Strategies for Enzymatic Inactivation of the Veterinary Antibiotic Florfenicol
Abstract
:1. Introduction
2. Results
2.1. Ability of the Hydrolase Estdl136 to Confer Resistance to Chloramphenicol and Florfenicol
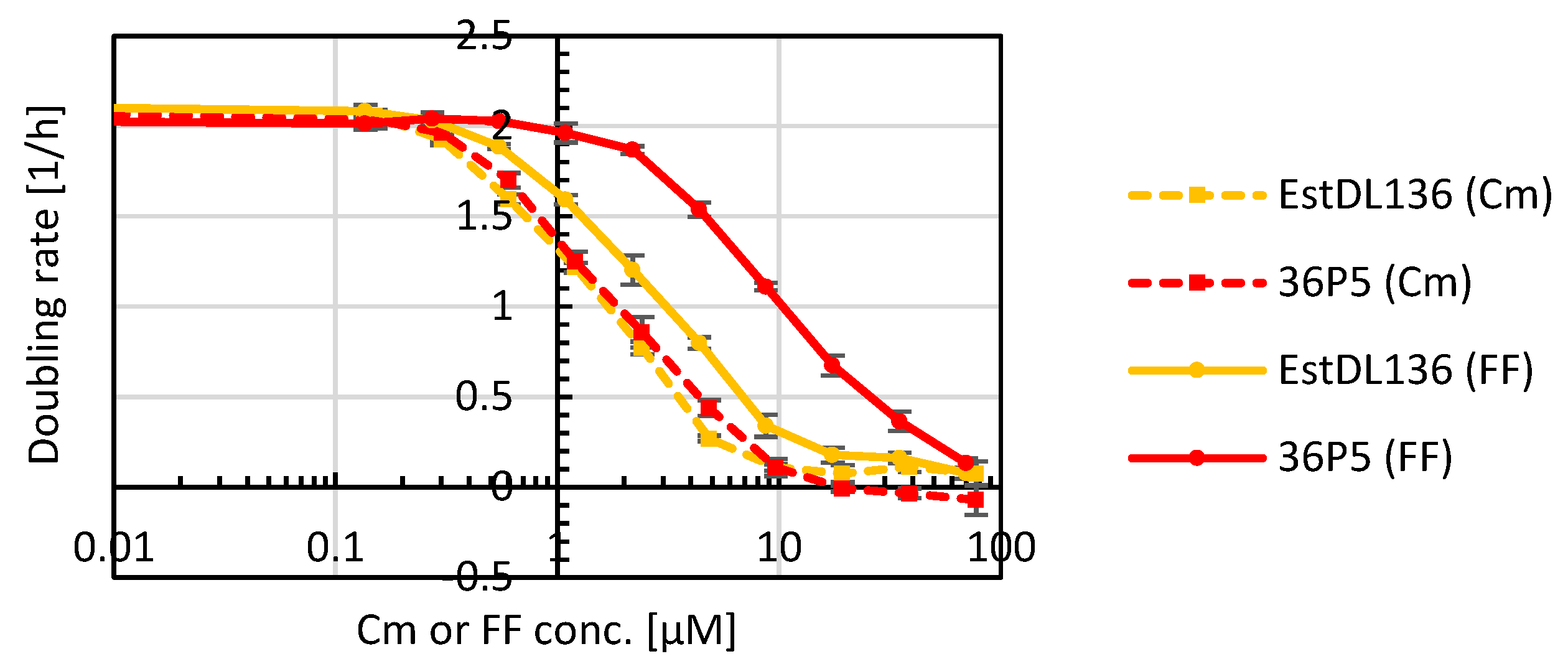
2.2. Optimization of the Hydrolase Estdl136 for Increased Florfenicol Inactivation
2.3. Characterization of the Hydrolase Mutant 36P5
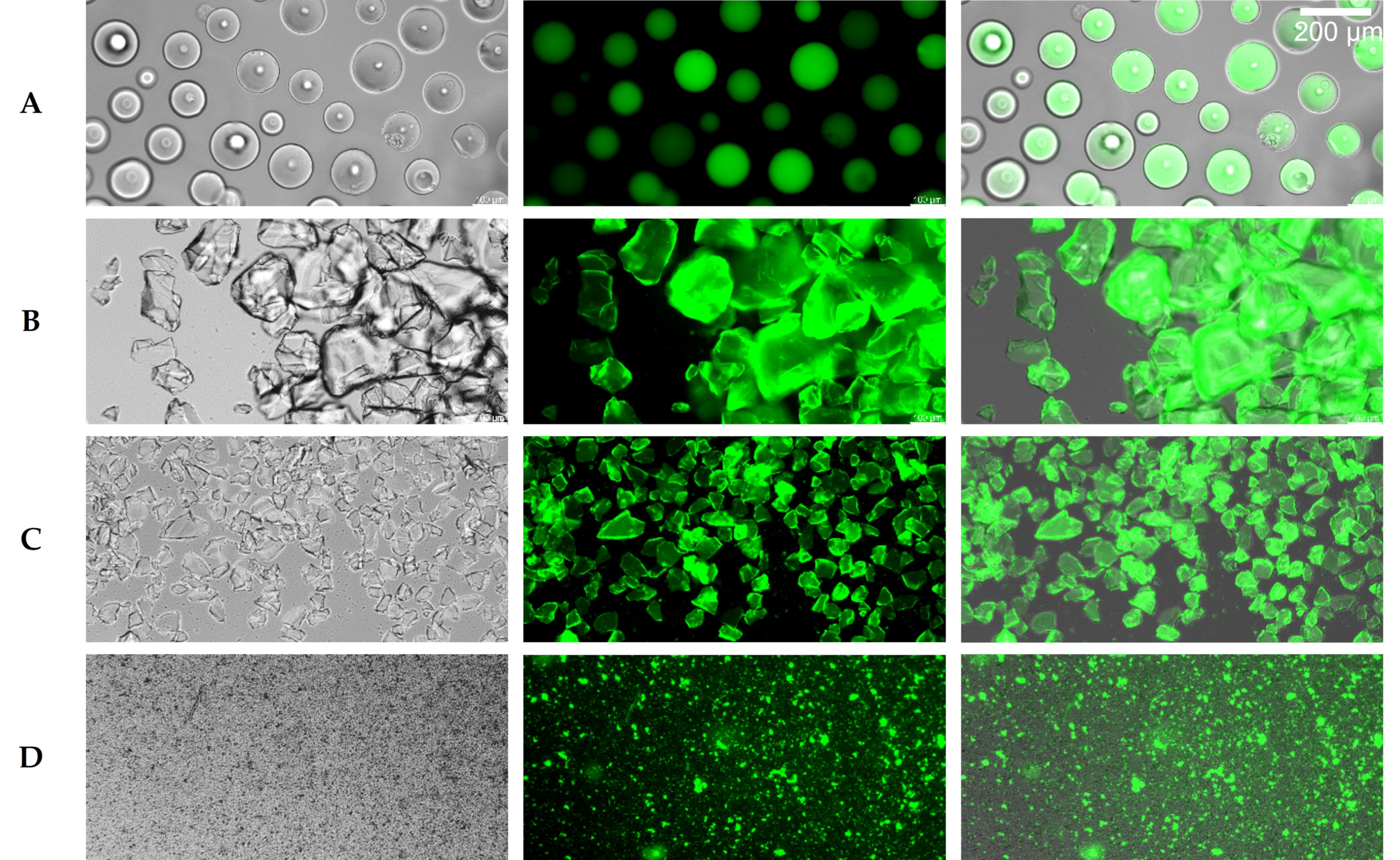
2.4. Hydrolyase Immobilization
2.5. Enzymatic Inactivation of Florfenicol in Salt Solutions
2.6. Enzymatic Inactivation of Florfenicol in Milk
3. Discussion
4. Materials and Methods
4.1. Plasmids
4.2. Strains
4.3. Library Generation and Selection
4.4. Bacterial Growth Assays
4.5. Protein Expression
4.6. Protein Purification
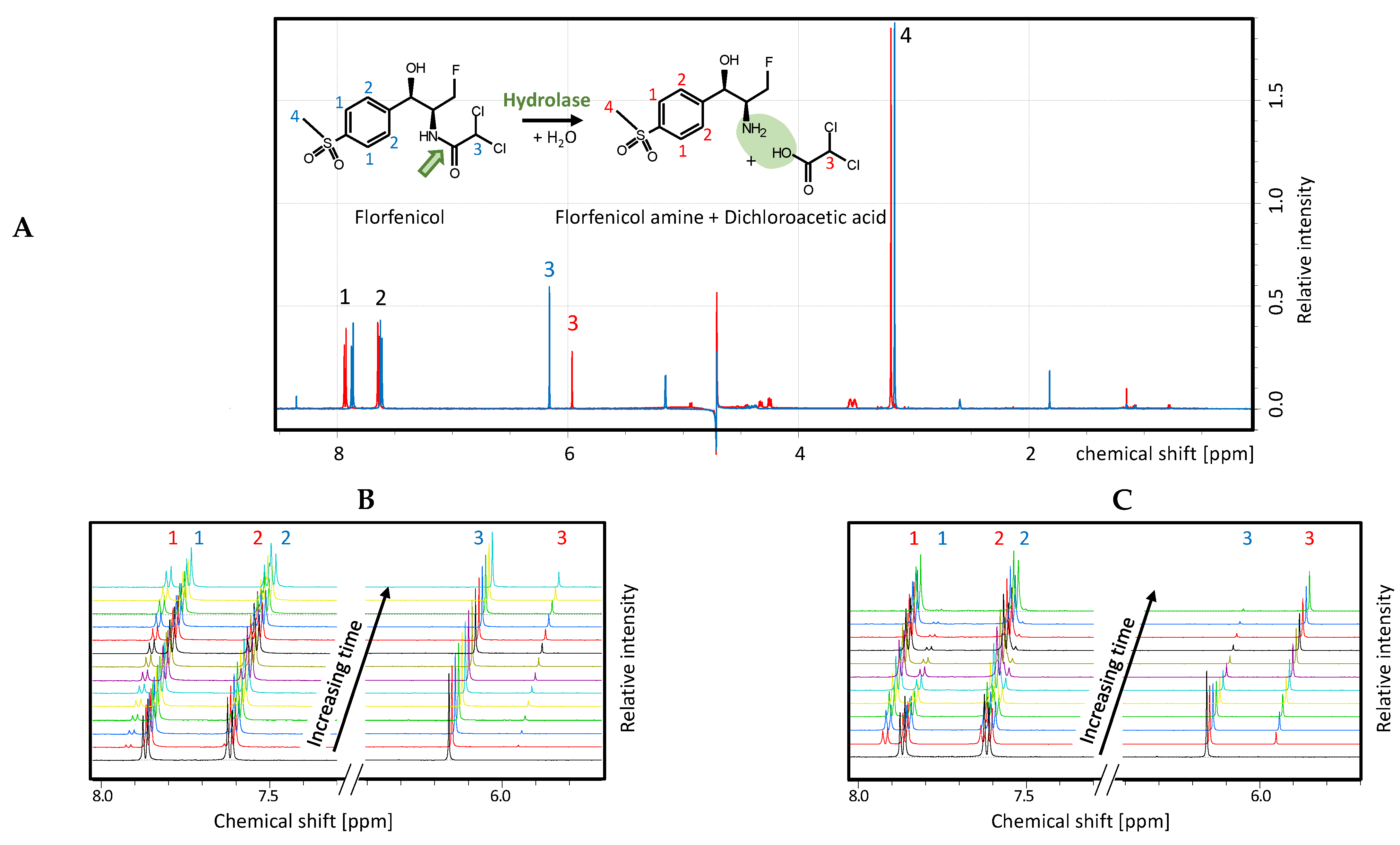
4.7. NMR Measurements
4.8. Immobilization
4.9. Florfenicol Inactivation Assay
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- O’Neill, J. Antimicrobial Resistance: Tackling a crisis for the health and wealth of nations. Review on Antimicrobial Resistance, Commissioned by the United Kingdom Government; Wellcome Collection: London, UK, 2014. [Google Scholar]
- Klein, E.Y.; Van Boeckel, T.P.; Martinez, E.M.; Pant, S.; Gandra, S.; Levin, S.A. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA 2018, 115, 3463–3470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Van Hoek, A.H.A.M.; Mevius, D.; Guerra, B.; Mullany, P.; Roberts, A.P.; Aarts, H.J.M. Acquired antibiotic resistance genes: An overview. Front. Microbiol. 2011, 2, 203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Zhu, T.; Zhou, D.; Lu, W.; Liu, H.; Sun, Z.; Ying, J.; Lu, J.; Lin, X.; Li, K.; et al. Analysis of Resistance to Florfenicol and the Related Mechanism of Dissemination in Different Animal-Derived Bacteria. Front. Cell. Infect. Microbiol. 2020, 10, 369. [Google Scholar] [CrossRef]
- Van Cuong, N.; Nhung, N.T.; Nghia, N.H.; Mai Hoa, N.T.; Trung, N.V.; Thwaites, G.; Carrique-Mas, J. Antimicrobial Consumption in Medicated Feeds in Vietnamese Pig and Poultry Production. Ecohealth 2016, 13, 490–498. [Google Scholar] [CrossRef] [Green Version]
- Higuera-Llantén, S.; Vásquez-Ponce, F.; Barrientos-Espinoza, B.; Mardones, F.O.; Marshall, S.H.; Olivares-Pacheco, J. Extended antibiotic treatment in salmon farms select multiresistant gut bacteria with a high prevalence of antibiotic resistance genes. PLoS ONE 2018, 13, e0203641. [Google Scholar] [CrossRef] [Green Version]
- Lozano, I.; Díaz, N.F.; Muñoz, S.; Lozano, I.; Díaz, N.F.; Muñoz, S.; Riquelme, C. Antibiotics in Chilean Aquaculture: A Review. In Antibiotic Use in Animals; Savic, S., Ed.; IntechOpen Limited: London, UK, 2018; pp. 25–44. [Google Scholar]
- Miranda, C.D.; Godoy, F.A.; Lee, M.R. Current Status of the Use of Antibiotics and the Antimicrobial Resistance in the Chilean Salmon Farms. Front. Microbiol. 2018, 9, 1284. [Google Scholar] [CrossRef]
- Subbiah, M.; Mitchell, S.M.; Ullman, J.L.; Call, D.R. Beta-Lactams and Florfenicol Antibiotics Remain Bioactive in Soils while Ciprofloxacin, Neomycin, and Tetracycline Are Neutralized. Appl. Environ. Microbiol. 2011, 77, 7255–7260. [Google Scholar] [CrossRef] [Green Version]
- Miranda, C.D.; Rojas, R. Occurrence of florfenicol resistance in bacteria associated with two Chilean salmon farms with different history of antibacterial usage. Aquaculture 2007, 266, 39–46. [Google Scholar] [CrossRef]
- Zeng, Q.; Liao, C.; Terhune, J.; Wang, L. Impacts of florfenicol on the microbiota landscape and resistome as revealed by metagenomic analysis. Microbiome 2019, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- FDA 2018 Summary Report on Antimicrobials Sold or Distributed for Use in Food Producing Animals; U.S. Food Drug Administration, Center for Veterinary Medicine: Silver Spring, MD, USA, 2019.
- Abdelfattah, E.M.; Ekong, P.S.; Okello, E.; Chamchoy, T.; Karle, B.M.; Black, R.A.; Sheedy, D.; ElAshmawy, W.R.; Williams, D.R.; Califano, D.; et al. Epidemiology of antimicrobial resistance (AMR) on California dairies: Descriptive and cluster analyses of AMR phenotype of fecal commensal bacteria isolated from adult cows. PeerJ 2021, 9, e11108. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liao, X.; Xiong, Z.; Lin, Q.; Wen, J.; Xu, C.; Qu, X.; Chen, K.; Zhang, J. Characterization of the emerging multidrug- resistant Salmonella enterica serotype Kentucky ST314 in China. Zoonoses Public Health 2021, 68, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Wallmann, J.; Bode, C.; Heberer, T. Abgabemengenerfassung von Antibiotika in Deutschland 2019. Dtsch. Tierärzteblatt 2020, 68, 1102–1109. [Google Scholar]
- Power, C.; Sayers, R.; Danaher, M.; Moloney, M.; O’Brien, B.; Furey, A.; Jordan, K. Investigation of the persistence of florfenicol residues in bovine milk and fate during processing. Int. Dairy J. 2014, 39, 270–275. [Google Scholar] [CrossRef]
- Firth, C.L.; Kremer, K.; Werner, T.; Käsbohrer, A. The effects of feeding waste milk containing antimicrobial residues on dairy calf health. Pathogens 2021, 10, 112. [Google Scholar] [CrossRef]
- Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Fernández Escámez, P.S.; Girones, R.; Koutsoumanis, K.; Lindqvist, R.; Nørrung, B.; et al. Risk for the development of Antimicrobial Resistance (AMR) due to feeding of calves with milk containing residues of antibiotics. EFSA J. 2017, 15, 4665. [Google Scholar] [CrossRef] [Green Version]
- Maynou, G.; Migura-Garcia, L.; Chester-Jones, H.; Ziegler, D.; Bach, A.; Terré, M. Effects of feeding pasteurized waste milk to dairy calves on phenotypes and genotypes of antimicrobial resistance in fecal Escherichia coli isolates before and after weaning. J. Dairy Sci. 2017, 100, 7967–7979. [Google Scholar] [CrossRef] [Green Version]
- Maynou, G.; Bach, A.; Terré, M. Feeding of waste milk to Holstein calves affects antimicrobial resistance of Escherichia coli and Pasteurella multocida isolated from fecal and nasal swabs. J. Dairy Sci. 2017, 100, 2682–2694. [Google Scholar] [CrossRef]
- Schwarz, S.; Kehrenberg, C.; Doublet, B.; Cloeckaert, A. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 2004, 28, 519–542. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Zhang, J.; Xu, L.; Liu, Y.; Li, P.; Zhu, T.; Cheng, C.; Lu, S. Spread of the florfenicol resistance floR gene among clinical Klebsiella pneumoniae isolates in China. Antimicrob. Resist. Infect. Control 2018, 7, 127. [Google Scholar] [CrossRef] [Green Version]
- Baucheron, S.; Tyler, S.; Boyd, D.; Mulvey, M.R.; Chaslus-Dancla, E.; Cloeckaert, A. AcrAB-TolC directs efflux-mediated multidrug resistance in Salmonella enterica serovar typhimurium DT104. Antimicrob. Agents Chemother. 2004, 48, 3729–3735. [Google Scholar] [CrossRef] [Green Version]
- Antonelli, A.; D’Andrea, M.M.; Brenciani, A.; Galeotti, C.L.; Morroni, G.; Pollini, S.; Varaldo, P.E.; Rossolini, G.M. Characterization of poxtA, a novel phenicol-oxazolidinone-tetracycline resistance gene from an MRSA of clinical origin. J. Antimicrob. Chemother. 2018, 73, 1763–1769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, K.S.; Poehlsgaard, J.; Kehrenberg, C.; Schwarz, S.; Vester, B. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 2006, 50, 2500–2505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharkey, L.K.R.; Edwards, T.A.; O’Neill, A.J. ABC-F proteins mediate antibiotic resistance through ribosomal protection. MBio 2016, 7, e01975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, W.; Lee, M.H.; Yoon, M.Y.; Kim, J.C.; Malhotra, S.; Wu, J.; Hwang, E.C.; Lee, S.W. Characterization of two metagenome-derived esterases that reactivate chloramphenicol by counteracting chloramphenicol acetyltransferase. J. Microbiol. Biotechnol. 2011, 21, 1203–1210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, W.; Lee, M.H.; Wu, J.; Kim, N.H.; Kim, J.C.; Chung, E.; Hwang, E.C.; Lee, S.W. Inactivation of chloramphenicol and florfenicol by a novel chloramphenicol hydrolase. Appl. Environ. Microbiol. 2012, 78, 6295–6301. [Google Scholar] [CrossRef] [Green Version]
- Zhan, Z.; Xu, X.; Shen, H.; Gao, Y.; Zeng, F.; Qu, X.; Zhang, H.; Liao, M.; Zhang, J. Rapid emergence of florfenicol-resistant invasive non-typhoidal salmonella in China: A potential threat to public health. Am. J. Trop. Med. Hyg. 2019, 101, 1282–1285. [Google Scholar] [CrossRef]
- Qian, C.; Liu, H.; Cao, J.; Ji, Y.; Lu, W.; Lu, J.; Li, A.; Zhu, X.; Shen, K.; Xu, H.; et al. Identification of floR Variants Associated With a Novel Tn4371-Like Integrative and Conjugative Element in Clinical Pseudomonas aeruginosa Isolates. Front. Cell. Infect. Microbiol. 2021, 11, 685068. [Google Scholar] [CrossRef]
- Ghebreyesus, T.A. Antimicrobial Resistance; World Health Assembly; World Health Organization: Geneva, Switzerland, 2019.
- Kim, S.H.; Kang, P.A.; Han, K.; Lee, S.W.; Rhee, S. Crystal structure of chloramphenicol-metabolizing enzyme EstDL136 from a metagenome. PLoS ONE 2019, 14, e0210298. [Google Scholar] [CrossRef] [Green Version]
- The PyMOL Molecular Graphics System; Version 2.3.4; Schrödinger, LLC: New York, NY, USA, 2020.
- Abdelhamid, M.A.A.; Motomura, K.; Ikeda, T.; Ishida, T.; Hirota, R.; Kuroda, A. Affinity purification of recombinant proteins using a novel silica-binding peptide as a fusion tag. Appl. Microbiol. Biotechnol. 2014, 98, 5677–5684. [Google Scholar] [CrossRef]
- Abdelhamid, M.A.A.; Ikeda, T.; Motomura, K.; Tanaka, T.; Ishida, T.; Hirota, R.; Kuroda, A. Application of volcanic ash particles for protein affinity purification with a minimized silica-binding tag. J. Biosci. Bioeng. 2016, 122, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, K.; Nomura, K.; Hata, Y.; Nishimura, T.; Asami, Y.; Kuroda, A. The Si-Tag for Immobilizing Proteins on a Silica Surface. Biotechnol. Bioeng. 2007, 96, 1023–1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeda, T.; Ninomiya, K.; Hirota, R.; Kuroda, A. Single-step affinity purification of recombinant proteins using the silica-binding Si-tag as a fusion partner. Protein Expr. Purif. 2010, 71, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, T.; Kuroda, A. Why does the silica-binding protein “Si-tag” bind strongly to silica surfaces? Implications of conformational adaptation of the intrinsically disordered polypeptide to solid surfaces. Colloids Surf. B Biointerfaces 2011, 86, 359–363. [Google Scholar] [CrossRef] [PubMed]
- Murray, M.; Salvatierra, G.; Dávila-Barclay, A.; Ayzanoa, B.; Castillo-Vilcahuaman, C.; Huang, M.; Pajuelo, M.J.; Lescano, A.G.; Cabrera, L.; Calderón, M.; et al. Market Chickens as a Source of Antibiotic-Resistant Escherichia coli in a Peri-Urban Community in Lima, Peru. Front. Microbiol. 2021, 12, 635871. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.M.; Eichman, J.; Katz, T.; Gilewicz, R. Stability of florfenicol in drinking water. J. AOAC Int. 2003, 86, 22–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, S.M.; Ullman, J.L.; Teel, A.L.; Watts, R.J. Chemosphere Hydrolysis of amphenicol and macrolide antibiotics: Chloramphenicol, florfenicol, spiramycin, and tylosin. Chemosphere 2015, 134, 504–511. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J.; Zhou, L.; Wang, G.; Feng, Y. Aqueous photodegradation of antibiotic florfenicol: Kinetics and degradation pathway studies. Environ. Sci. Pollut. Res. 2016, 23, 6982–6989. [Google Scholar] [CrossRef]
- Pouliquen, H.; Delépée, R.; Larhantec-Verdier, M.; Morvan, M.L.; Le Bris, H. Comparative hydrolysis and photolysis of four antibacterial agents (oxytetracycline oxolinic acid, flumequine and florfenicol) in deionised water, freshwater and seawater under abiotic conditions. Aquaculture 2007, 262, 23–28. [Google Scholar] [CrossRef]
- Boreen, A.L.; Arnold, W.A.; McNeill, K. Photodegradation of pharmaceuticals in the aquatic environment: A review. Aquat. Sci. 2003, 65, 320–341. [Google Scholar] [CrossRef]
- Hanna, N.; Sun, P.; Sun, Q.; Li, X.; Yang, X.; Ji, X.; Zou, H.; Ottoson, J.; Nilsson, L.E.; Berglund, B.; et al. Presence of antibiotic residues in various environmental compartments of Shandong province in eastern China: Its potential for resistance development and ecological and human risk. Environ. Int. 2018, 114, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Jin, K.Y.; Shahzad, M.; Corresp, A. The occurrence of pharmaceutical waste in different parts of the world: A scoping review. PeerJ Prepr. 2019, 7, e27951v1. [Google Scholar] [CrossRef]
- Feng, L.; Cheng, Y.; Zhang, Y.; Li, Z.; Yu, Y.; Feng, L.; Zhang, S.; Xu, L. Distribution and human health risk assessment of antibiotic residue s in large-scale drinking water sources in Chongqing area of the Yangtze River. Environ. Res. 2020, 185, 109386. [Google Scholar] [CrossRef] [PubMed]
- Barani, A.; Fallah, A.A. Occurrence of tetracyclines, sulfonamides, fluoroquinolones and florfenicol in farmed rainbow trout in Iran. Food Agric. Immunol. 2015, 26, 420–429. [Google Scholar] [CrossRef]
- Bortolotte, A.R.; Daniel, D.; Reyes, F.G.R. Occurrence of antimicrobial residues in tilapia (Oreochromis niloticus) fillets produced in Brazil and available at the retail market. Food Res. Int. 2021, 140, 109865. [Google Scholar] [CrossRef]
- Gürbüz, S.; Baydan, E.; Türe, M.; Taçbaş, E.; Akbulut, B.; Özcelep, T. The effect of cooking and cold storage processes on florfenicol residues in muscle tissues of sturgeon (Acipenser gueldenstaedtii) reared in black sea. Pakistan J. Agric. Sci. 2021, 58, 421–427. [Google Scholar] [CrossRef]
- Li, M.; Li, Z.; Zhong, Q.; Liu, J.; Han, G.; Li, Y.; Li, C. Antibiotic resistance of fecal carriage of Escherichia coli from pig farms in China: A meta-analysis. Environ. Sci. Pollut. Res. 2021, 1–12. [Google Scholar] [CrossRef]
- Wu, C.; Zhang, X.; Liang, J.; Li, Q.; Lin, H.; Lin, C.; Liu, H.; Zhou, D.; Lu, W.; Sun, Z.; et al. Characterization of florfenicol resistance genes in the coagulase-negative Staphylococcus (CoNS) isolates and genomic features of a multidrug-resistant Staphylococcus lentus strain H29. Antimicrob. Resist. Infect. Control 2021, 10, 9. [Google Scholar] [CrossRef]
- Ibrahim, R.A.; Cryer, T.L.; Lafi, S.Q.; Basha, E.; Good, L.; Tarazi, Y.H. Identification of Escherichia coli from broiler chickens in Jordan, their antimicrobial resistance, gene characterization and the associated risk factors. BMC Vet. Res. 2019, 15, 159. [Google Scholar] [CrossRef]
- Liao, C.Y.; Balasubramanian, B.; Peng, J.J.; Tao, S.R.; Liu, W.C.; Ma, Y. Antimicrobial Resistance of Escherichia coli From Aquaculture Farms and Their Environment in Zhanjiang, China. Front. Vet. Sci. 2021, 8, 806653. [Google Scholar] [CrossRef]
- Adesoji, A.T.; Call, D.R. Molecular analysis of florfenicol-resistant bacteria isolated from drinking water distribution systems in Southwestern Nigeria. J. Glob. Antimicrob. Resist. 2020, 23, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Fang, Y.; Zhu, J.; Xu, W.; Zhu, K. Characterization of Bacillus species from market foods in Beijing, China. Processes 2021, 9, 866. [Google Scholar] [CrossRef]
- Li, S.M.; Zhou, Y.F.; Li, L.; Fang, L.X.; Duan, J.H.; Liu, F.R.; Liang, H.Q.; Wu, Y.T.; Gu, W.Q.; Liao, X.P.; et al. Characterization of the multi-drug resistance gene cfr in methicillin-resistant staphylococcus aureus (MRSA) strains isolated from animals and humans in China. Front. Microbiol. 2018, 9, 2925. [Google Scholar] [CrossRef] [PubMed]
- Udo, E.E.; Boswihi, S.S.; Mathew, B.; Noronha, B.; Verghese, T. Resurgence of chloramphenicol resistance in methicillin-resistant staphylococcus aureus due to the acquisition of a variant florfenicol exporter (Fexav)-mediated chloramphenicol resistance in Kuwait hospitals. Antibiotics 2021, 10, 1250. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Sijak, S.; Zheng, M.; Tang, L.; Xu, G.; Wu, M. Aquatic photolysis of florfenicol and thiamphenicol under direct UV irradiation, UV/H2O2 and UV/Fe(II) processes. Chem. Eng. J. 2015, 260, 826–834. [Google Scholar] [CrossRef]
- Gao, Y.; Gao, N.; Deng, Y. Degradation of florfenicol in water by UV/Na2S2O8 process. Environ. Sci. Pollut. Res. 2015, 22, 8693–8701. [Google Scholar] [CrossRef]
- Maurer, R.; Meyer, B.J.; Ptashne, M. Gene regulation at the right operator (OR) of bacteriophage λ. I. OR3 and autogenous negative control by repressor. J. Mol. Biol. 1980, 139, 147–161. [Google Scholar] [CrossRef]
- Marisch, K.; Bayer, K.; Cserjan-Puschmann, M.; Luchner, M.; Striedner, G. Evaluation of three industrial Escherichia coli strains in fed-batch cultivations during high-level SOD protein production. Microb. Cell Fact. 2013, 12, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Waldo, G.S. Genetic screens and directed evolution for protein solubility. Curr. Opin. Chem. Biol. 2003, 7, 33–38. [Google Scholar] [CrossRef]
- Roodveldt, C.; Aharoni, A.; Tawfik, D.S. Directed evolution of proteins for heterologous expression and stability. Curr. Opin. Struct. Biol. 2005, 15, 50–56. [Google Scholar] [CrossRef]
- Potrykus, J.; Wegrzyn, G. Chloramphenicol-sensitive Escherichia coli strain expressing the chloramphenicol acetyltransferase (cat) gene. Antimicrob. Agents Chemother. 2001, 45, 3610–3612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takamura, Y.; Nomura, G. Changes in the Intracellular Concentration of Acetyl-CoA and Malonyl-CoA in Relation to the Carbon and Energy Metabolism of Escherichia coli K12. Microbiology 1988, 134, 2249–2253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imperial, S.; Lin, T.; Posarac, V. Intracellular acetyl-CoA depletion by the cat gene is responsible for growth inhibition of Escherichia coli C584 on M9 minimal media. J. Exp. Microbiol. Immun. 2005, 7, 49–56. [Google Scholar]
- Derrick, J.P.; Lian, L.-Y.; Roberts, G.C.K.; Shaw, W.V. Analysis of the Binding of 1,3-Diacetylchloramphenicol to Chloramphenicol Acetyltransferase by Isotope-Edited lH NMR and Site-Directed Mutagenesis. Biochemistry 1992, 31, 8191–8195. [Google Scholar] [CrossRef]
- Crabb, D.W.; Dixon, J.E. A method for increasing the sensitivity of chloramphenicol acetyltransferase assays in extracts of transfected cultured cells. Anal. Biochem. 1987, 163, 88–92. [Google Scholar] [CrossRef]
- Chapman, J.; Ismail, A.E.; Dinu, C.Z. Industrial applications of enzymes: Recent advances, techniques, and outlooks. Catalysts 2018, 8, 238. [Google Scholar] [CrossRef] [Green Version]
- Gaberc-Porekar, V.; Menart, V. Potential for using histidine tags in purification of proteins at large scale. Chem. Eng. Technol. 2005, 28, 1306–1314. [Google Scholar] [CrossRef]
- Imam, H.T.; Marr, P.C.; Marr, A.C. Enzyme entrapment, biocatalyst immobilization without covalent attachment. Green Chem. 2021, 23, 4980–5005. [Google Scholar] [CrossRef]
- Kitazono, Y.; Ihara, I.; Toyoda, K.; Umetsu, K. Antibiotic removal from waste milk by electrochemical process: Degradation characteristics in concentrated organic solution. J. Mater. Cycles Waste Manag. 2017, 19, 1261–1269. [Google Scholar] [CrossRef]
- Zhang, Y.; Werling, U.; Edelmann, W. SLiCE: A novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res. 2012, 40, e55. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]

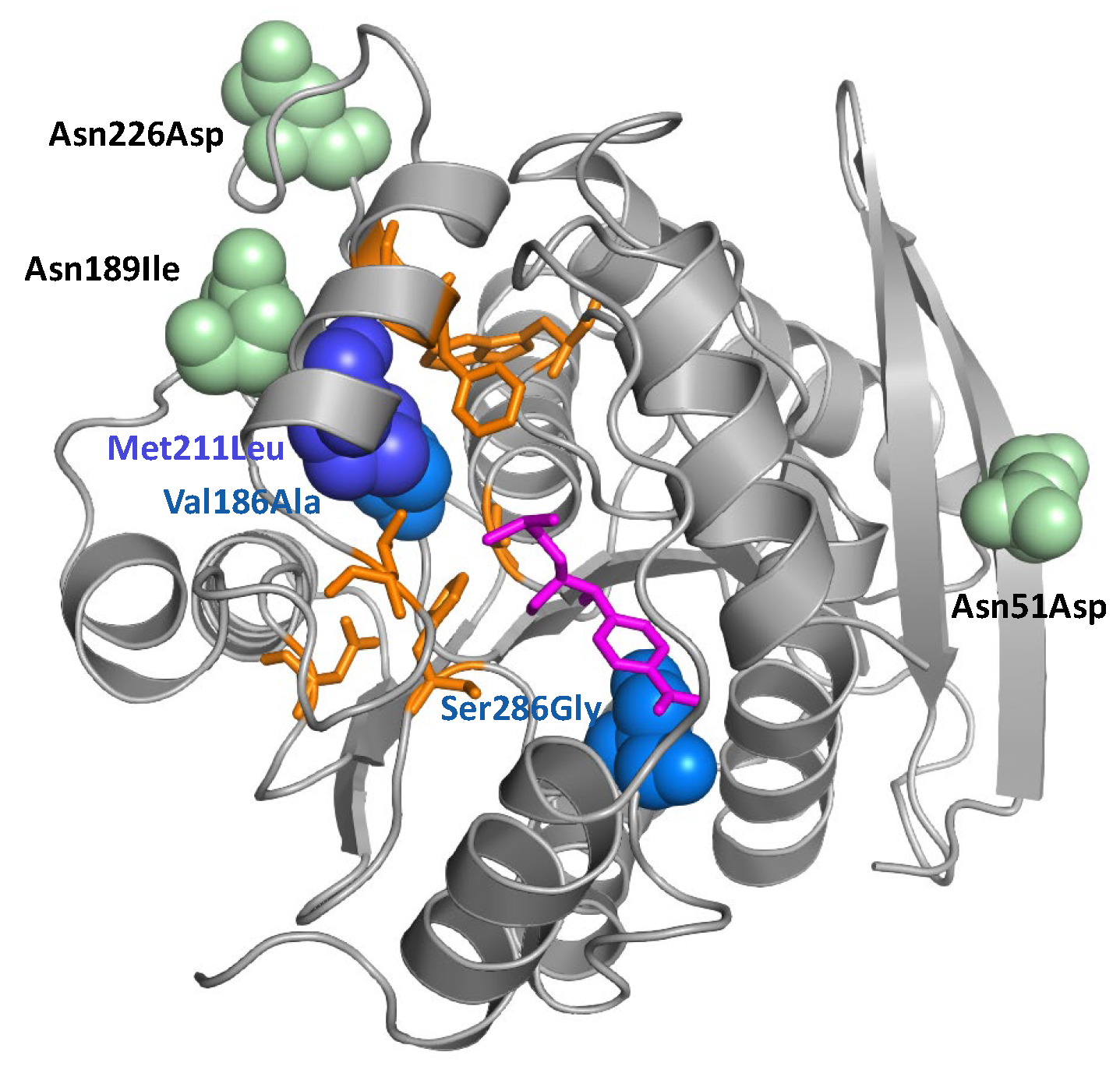



| Amino Acid Position 1 | Location in the Protein 2 | Wt Hydrolase | Mutant 33P5 (1×) 3 | Mutant 36P4 (2×) | Mutant 36P5 (2×) |
|---|---|---|---|---|---|
| 51 | surface | Asn (AAT) | Asp (GAT) | ||
| 186 | near Cm | Val (GTA) | Ala (GCA) | Ala (GCA) | Ala (GCA) |
| 189 | surface | Asn (AAT) | Ile (ATT) | ||
| 211 | near Cm | Met (ATG) | Leu (TTG) | Leu (TTG) | |
| 226 | surface | Asn (AAT) | Asp (GAT) | ||
| 286 | near Cm | Ser (AGC) | Gly (GGC) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Müller, M.M.; Nedielkov, R.; Arndt, K.M. Strategies for Enzymatic Inactivation of the Veterinary Antibiotic Florfenicol. Antibiotics 2022, 11, 443. https://doi.org/10.3390/antibiotics11040443
Müller MM, Nedielkov R, Arndt KM. Strategies for Enzymatic Inactivation of the Veterinary Antibiotic Florfenicol. Antibiotics. 2022; 11(4):443. https://doi.org/10.3390/antibiotics11040443
Chicago/Turabian StyleMüller, Marik M., Ruslan Nedielkov, and Katja M. Arndt. 2022. "Strategies for Enzymatic Inactivation of the Veterinary Antibiotic Florfenicol" Antibiotics 11, no. 4: 443. https://doi.org/10.3390/antibiotics11040443
APA StyleMüller, M. M., Nedielkov, R., & Arndt, K. M. (2022). Strategies for Enzymatic Inactivation of the Veterinary Antibiotic Florfenicol. Antibiotics, 11(4), 443. https://doi.org/10.3390/antibiotics11040443







