A.; Elbestawy, A.R.; El-Saadony, M.T.; Hussein, E.O.S.; Alhotan, R.; Suliman, G.M.; Taha, A.E.; Ba-Awadh, H.; El-Tarabily, K.A.; El-Hack, M.E.A. Ways to minimize bacterial infections, with special reference to Escherichia coli, to cope with the first-week mortality in chicks: An updated overview. Poult. Sci. 2021, 100, 101039. [Google Scholar] [CrossRef] [PubMed]
- Durso, L.M.; Cook, K.L. One Health and antibiotic resistance in agroecosystems. EcoHealth 2019, 16, 414–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, T.; Chen, T.; Cao, Z.; Zhong, S.; Wen, X.; Mi, J.; Ma, B.; Zou, Y.; Zhang, N.; Liao, X.; et al. Antibiotic resistance genes in layer farms and their correlation with environmental samples. Poult. Sci. 2021, 100, 101485. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.; Hur, H.G.; Sadowsky, M.J.; Byappanahalli, M.N.; Yan, T.; Ishii, S. Environmental Escherichia coli: Ecology and public health implications—A review. J. Appl. Microbiol. 2017, 123, 570–581. [Google Scholar] [CrossRef] [Green Version]
- Vila, J.; Sáez-López, E.; Johnson, J.R.; Römling, U.; Dobrindt, U.; Cantón, R.; Giske, C.G.; Naas, T.; Carattoli, A.; Martínez-Medina, M.; et al. Escherichia coli: An old friend with new tidings. FEMS Microbiol. Rev. 2016, 40, 437–463. [Google Scholar] [CrossRef] [Green Version]
- Canton, R.; Gonzalez-Alba, J.M.; Galan, J.C. CTX-M enzymes: Origin and diffusion. Front. Microbiol. 2012, 3, 110. [Google Scholar] [CrossRef] [Green Version]
- Dandachi, I.; Chabou, S.; Daoud, Z.; Rolain, J.M. Prevalence and emergence of extended-spectrum cephalosporin-, carbapenem- and colistin resistant Gram-negative bacteria of animal origin in the Mediterranean Basin. Front. Microbiol. 2018, 9, 2299. [Google Scholar] [CrossRef]
- Alonso, C.A.; Michael, G.B.; Li, J.; Somalo, S.; Simón, C.; Wang, Y.; Kaspar, H.; Kadlec, K.; Torres, C.; Schwarz, S. Analysis of blaSHV-12-carrying Escherichia coli clones and plasmids from human, animal, and food sources. J. Antimicrob. Chemother. 2017, 72, 1589–1596. [Google Scholar] [CrossRef]
- Vinué, L.; Sáenz, Y.; Somalo, S.; Escudero, E.; Moreno, M.Á.; Ruiz-Larrea, F.; Torres, C. Prevalence and diversity of integrons and associated resistance genes in faecal Escherichia coli isolates of healthy humans in Spain. J. Antimicrob. Chemother. 2008, 62, 934–937. [Google Scholar] [CrossRef] [Green Version]
- Briñas, L.; Moreno, M.A.; Zarazaga, M.; Porrero, C.; Sáenz, Y.; García, M.; Dominguez, L.; Torres, C. Detection of CMY-2, CTX-M-14, and SHV-12 β-lactamases in Escherichia coli fecal-sample isolates from healthy chickens. Antimicrob. Agents Chemother. 2003, 47, 2056–2058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coque, T.M.; Baquero, F.; Canton, R. Increasing prevalence of ESBL—Producing Enterobacteriaceae in Europe. Eurosurveillance 2008, 13, 19044. [Google Scholar] [CrossRef] [PubMed]
- Diestra, K.; Coque, T.M.; Miró, E.; Oteo, J.; Nicolau, C.J.; Campos, J.; Moyá, B.; Curiao, T.; Pérez-Vázquez, M.; Cantón, R.; et al. Caracterización y epidemiología molecular de betalactamasas de espectro extendido en Escherichia coli y Klebsiella pneumoniae en once hospitales españoles (2004). Enferm. Infecc. Microbiol. Clin. 2008, 26, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Costa, D.; Vinué, L.; Poeta, P.; Coelho, A.C.; Matos, M.; Sáenz, Y.; Somalo, S.; Zarazaga, M.; Rodrigues, J.; Torres, C. Prevalence of extended-spectrum beta-lactamase-producing Escherichia coli isolates in faecal samples of broilers. Vet. Microbiol. 2009, 138, 339–344. [Google Scholar] [CrossRef]
- Galler, H.; Luxner, J.; Petternel, C.; Reinthaler, F.F.; Habib, J.; Haas, D.; Kittinger, C.; Pless, P.; Feierl, G.; Zarfel, G. Multiresistant bacteria isolated from intestinal faeces of farm animals in Austria. Antibiotics 2021, 10, 466. [Google Scholar] [CrossRef]
- Ewers, C.; Jong, A.; de Prenger-Berninghoff, E.; Garch, F.; el Leidner, U.; Tiwari, S.K.; Semmler, T. Genomic diversity and virulence potential of ESBL- and AmpC-β-lactamase-producing Escherichia coli strains from healthy food animals across Europe. Front. Microbiol. 2021, 12, 626774. [Google Scholar] [CrossRef]
- Gillings, M.R. Integrons: Past, present, and future. Microbiol. Mol. Biol. Rev. 2014, 78, 257–277. [Google Scholar] [CrossRef] [Green Version]
- Kalantari, M.; Sharifiyazdi, H.; Asasi, K.; Abdi-Hachesoo, B. High incidence of multidrug resistance and class 1 and 2 integrons in Escherichia coli isolated from broiler chickens in South of Iran. Vet. Res. Forum. 2021, 12, 101–107. [Google Scholar] [CrossRef]
- Pérez-Etayo, L.; Berzosa, M.; González, D.; Vitas, A.I. Prevalence of integrons and insertion sequences in ESBL-producing E. coli isolated from different sources in Navarra, Spain. Int. J. Environ. Res. Public Health 2018, 15, 2308. [Google Scholar] [CrossRef] [Green Version]
- Sanz, S.; Olarte, C.; Hidalgo-Sanz, R.; Ruiz-Ripa, L.; Fernández-Fernández, R.; García-Vela, S.; Martínez-Álvarez, S.; Torres, C. Airborne dissemination of bacteria (enterococci, staphylococci and Enterobacteriaceae) in a modern broiler farm and its environment. Animals 2021, 11, 1783. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement, M100-Ed31; National Committee for Clinical Laboratory Standards: Wayne, PA, USA, 2021. [Google Scholar]
- Ruiz, E.; Sáenz, Y.; Zarazaga, M.; Rocha-Gracia, R.; Martínez-Martínez, L.; Arlet, G.; Torres, C. qnr, aac(6′)-Ib-cr and qepA genes in Escherichia coli and Klebsiella spp.: Genetic environments and plasmid and chromosomal location. J. Antimicrob. Chemother. 2012, 67, 886–897. [Google Scholar] [CrossRef] [PubMed]
- Vinué, L.; Sáenz, Y.; Martínez, S.; Somalo, S.; Moreno, M.A.; Torres, C.; Zarazaga, M. Prevalence and diversity of extended-spectrum ß-lactamases in faecal Escherichia coli isolates from healthy humans in Spain. Clin. Microbiol. Infect. 2009, 15, 954–957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sáenz, Y.; Vinué, L.; Ruiz, E.; Somalo, S.; Martínez, S.; Rojo-Bezares, B.; Zarazaga, M.; Torres, C. Class 1 integrons lacking qacEΔ1 and sul1 genes in Escherichia coli isolates of food, animal and human origins. Vet. Microbiol. 2010, 144, 493–497. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Bertini, A.; Villa, L.; Falbo, V.; Hopkins, K.L.; Threlfall, E.J. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 2005, 63, 219–228. [Google Scholar] [CrossRef]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- Weissman, S.J.; Johnson, J.R.; Tchesnokova, V.; Billig, M.; Dykhuizen, D.; Riddell, K.; Rogers, P.; Qin, X.; Butler-Wu, S.; Cookson, B.T.; et al. High-resolution two-locus clonal typing of extraintestinal pathogenic Escherichia coli. Appl. Environ. Microbiol. 2012, 78, 1353–1360. [Google Scholar] [CrossRef] [Green Version]
- Egea, P.; López-Cerero, L.; Torres, E.; Gómez-Sánchez, M.d.; Serrano, L.; Sánchez-Ortiz, M.D.N.; Rodriguez-Baño, J.; Pascual, A. Increased raw poultry meat colonization by extended spectrum beta-lactamase-producing Escherichia coli in the south of Spain. Int. J. Food Microbiol. 2012, 159, 69–73. [Google Scholar] [CrossRef]
- Irrgang, A.; Zhao, G.; Juraschek, K.; Kaesbohrer, A.; Hammerl, J.A. Characterization of E. coli isolates producing extended spectrum beta-lactamase SHV-variants from the food chain in Germany. Microorganisms 2021, 9, 1926. [Google Scholar] [CrossRef]
- Briñas, L.; Moreno, M.A.; Teshager, T.; Sáenz, Y.; Porrero, M.C.; Domínguez, L.; Torres, C. Monitoring and characterization of extended-spectrum β-lactamases in Escherichia coli strains from healthy and sick animals in Spain in 2003. Antimicrob. Agents Chemother. 2005, 49, 1262–1264. [Google Scholar] [CrossRef] [Green Version]
- Ejaz, H.; Younas, S.; Abosalif, K.O.A.; Junaid, K.; Alzahrani, B.; Alsrhani, A.; Abdalla, A.E.; Ullah, M.I.; Qamar, M.U.; Hamam, S.S.M. Molecular analysis of blaSHV, blaTEM, and blaCTX-M in extended-spectrum β-lactamase producing Enterobacteriaceae recovered from fecal specimens of animals. PLoS ONE 2021, 16, e0245126. [Google Scholar] [CrossRef]
- Osman, K.M.; Kappell, A.D.; Elhadidy, M.; ElMougy, F.; El-Ghany, W.A.A.; Orabi, A.; Mubarak, A.S.; Dawoud, T.M.; Hemeg, H.A.; Moussa, I.M.I.; et al. Poultry hatcheries as potential reservoirs for antimicrobial-resistant Escherichia coli: A risk to public health and food safety. Sci. Rep. 2018, 8, 5859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savin, M.; Bierbaum, G.; Hammerl, J.A.; Heinemann, C.; Parcina, M.; Sib, E.; Voigt, A.; Kreyenschmidt, J. ESKAPE bacteria and extended-spectrum-β-lactamase-producing Escherichia coli isolated from wastewater and process water from German poultry slaughterhouses. Appl. Environ. Microbiol. 2020, 86, e02748-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flament-Simon, S.C.; García, V.; Duprilot, M.; Mayer, N.; Alonso, M.P.; García-Meniño, I.; Blanco, J.E.; Blanco, M.; Nicolas-Chanoine, M.H.; Blanco, J. High prevalence of ST131 subclades C2-H30Rx and C1-M27 among extended-spectrum β-lactamase-producing Escherichia coli causing human extraintestinal infections in patients from two hospitals of Spain and France during 2015. Front. Cell. Infect. Microbiol. 2020, 10, 125. [Google Scholar] [CrossRef] [PubMed]
- Mamani, R.; Flament-Simon, S.C.; García, V.; Mora, A.; Alonso, M.P.; López, C.; García-Meniño, I.; Díaz-Jiménez, D.; Blanco, J.E.; Blanco, M.; et al. Sequence types, clonotypes, serotypes, and virotypes of extended-spectrum β-lactamase-producing Escherichia coli causing bacteraemia in a Spanish Hospital over a 12-Year Period (2000 to 2011). Front. Microbiol. 2019, 10, 1530. [Google Scholar] [CrossRef] [Green Version]
- Leverstein-van Hall, M.A.; Dierikx, C.M.; Stuart, J.C.; Voets, G.M.; van den Munckhof, M.P.; van Essen-Zandbergen, A.; Platteel, T.; Fluit, A.C.; van de Sande-Bruinsma, N.; Scharinga, J.; et al. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin. Microbiol. Infect. 2011, 17, 873–880. [Google Scholar] [CrossRef] [Green Version]
- Bortolaia, V.; Guardabassi, L.; Trevisani, M.; Bisgaard, M.; Venturi, L.; Bojesen, A.M. High diversity of extended-spectrum β-lactamases in Escherichia coli isolates from Italian broiler flocks. Antimicrob. Agents Chemother. 2010, 54, 1623–1626. [Google Scholar] [CrossRef] [Green Version]
- Dierikx, C.; van der Goot, J.; Fabri, T.; van Essen-Zandbergen, A.; Smith, H.; Mevius, D. Extended-spectrum-β-lactamase- and AmpC-β-lactamase-producing Escherichia coli in Dutch broilers and broiler farmers. J. Antimicrob. Chemother. 2013, 68, 60–67. [Google Scholar] [CrossRef]
- De Been, M.; Lanza, V.F.; de Toro, M.; Scharringa, J.; Dohmen, W.; Du, Y.; Hu, J.; Lei, Y.; Li, N.; Tooming-Klunderud, A.; et al. Dissemination of cephalosporin resistance genes between Escherichia coli strains from farm animals and humans by specific plasmid lineages. PLoS Genet. 2014, 10, e1004776. [Google Scholar] [CrossRef]
- Börjesson, S.; Egervärn, M.; Lindblad, M.; Englund, S. Frequent occurrence of extended-spectrum beta-lactamase- and transferable AmpC beta-lactamase-producing Escherichia coli on domestic chicken meat in Sweden. Appl. Environ. Microbiol. 2013, 79, 2463–2466. [Google Scholar] [CrossRef] [Green Version]
- Papouskova, A.; Masarikova, M.; Valcek, A.; Senk, D.; Cejkova, D.; Jahodarova, E.; Cizek, A. Genomic analysis of Escherichia coli strains isolated from diseased chicken in the Czech Republic. BMC Vet. Res. 2020, 16, 189. [Google Scholar] [CrossRef]
- Valverde, R.; Cantón, R.; Garcillán-Barcia, M.P.; Novais, Â.; Galán, J.C.; Alvarado, A.; de la Cruz, F.; Baquero, F.; Coque, T.M. Spread of blaCTX-M-14 is driven mainly by IncK plasmids disseminated among Escherichia coli phylogroups A, B1, and D in Spain. Antimicrob. Agents Chemother. 2009, 53, 5204–5212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siller, P.; Daehre, K.; Thiel, N.; Nübel, U.; Roesler, U. Impact of short-term storage on the quantity of extended-spectrum beta-lactamase–producing Escherichia coli in broiler litter under practical conditions. Poult. Sci. 2020, 99, 2125–2135. [Google Scholar] [CrossRef] [PubMed]
- Rumi, M.V.; Mas, J.; Elena, A.; Cerdeira, L.; Muñoz, M.E.; Lincopan, N.; Gentilini, É.R.; di Conza, J.; Gutkind, G. Co-occurrence of clinically relevant β-lactamases and mcr-1 encoding genes in Escherichia coli from companion animals in Argentina. Vet. Microbiol. 2019, 230, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Salgado-Caxito, M.; Benavides, J.A.; Adell, A.D.; Paes, A.C.; Moreno-Switt, A.I. Global prevalence and molecular characterization of extended-spectrum β-lactamase producing-Escherichia coli in dogs and cats—A scoping review and meta-analysis. One Health 2021, 12, 100236. [Google Scholar] [CrossRef]
- Wang, J.; Ma, Z.B.; Zeng, Z.L.; Yang, X.W.; Huang, Y.; Liu, J.H. The role of wildlife (wild birds) in the global transmission of antimicrobial resistance genes. Zool. Res. 2017, 38, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beyrouthy, R.; Sabença, C.; Robin, F.; Poeta, P.; Igrejas, G.; Bonnet, R. Successful dissemination of plasmid-mediated extended-spectrum β-lactamases in enterobacterales over humans to wild fauna. Microorganisms 2021, 9, 1471. [Google Scholar] [CrossRef]
- Ruiz-Garbajosa, P.; Hernández-García, M.; Beatobe, L.; Tato, M.; Méndez, M.I.; Grandal, M.; Aranzábal, L.; Alonso, S.; Lópaz, M.A.; Astray, J.; et al. A single-day point-prevalence study of faecal carriers in long-term care hospitals in Madrid (Spain) depicts a complex clonal and polyclonal dissemination of carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 2016, 71, 348–352. [Google Scholar] [CrossRef] [Green Version]
- Apostolakos, I.; Feudi, C.; Eichhorn, I.; Palmieri, N.; Fasolato, L.; Schwarz, S.; Piccirillo, A. High-resolution characterisation of ESBL/pAmpC-producing Escherichia coli isolated from the broiler production pyramid. Sci. Rep. 2020, 10, 11123. [Google Scholar] [CrossRef]
- Roedel, A.; Vincze, S.; Projahn, M.; Roesler, U.; Robé, C.; Hammerl, J.A.; Noll, M.; Al Dahouk, S.; Dieckmann, R. Genetic but no phenotypic associations between biocide tolerance and antibiotic resistance in Escherichia coli from German broiler fattening farms. Microorganisms 2021, 9, 651. [Google Scholar] [CrossRef]
- Cummins, M.L.; Reid, C.J.; Chowdhury, P.R.; Bushell, R.N.; Esbert, N.; Tivendale, K.A.; Noormohammadi, A.H.; Islam, S.; Marenda, M.S.; Browning, G.F.; et al. Whole genome sequence analysis of Australian avian pathogenic Escherichia coli that carry the class 1 integrase gene. Microb. Genom. 2019, 5, e000250. [Google Scholar] [CrossRef]
- Solà-Ginés, M.; Cameron-Veas, K.; Badiola, I.; Dolz, R.; Majó, N.; Dahbi, G.; Viso, S.; Mora, A.; Blanco, J.; Piedra-Carrasco, N.; et al. Diversity of multi-drug resistant avian pathogenic Escherichia coli (APEC) causing outbreaks of colibacillosis in broilers during 2012 in Spain. PLoS ONE 2015, 10, e143191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reich, F.; Atanassova, V.; Klein, G. Extended-spectrum β-lactamase- and AmpC-producing enterobacteria in healthy broiler chickens, Germany. Emerg. Infect. Dis. 2013, 19, 1253–1259. [Google Scholar] [CrossRef] [PubMed]
- Vounba, P.; Arsenault, J.; Bada-Alambédji, R.; Fairbrother, J.M. Prevalence of antimicrobial resistance and potential pathogenicity, and possible spread of third generation cephalosporin resistance, in Escherichia coli isolated from healthy chicken farms in the region of Dakar, Senegal. PLoS ONE 2019, 14, e0214304. [Google Scholar] [CrossRef] [PubMed]
- Moser, K.A.; Zhang, L.; Spicknall, I.; Braykov, N.P.; Levy, K.; Marrs, C.F.; Foxman, B.; Trueba, G.; Cevallos, W.; Goldstick, J.; et al. The role of mobile genetic elements in the spread of antimicrobial-resistant Escherichia coli from chickens to humans in small-scale production poultry operations in rural Ecuador. Am. J. Epidemiol. 2018, 187, 558–567. [Google Scholar] [CrossRef]
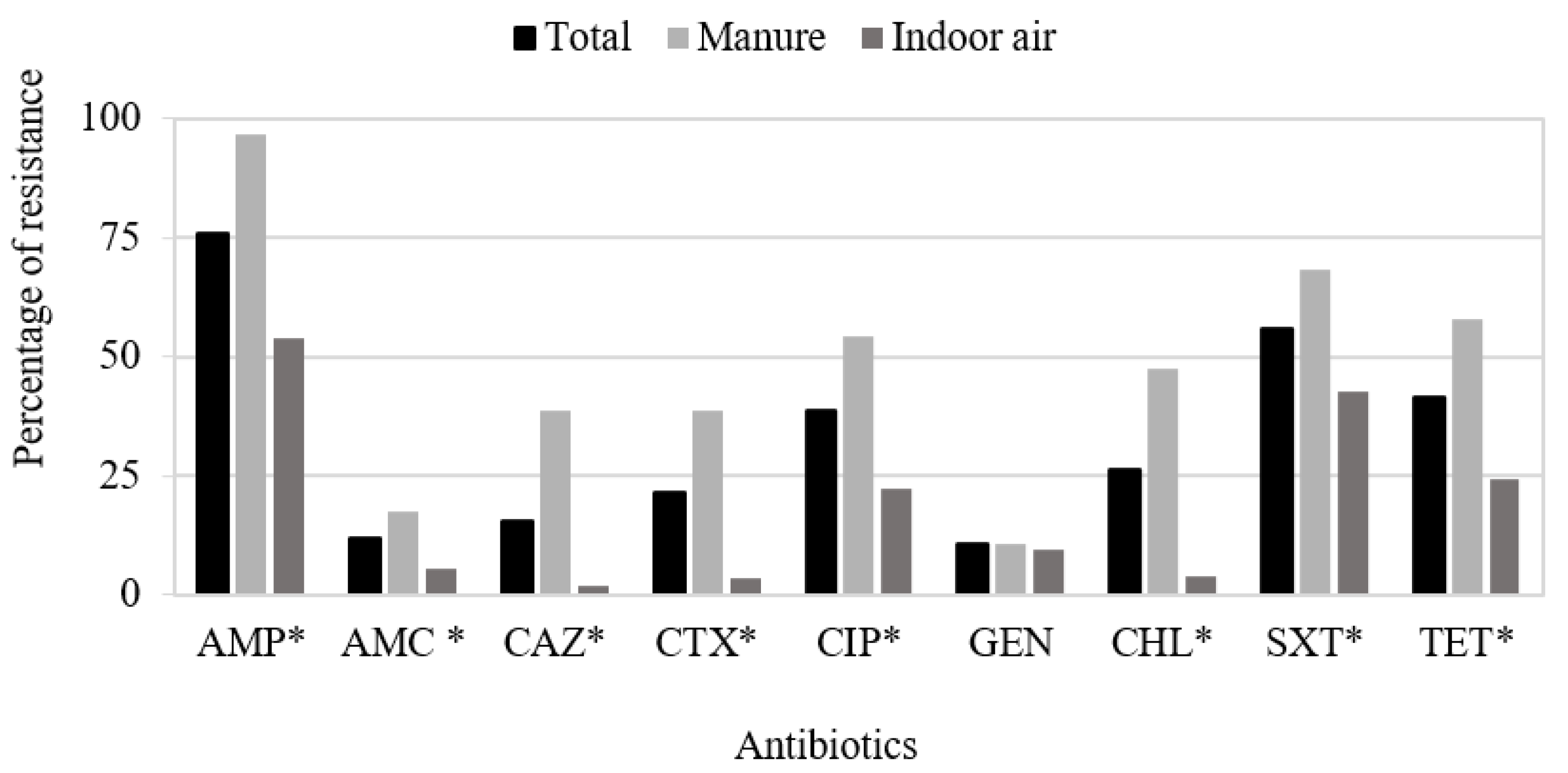

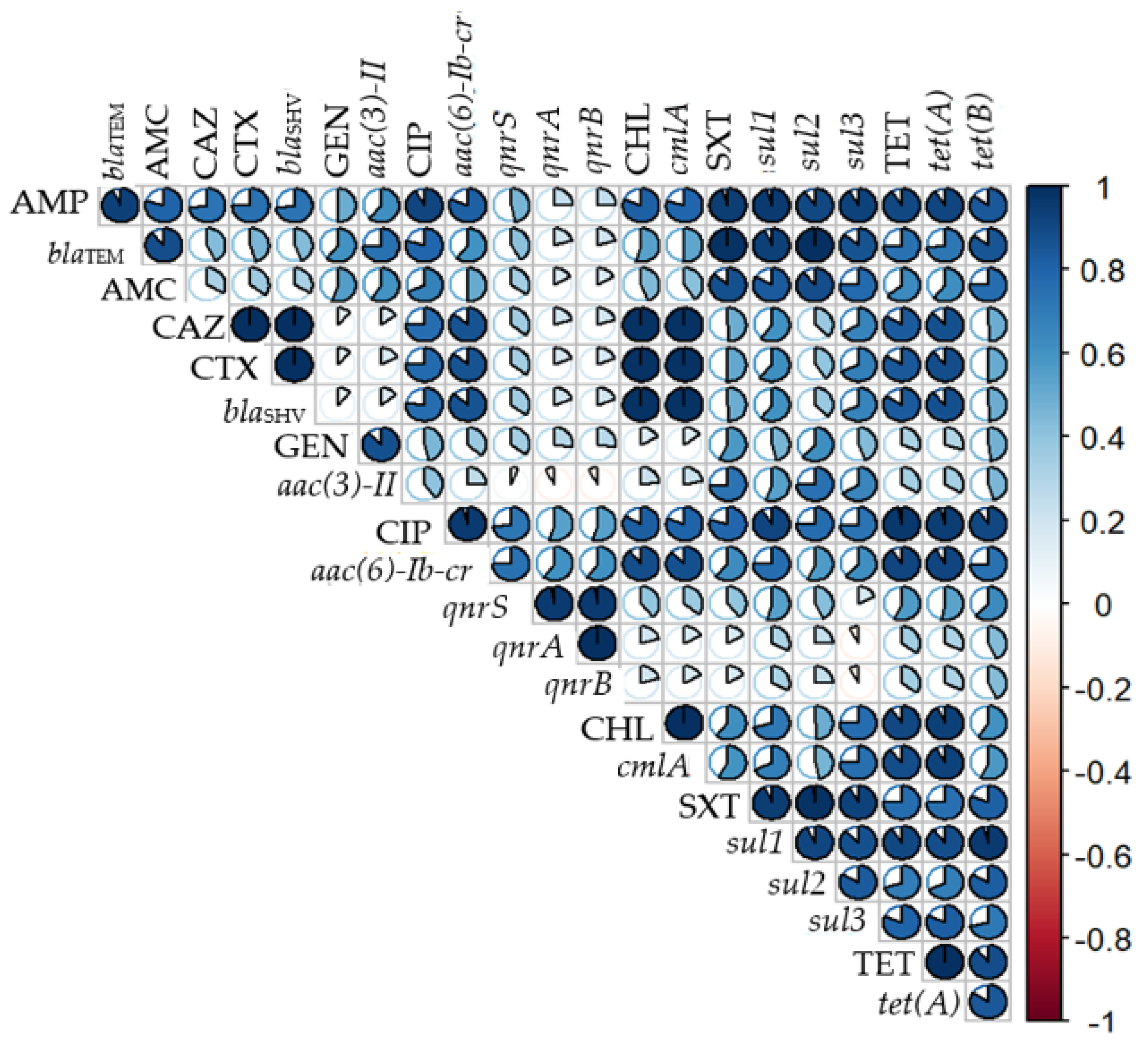
| E. coli Isolate | Origin | Resistance Phenotype a | Resistance Genotype | Phylogroup, Sequence Type (MLST), and Clonotype | Replicon Type |
|---|---|---|---|---|---|
| X2583 | Airborne | AMP, CAZ, CTX, CHL, CIP, TET | blaSHV-12, cmlA, tet(A), aac(6′)-Ib-cr | E-ST770-CH116–552 | IncF, IncK, IncB/O |
| X2685 | Manure | AMP, CAZ, CTX, CHL, CIP, TET | blaSHV-12, blaTEM-1, cmlA, tet(A), aac(6′)-Ib-cr | E-ST770-CH116–552 | IncFIB, IncF, IncK |
| X2636 | Manure | AMP, CAZ, CTX, CHL, CIP, TET | blaSHV-12, cmlA, tet(A), aac(6′)-Ib-cr | E-ST770-CH116–552 | IncFIB, IncF, IncK |
| X2630 | Manure | AMP, CAZ, CTX, CHL, CIP, TET | blaSHV-12, cmlA, tet(A), aac(6′)-Ib-cr | E-ST770-CH116–552 | IncFIB, IncF, IncB/O |
| X2686 | Manure | AMP, CAZ, CTX, CHL, CIP, TET | blaSHV-12, cmlA, tet(A), aac(6′)-Ib-cr | E-ST770-CH116–552 | IncF, IncK |
| X2635 | Manure | AMP, CAZ, CTX, CHL, CIP, TET | blaSHV-12, cmlA, tet(A), aac(6′)-Ib-cr | E-ST770-CH116–552 | IncFIB, IncF, IncK |
| X2637 | Manure | AMP, CAZ, CTX, CHL, CIP, TET | blaSHV-12, cmlA, tet(A), aac(6′)-Ib-cr | E-ST770-CH116–552 | IncFIB, IncF, IncK |
| X2639 | Manure | AMP, CAZ, CTX, CHL, CIP, TET | blaSHV-12, cmlA, tet(A), aac(6′)-Ib-cr | E-ST770-CH116–552 | IncI1, IncFIB, IncF, IncK |
| X2633 | Manure | AMP, CAZ, CTX, CHL, CIP, TET | blaSHV-12, cmlA, tet(A), aac(6′)-Ib-cr | E-ST770-CH116–552 | IncFIB, IncF, IncK |
| X2683 | Manure | AMP, CAZ, CTX, CHL, CIP, TET | blaSHV-12, cmlA, tet(A), aac(6′)-Ib-cr | E-ST770-CH116–552 | IncF, IncK |
| X2684 | Manure | AMP, CAZ, CTX, CHL, CIP, TET | blaSHV-12, cmlA, tet(A), aac(6′)-Ib-cr | E-ST770-CH116–552 | IncFIB, IncF, IncK |
| X2638 | Manure | AMP, CAZ, CTX, CHL, CIP b, TET | blaSHV-12, cmlA, tet(A) | E-ST770-CH116–552 | IncFIB, IncF, IncK |
| X2632 | Manure | AMP, CAZ, CTX, CHL, CIP b, TET | blaSHV-12, cmlA, tet(A) | E-ST68-CH26–382 | IncFIB, IncF |
| X2634 | Manure | AMP, CAZ, CTX, CHL | blaSHV-12, cmlA, tet(A) | E-ST68-CH26–382 | IncFIB, IncF |
| X2682 | Manure | AMP, CAZ, CTX, CHL, CIP | blaSHV-12, cmlA | E-ST68-CH26–49 | IncI1, IncP, IncF |
| X2631 | Manure | AMP, CAZ, CTX, CHL, SXT, TET | blaSHV-12, cmlA, tet(A), sul1, dfrA1, aadA1 | B2-ST117-CH45–97 | IncFIB, IncF, IncB/O |
| X2640 | Manure | AMP, CAZ, CTX, CHL | blaSHV-12, cmlA | B2-ST117- CH45–97 | IncFIB, IncF, IncK |
| X2641 | Manure | AMP, CAZ, CTX, CHL | blaSHV-12, cmlA | B2-ST117-CH45–97 | IncFIB, IncF, IncK |
| X2642 | Manure | AMP, CAZ, CTX, CHL | blaSHV-12, cmlA | B2-ST117-CH45–97 | IncFIB, IncF, IncK |
| X2643 | Manure | AMP, CAZ, CTX, CHL, SXT, TET | blaSHV-12, cmlA, tet(A), sul1, sul3, dfrA1, aadA1 | A-ST10992-CH11–23 | IncFIB, IncF |
| X2644 | Manure | AMP, CAZ, CTX, CHL, SXT, TET | blaSHV-12, cmlA, tet(A), sul3 | A-ST10992-CH11–41 | IncFIB |
| X2646 | Manure | AMP, CAZ, CTX, CHL, SXT, TET | blaSHV-12, cmlA, tet(A), sul3 | B1-ST10992-CH11–41 | IncP, IncI1 |
| X2645 | Manure | AMP, CAZ, CTX, CHL, SXT, TET | blaSHV-12, cmlA, tet(A), sul3 | B1-ST10992-CH11–580 | IncP, IncK |
| Antibiotic | Number of Resistant Isolates | Resistance Genes Detected (Number of Isolates) | Integrase of Class 1/2 Integrons (Number of Isolates) |
|---|---|---|---|
| Ampicillin | 61 | blaTEM (58) | - |
| Ciprofloxacin | 29 | aac6′-Ib-cr (6), qnrS (2) | - |
| Gentamicin | 11 | aac(3)-II (2) | - |
| Chloramphenicol | 7 | cmlA (6) | - |
| SXT a | 57 | sul1 (2), sul2 (31), sul3 (7), sul1 + sul2 (9), sul2 + sul3 (2) | int1 (26), int2 (7), int1 + int2 (5) |
| Tetracycline | 28 | tet(A) (12), tet(B) (3), tet(A) + tet(B) (7) |
| E. coli Isolate | Origin | Resistance Phenotype a | ESBL Phenotype b | Resistance Genotype | Phylogroup (MLST) | Class 1 Integron | Class 2 Integron | ||
|---|---|---|---|---|---|---|---|---|---|
| intl1/3′CS | VR c | intl2 | VR c | ||||||
| X2631 | Manure | AMP, CAZ, CTX, CHL, SXT, TET | + | blaSHV-12, cmlA, tet(A) | B2 (ST117) | +/+ | dfrA1-aadA1 | − | |
| X2643 | Manure | AMP, CAZ, CTX, CHL, SXT, TET | + | blaSHV-12, cmlA, tet(A), sul3 | A (ST10992) | +/+ | dfrA1-aadA1 | − | |
| X2576 | Airborne | AMP, SXT, TET | − | blaTEM-1, tet(A), sul2 | A (ST5766) | +/+ | dfrA1-aadA1 | − | |
| X2671 | Manure | AMP, CIP, SXT, TET | − | blaTEM-1, tet(A), sul2 | D (ST69) | +/+ | dfrA1-aadA1 | − | |
| X2579 | Airborne | AMP, CIP, SXT, TET | − | blaTEM-1, tet(A), sul2 | A | +/+ | dfrA1-aadA1 | − | |
| X2580 | Airborne | AMP, CIP, SXT, TET | − | blaTEM-1, tet(A), sul2 | E | +/+ | dfrA1-aadA1 | − | |
| X2675 | Manure | AMP, AMC, CIP, SXT, TET | − | blaTEM-1, tet(A), sul2 | A | +/+ | dfrA1-aadA1 | − | |
| X2679 | Manure | AMP, SXT | − | blaTEM-1, sul2 | A | +/+ | dfrA1-aadA1 | − | |
| X2674 | Manure | AMP, AMC, CIP, SXT, TET | − | blaTEM-1, tet(B), sul2 | A | +/+ | dfrA12-aadA2 | + | sat2-aadA1 |
| X2680 | Manure | AMP, CHL, CIP, SXT, TET | − | tet(A), aac(6′)-Ib-cr, qnrS | E | +/+ | dfrA12-aadA2 | − | |
| X2681 | Manure | AMP, CHL, CIP, SXT, TET | − | tet(B), cmlA, aac(6′)-Ib-cr, sul2, qnrS | A | +/+ | dfrA12-orfX-aadA2-cmlA | − | |
| X2665 | Manure | AMP, AMC, TET, SXT, CHL | − | blaTEM-1, cmlA, tet(A), sul2, sul3 | C | +/− | + | sat2-aadA1 | |
| X2662 | Manure | AMP, AMC, TET, SXT, CHL | − | blaTEM-1, cmlA, sul3 | A | +/− | + | sat2-aadA1 | |
| X2658 | Manure | AMP, TET, SXT, CHL | − | blaTEM-1, tet(A), sul3, cmlA | A | +/− | + | sat2-aadA1 | |
| X2666 | Manure | AMP, TET, SXT, CHL | blaTEM-1, cmlA, tet(A), sul3 | C | +/− | + | sat2-aadA1 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Álvarez, S.; Sanz, S.; Olarte, C.; Hidalgo-Sanz, R.; Carvalho, I.; Fernández-Fernández, R.; Campaña-Burguet, A.; Latorre-Fernández, J.; Zarazaga, M.; Torres, C. Antimicrobial Resistance in Escherichia coli from the Broiler Farm Environment, with Detection of SHV-12-Producing Isolates. Antibiotics 2022, 11, 444. https://doi.org/10.3390/antibiotics11040444
Martínez-Álvarez S, Sanz S, Olarte C, Hidalgo-Sanz R, Carvalho I, Fernández-Fernández R, Campaña-Burguet A, Latorre-Fernández J, Zarazaga M, Torres C. Antimicrobial Resistance in Escherichia coli from the Broiler Farm Environment, with Detection of SHV-12-Producing Isolates. Antibiotics. 2022; 11(4):444. https://doi.org/10.3390/antibiotics11040444
Chicago/Turabian StyleMartínez-Álvarez, Sandra, Susana Sanz, Carmen Olarte, Raquel Hidalgo-Sanz, Isabel Carvalho, Rosa Fernández-Fernández, Allelen Campaña-Burguet, Javier Latorre-Fernández, Myriam Zarazaga, and Carmen Torres. 2022. "Antimicrobial Resistance in Escherichia coli from the Broiler Farm Environment, with Detection of SHV-12-Producing Isolates" Antibiotics 11, no. 4: 444. https://doi.org/10.3390/antibiotics11040444
APA StyleMartínez-Álvarez, S., Sanz, S., Olarte, C., Hidalgo-Sanz, R., Carvalho, I., Fernández-Fernández, R., Campaña-Burguet, A., Latorre-Fernández, J., Zarazaga, M., & Torres, C. (2022). Antimicrobial Resistance in Escherichia coli from the Broiler Farm Environment, with Detection of SHV-12-Producing Isolates. Antibiotics, 11(4), 444. https://doi.org/10.3390/antibiotics11040444








