Combination of Amphiphilic Cyclic Peptide [R4W4] and Levofloxacin against Multidrug-Resistant Bacteria
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis and Purification of the Compounds
2.2. Antibacterial Activity
2.3. Combination Studies Using Levo and Peptides
2.4. Hemolytic Study
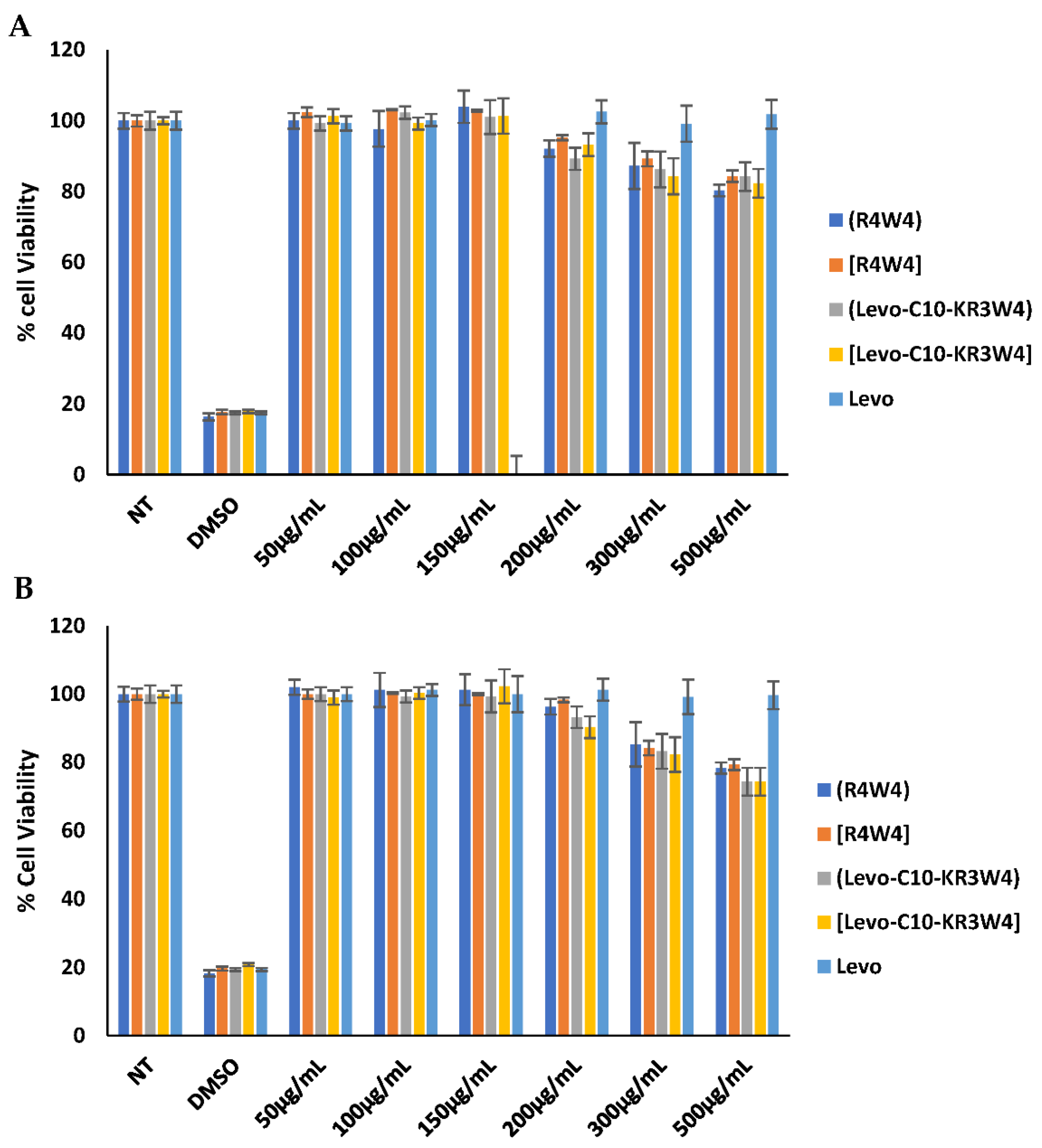
2.5. Cytotoxicity Assay
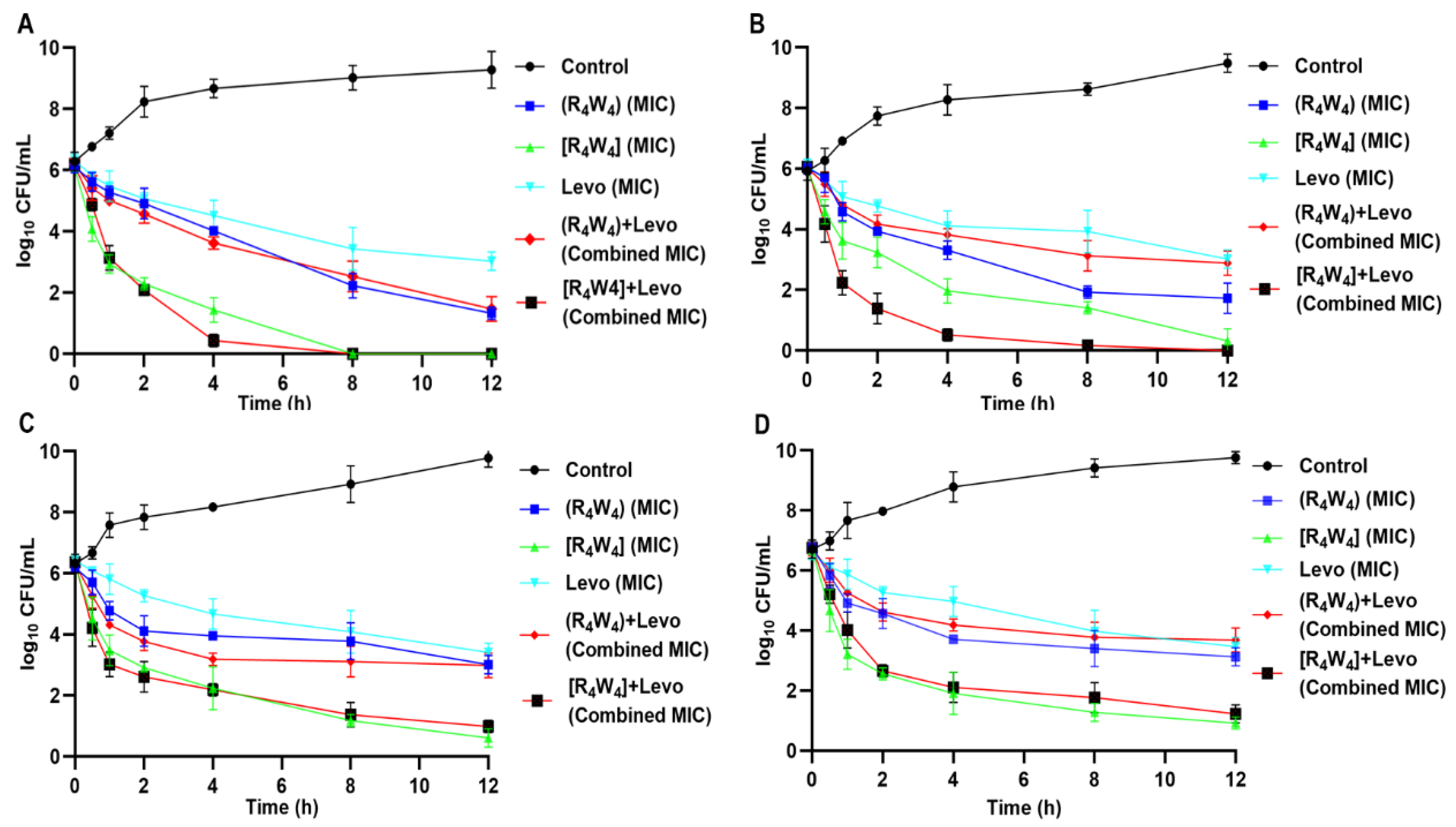
2.6. Time Kill Kinetics
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Design and Synthesis of Levo-[W4R3K] Conjugate
3.3. Antibacterial Activity
3.3.1. Bacterial Strains
3.3.2. Determination of Minimal Inhibitory Concentrations (MICs)
3.3.3. Checkerboard Assay
3.4. Hemolysis Assay
3.5. Cytotoxicity Assay
3.6. Time-Kill Kinetics Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hu, X.-Y.; Logue, M.; Robinson, N. Antimicrobial resistance is a global problem—A UK perspective. Eur. J. Integr. Med. 2020, 36, 101136. [Google Scholar] [CrossRef] [PubMed]
- Denny, K.J.; De Wale, J.; Laupland, K.B.; Harris, P.N.; Lipman, J. When not to start antibiotics: Avoiding antibiotic overuse in the intensive care unit. Clin. Microbiol. Infect. 2020, 26, 35–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jubeh, B.; Breijyeh, Z.; Karaman, R. Resistance of gram-positive bacteria to current antibacterial agents and overcoming approaches. Molecules 2020, 25, 2888. [Google Scholar] [CrossRef]
- Bhuiyan, M.S.; Jiang, J.-H.; Kostoulias, X.; Theegala, R.; Lieschke, G.J.; Peleg, A.Y. The Resistance to Host Antimicrobial Peptides in Infections Caused by Daptomycin-Resistant Staphylococcus aureus. Antibiotics 2021, 10, 96. [Google Scholar] [CrossRef] [PubMed]
- Hurdle, J.G.; O’neill, A.J.; Chopra, I.; Lee, R.E. Targeting bacterial membrane function: An underexploited mechanism for treating persistent infections. Nat. Rev. Microbiol. 2011, 9, 62–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nuding, S.; Frasch, T.; Schaller, M.; Stange, E.F.; Zabel, L.T. Synergistic effects of antimicrobial peptides and antibiotics against Clostridium difficile. Antimicrob. Agents Chemother. 2014, 58, 5719–5725. [Google Scholar] [CrossRef] [Green Version]
- Jahangiri, A.; Neshani, A.; Mirhosseini, S.A.; Ghazvini, K.; Zare, H.; Sedighian, H. Synergistic effect of two antimicrobial peptides, Nisin and P10 with conventional antibiotics against extensively drug-resistant Acinetobacter baumannii and colistin-resistant Pseudomonas aeruginosa isolates. Microb. Pathog. 2021, 150, 104700. [Google Scholar] [CrossRef]
- Choi, H.; Lee, D.G. Synergistic effect of antimicrobial peptide arenicin-1 in combination with antibiotics against pathogenic bacteria. Res. Microbiol. 2012, 163, 479–486. [Google Scholar] [CrossRef]
- Di Li, Y.Y.; Tian, Z.; Lv, J.; Sun, F.; Wang, Q.; Liu, Y.; Xia, P. Synergistic antibiotic effect of looped antimicrobial peptide CLP-19 with bactericidal and bacteriostatic agents. Oncotarget 2017, 8, 55958. [Google Scholar] [CrossRef] [Green Version]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Haney, E.F.; Vogel, H.J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011, 29, 464–472. [Google Scholar] [CrossRef]
- Hancock, R.E.; Sahl, H.-G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef] [PubMed]
- Lázár, V.; Martins, A.; Spohn, R.; Daruka, L.; Grézal, G.; Fekete, G.; Számel, M.; Jangir, P.K.; Kintses, B.; Csörgő, B. Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides. Nat. Microbiol. 2018, 3, 718–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuzaki, K. Control of cell selectivity of antimicrobial peptides. Biochim. Biophys. Acta 2009, 1788, 1687–1692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haney, E.F.; Hancock, R.E. Peptide design for antimicrobial and immunomodulatory applications. Pept. Sci. 2013, 100, 572–583. [Google Scholar] [CrossRef]
- Dijksteel, G.S.; Ulrich, M.M.W.; Middelkoop, E.; Boekema, B. Review: Lessons Learned From Clinical Trials Using Antimicrobial Peptides (AMPs). Front. Microbiol. 2021, 12, 616979. [Google Scholar] [CrossRef]
- Greber, K.E.; Dawgul, M. Antimicrobial Peptides Under Clinical Trials. Curr. Top. Med. Chem. 2017, 17, 620–628. [Google Scholar] [CrossRef]
- Pfalzgraff, A.; Brandenburg, K.; Weindl, G. Antimicrobial peptides and their therapeutic potential for bacterial skin infections and wounds. Front. Pharmacol. 2018, 9, 281. [Google Scholar] [CrossRef]
- Biswaro, L.S.; da Costa Sousa, M.G.; Rezende, T.; Dias, S.C.; Franco, O.L. Antimicrobial peptides and nanotechnology, recent advances and challenges. Front. Microbiol. 2018, 9, 855. [Google Scholar] [CrossRef] [Green Version]
- Lopes, F.E.; da Costa, H.P.; Souza, P.F.; Oliveira, J.P.; Ramos, M.V.; Freire, J.E.; Jucá, T.L.; Freitas, C.D. Peptide from thaumatin plant protein exhibits selective anticandidal activity by inducing apoptosis via membrane receptor. Phytochemistry 2019, 159, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, J.T.; Souza, P.F.; Vasconcelos, I.M.; Dias, L.P.; Martins, T.F.; Van Tilburg, M.F.; Guedes, M.I.; Sousa, D.O. Mo-CBP3-PepI, Mo-CBP3-PepII, and Mo-CBP3-PepIII are synthetic antimicrobial peptides active against human pathogens by stimulating ROS generation and increasing plasma membrane permeability. Biochimie 2019, 157, 10–21. [Google Scholar] [CrossRef]
- Riahifard, N.; Mozaffari, S.; Aldakhil, T.; Nunez, F.; Alshammari, Q.; Alshammari, S.; Yamaki, J.; Parang, K.; Tiwari, R.K. Design, synthesis, and evaluation of amphiphilic cyclic and linear peptides composed of hydrophobic and positively-charged amino acids as antibacterial agents. Molecules 2018, 23, 2722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, D.; Sun, J.; Nasrolahi Shirazi, A.; LaPlante, K.L.; Rowley, D.C.; Parang, K. Antibacterial activities of amphiphilic cyclic cell-penetrating peptides against multidrug-resistant pathogens. Mol. Pharm. 2014, 11, 3528–3536. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, J.; Ashley, D.; Cao, R.; Abrahem, R.; Nguyen, T.; To, K.; Yegiazaryan, A.; Akinwale David, A.; Kumar Tiwari, R.; Venketaraman, V. Cyclic Peptide [R4W4] in Improving the Ability of First-Line Antibiotics to Inhibit Mycobacterium tuberculosis Inside in vitro Human Granulomas. Front. Immunol. 2020, 11, 1677. [Google Scholar] [CrossRef]
- Zeiders, S.M.; Chmielewski, J. Antibiotic-cell-penetrating peptide conjugates targeting challenging drug-resistant and intracellular pathogenic bacteria. Chem. Biol. Drug Des. 2021, 98, 762–778. [Google Scholar] [CrossRef]
- Surur, A.S.; Sun, D. Macrocycle-Antibiotic Hybrids: A Path to Clinical Candidates. Front. Chem. 2021, 9, 659845. [Google Scholar] [CrossRef]
- Drlica, K.; Zhao, X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 1997, 61, 377–392. [Google Scholar]
- Anderson, V.R.; Perry, C.M. Levofloxacin. Drugs 2008, 68, 535–565. [Google Scholar] [CrossRef]
- Riahifard, N.; Tavakoli, K.; Yamaki, J.; Parang, K.; Tiwari, R. Synthesis and evaluation of antimicrobial activity of [R4W4K]-Levofloxacin and [R4W4K]-Levofloxacin-Q conjugates. Molecules 2017, 22, 957. [Google Scholar] [CrossRef]
- Abdul Ghaffar, K.; Hussein, W.M.; Khalil, Z.G.; Capon, R.J.; Skwarczynski, M.; Toth, I. Levofloxacin and indolicidin for combination antimicrobial therapy. Curr. Drug Deliv. 2015, 12, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Azad, Z.M.; Moravej, H.; Fasihi-Ramandi, M.; Masjedian, F.; Nazari, R.; Mirnejad, R.; Moghaddam, M.M. In Vitro synergistic effects of a short cationic peptide and clinically used antibiotics against drug-resistant isolates of Brucella melitensis. J. Med. Microbiol. 2017, 66, 919–926. [Google Scholar] [CrossRef] [PubMed]
- Kampshoff, F.; Willcox, M.D.; Dutta, D. A pilot study of the synergy between two antimicrobial peptides and two common antibiotics. Antibiotics 2019, 8, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, C.; Miller, M.J. Chemical syntheses and in vitro antibacterial activity of two desferrioxamine B-ciprofloxacin conjugates with potential esterase and phosphatase triggered drug release linkers. Bioorg. Med. Chem. 2012, 20, 3828–3836. [Google Scholar] [CrossRef] [Green Version]
- Elshikh, M.; Ahmed, S.; Funston, S.; Dunlop, P.; McGaw, M.; Marchant, R.; Banat, I.M. Resazurin-based 96-well plate microdilution method for the determination of minimum inhibitory concentration of biosurfactants. Biotechnol. Lett. 2016, 38, 1015–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaara, M.; Siikanen, O.; Apajalahti, J.; Fox, J.; Frimodt-Møller, N.; He, H.; Poudyal, A.; Li, J.; Nation, R.L.; Vaara, T. A novel polymyxin derivative that lacks the fatty acid tail and carries only three positive charges has strong synergism with agents excluded by the intact outer membrane. Antimicrob. Agents Chemother. 2010, 54, 3341–3346. [Google Scholar] [CrossRef] [Green Version]
- Sopirala, M.M.; Mangino, J.E.; Gebreyes, W.A.; Biller, B.; Bannerman, T.; Balada-Llasat, J.-M.; Pancholi, P. Synergy testing by Etest, microdilution checkerboard, and time-kill methods for pan-drug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2010, 54, 4678–4683. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.; Urban, C.; Terzian, C.; Mariano, N.; Rahal, J.J. In Vitro double and triple synergistic activities of polymyxin B, imipenem, and rifampin against multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2004, 48, 753–757. [Google Scholar] [CrossRef] [Green Version]
- Den Hollander, J.G.; Mouton, J.W.; Verbrugh, H.A. Use of pharmacodynamic parameters to predict efficacy of combination therapy by using fractional inhibitory concentration kinetics. Antimicrob. Agents Chemother. 1998, 42, 744–748. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Baeder, D.Y.; Regoes, R.R.; Rolff, J. Combination effects of antimicrobial peptides. Antimicrob. Agents Chemother. 2016, 60, 1717–1724. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Li, Z.; Li, X.; Tian, Y.; Fan, Y.; Yu, C.; Zhou, B.; Liu, Y.; Xiang, R.; Yang, L. Synergistic effects of antimicrobial peptide DP7 combined with antibiotics against multidrug-resistant bacteria. Drug Des. Dev. Ther. 2017, 11, 939. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Bacterial Strain | MIC (µg/mL) (µM) a | ||||||
|---|---|---|---|---|---|---|---|
| Levo | (R4W4) | [R4W4] | (Levo-C10-KR3W4) | [Levo-C10-KR3W4] | Levo + (R4W4) b | Levo + [R4W4] b | |
| S. aureus (ATCC 29213) | 3.13 | 6.25 | 3.13 | 12.5 | 25 | 3.13 | 3.13 |
| (8.66) | (4.51) | (2.28) | (6.67) | (13.45) | |||
| S. aureus (ATCC BAA-1556) c | 3.13 | 6.25 | 3.13 | 12.5 | 50 | 3.13 | 6.25 |
| (8.66) | (4.51) | (2.28) | (6.67) | (26.90) | |||
| E. coli (ATCC 25922) | 50 | 25 | 6.25 | 50 | 50 | 12.5 | 12.5 |
| (138.36) | (18.03) | (4.55) | (26.68) | (26.90) | |||
| E. coli (ATCC BAA-2452) d,e | 50 | 50 | 12.5 | 50 | 100 | 25 | 12.5 |
| (138.36) | (36.06) | (9.11) | (26.68) | (53.79) | |||
| P. aeruginosa (ATCC 27883) | 0.78 | 25 | 12.5 | 100 | 100 | 1.56 | 1.56 |
| (2.16) | (18.03) | (9.11) | (53.36) | (53.79) | |||
| P. aeruginosa (ATCC BAA-1744) e | 0.78 | 25 | 12.5 | 100 | >100 | 1.56 | 0.78 |
| (2.16) | (18.03) | (9.11) | (53.36) | (>53.79) | |||
| K. pneumoniae (ATCC 13883) | 25 | 50 | 12.5 | 25 | 50 | 25 | 12.5 |
| (69.18) | (36.06) | (9.11) | (13.34) | (26.90) | |||
| K. pneumoniae (ATCC BAA-1705) f | 50 | 25 | 12.5 | 50 | 50 | 12.5 | 12.5 |
| (138.36) | (18.03) | (9.11) | (26.68) | (26.90) | |||
| Bacterial Strain | MIC in Combination (µg/mL) | FICI a | ||||
|---|---|---|---|---|---|---|
| Levo/(R4W4) | Levo/[R4W4] | Levo + (R4W4) | Interactive Category | Levo + [R4W4] | Interactive Category | |
| S. aureus (ATCC 29213) | 1.56/1.56 | 0.78/1.56 | 0.748 | Partial synergy | 0.748 | Partial synergy |
| S. aureus (ATCC BAA-1556) | 1.56/3.13 | 0.78/0.78 | 0.999 | Partial synergy | 0.498 | Synergy |
| E. coli (ATCC 25922) | 25/12.5 | 6.25/1.56 | 1 | Indifference | 0.374 | Synergy |
| E. coli (ATCC BAA-2452) | 25/12.5 | 3.13/3.13 | 0.75 | Partial synergy | 0.313 | Synergy |
| P. aeruginosa (ATCC 27883) | 0.78/1.56 | 0.15/1.56 | 1.062 | Indifference | 0.317 | Synergy |
| P. aeruginosa (ATCC BAA-1744) | 0.39/1.56 | 0.07/1.56 | 0.562 | Partial Synergy | 0.214 | Synergy |
| K. pneumoniae (ATCC 13883) | 25/12.5 | 6.25/3.13 | 1.25 | Indifference | 0.500 | Synergy |
| K. pneumoniae (ATCC BAA-1705) | 25/12.5 | 12.5/1.56 | 1 | Indifference | 0.374 | Synergy |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sajid, M.I.; Lohan, S.; Kato, S.; Tiwari, R.K. Combination of Amphiphilic Cyclic Peptide [R4W4] and Levofloxacin against Multidrug-Resistant Bacteria. Antibiotics 2022, 11, 416. https://doi.org/10.3390/antibiotics11030416
Sajid MI, Lohan S, Kato S, Tiwari RK. Combination of Amphiphilic Cyclic Peptide [R4W4] and Levofloxacin against Multidrug-Resistant Bacteria. Antibiotics. 2022; 11(3):416. https://doi.org/10.3390/antibiotics11030416
Chicago/Turabian StyleSajid, Muhammad Imran, Sandeep Lohan, Shun Kato, and Rakesh Kumar Tiwari. 2022. "Combination of Amphiphilic Cyclic Peptide [R4W4] and Levofloxacin against Multidrug-Resistant Bacteria" Antibiotics 11, no. 3: 416. https://doi.org/10.3390/antibiotics11030416
APA StyleSajid, M. I., Lohan, S., Kato, S., & Tiwari, R. K. (2022). Combination of Amphiphilic Cyclic Peptide [R4W4] and Levofloxacin against Multidrug-Resistant Bacteria. Antibiotics, 11(3), 416. https://doi.org/10.3390/antibiotics11030416








