Antimicrobial Susceptibility of Streptococcus suis Isolated from Diseased Pigs in Thailand, 2018–2020
Abstract
1. Introduction
2. Results
2.1. Bacterial Sampling
2.2. Serotyping
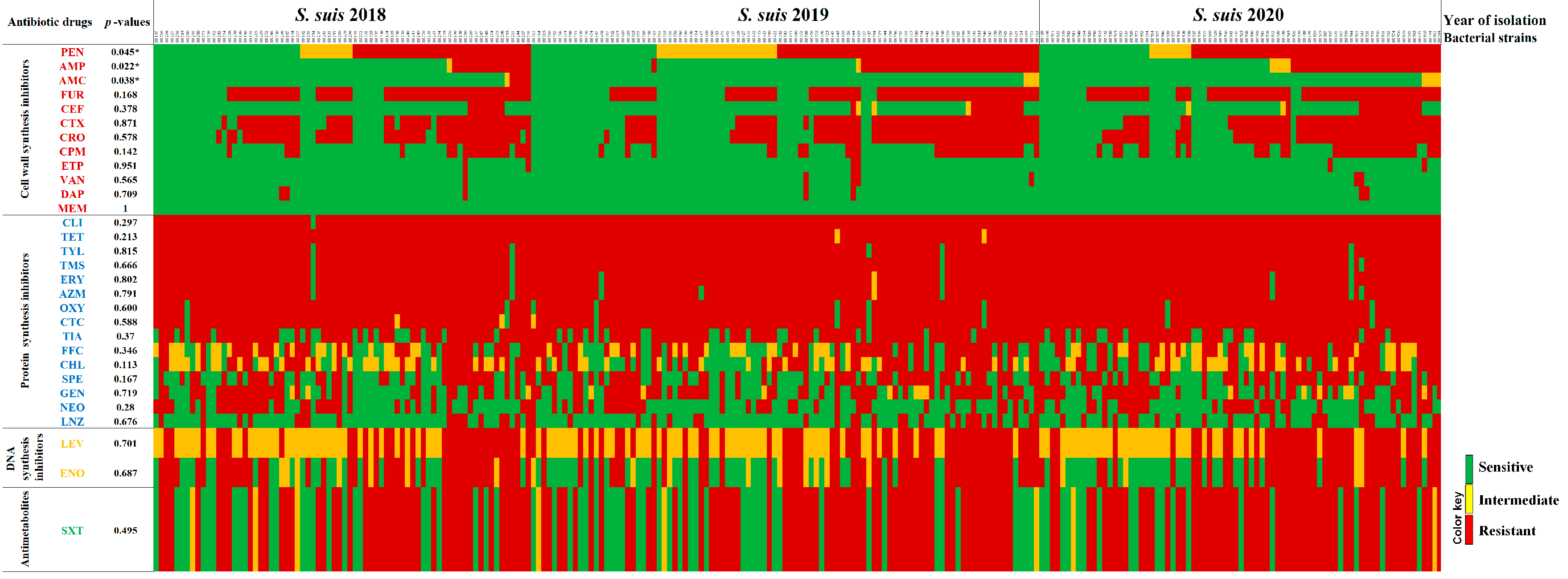
2.3. Antimicrobial Susceptibility Profiles
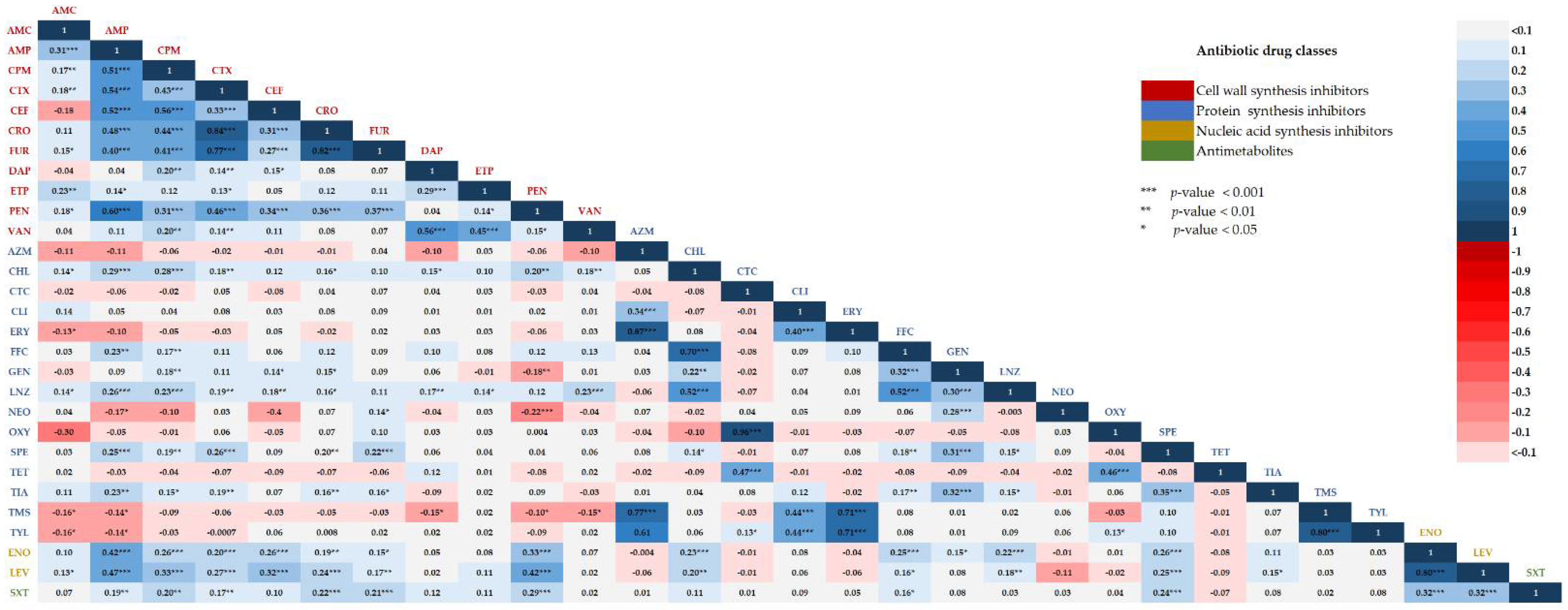
2.4. Correlations between Two Different Antibiotic Susceptibility Statuses among Isolates
3. Discussion
4. Materials and Methods
4.1. Bacterial Collection
4.2. Multiplex PCR-Based Serotyping
4.3. Antimicrobial Susceptibility Testing
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lun, Z.R.; Wang, Q.P.; Chen, X.G.; Li, A.X.; Zhu, X.Q. Streptococcus suis: An emerging zoonotic pathogen. Lancet Infect. Dis. 2007, 7, 201–209. [Google Scholar] [CrossRef]
- Segura, M.; Aragon, V.; Brockmeier, S.L.; Gebhart, C.; Greeff, A.; Kerdsin, A.; O’Dea, M.A.; Okura, M.; Saléry, M.; Schultsz, C.; et al. Update on Streptococcus suis research and prevention in the era of antimicrobial restriction: 4th International Workshop on S. suis. Pathogens 2020, 9, 374. [Google Scholar] [CrossRef] [PubMed]
- Kerdsin, A.; Akeda, Y.; Hatrongjit, R.; Detchawna, U.; Sekizaki, T.; Hamada, S.; Gottschalk, M.; Oishi, K. Streptococcus suis serotyping by a new multiplex PCR. J. Med. Microbiol. 2014, 63, 824–830. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.I.; Jeon, A.B.; Jung, B.Y.; Byun, J.W.; Gottschalk, M.; Kim, A.; Kim, J.W.; Kim, H.Y. Capsular serotypes, virulence-associated genes and antimicrobial susceptibility of Streptococcus suis isolates from pigs in Korea. J. Vet. Med. Sci. 2017, 79, 780–787. [Google Scholar] [CrossRef] [PubMed]
- Lekagul, A.; Tangcharoensathien, V.; Mills, A.; Rushton, J.; Yeung, S. How antibiotics are used in pig farming: A mixed-methods study of pig farmers, feed mills and veterinarians in Thailand. BMJ Glob. Health 2020, 5, e001918. [Google Scholar] [CrossRef] [PubMed]
- Petrocchi-Rilo, M.; Martínez-Martínez, S.; Aguarón-Turrientes, Á.; Roca-Martínez, E.; García-Iglesias, M.J.; Pérez-Fernández, E.; González-Fernández, A.; Herencia-Lagunar, E.; Gutiérrez-Martín, C.B. Anatomical site, typing, virulence gene profiling, antimicrobial susceptibility and resistance genes of Streptococcus suis isolates recovered from pigs in Spain. Antibiotics 2021, 10, 707. [Google Scholar] [CrossRef]
- Tan, M.F.; Tan, J.; Zeng, Y.B.; Li, H.Q.; Yang, Q.; Zhou, R. Antimicrobial resistance phenotypes and genotypes of Streptococcus suis isolated from clinically healthy pigs from 2017 to 2019 in Jiangxi Province, China. J. Appl. Microbiol. 2020, 130, 797–806. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, P.; Wang, Y.; Fu, L.; Liu, L.; Xu, D.; Hou, Y.; Li, Y.; Fu, M.; Wang, X.; et al. Capsular serotypes, antimicrobial susceptibility, and the presence of transferable oxazolidinone resistance genes in Streptococcus suis isolated from healthy pigs in China. Vet. Microbiol. 2020, 247, 108750. [Google Scholar] [CrossRef]
- Yongkiettrakul, S.; Maneerat, K.; Arechanajan, B.; Malila, Y.; Srimanote, P.; Gottschalk, M.; Visessanguan, W. Antimicrobial susceptibility of Streptococcus suis isolated from diseased pigs, asymptomatic pigs, and human patients in Thailand. BMC Vet. Res. 2019, 15, 5. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Hoa, N.T.; Chieu, T.T.; Nghia, H.D.; Mai, N.T.; Anh, P.H.; Wolbers, M.; Baker, S.; Campbell, J.I.; Chau, N.V.; Hien, T.T.; et al. The antimicrobial resistance patterns and associated determinants in Streptococcus suis isolated from humans in southern Vietnam, 1997–2008. BMC Infect. Dis. 2011, 11, 6. [Google Scholar] [CrossRef] [PubMed]
- Prüfer, T.L.; Rohde, J.; Verspohl, J.; Rohde, M.; de Greeff, A.; Willenborg, J.; Valentin-Weigand, P. Molecular typing of Streptococcus suis strains isolated from diseased and healthy pigs between 1996–2016. PLoS ONE 2019, 14, e0210801. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, Z.; Song, L.; Fan, X.; Wen, F.; Xu, S.; Ning, Y. Antimicrobial resistance profile and genotypic characteristics of Streptococcus suis capsular type 2 isolated from clinical carrier sows and diseased pigs in China. Biomed. Res. Int. 2015, 2015, 284303. [Google Scholar] [PubMed]
- Kerdsin, A.; Takeuchi, D.; Nuangmek, A.; Akeda, Y.; Gottschalk, M.; Oishi, K. Genotypic comparison between Streptococcus suis isolated from pigs and humans in Thailand. Pathogens 2020, 9, 50. [Google Scholar] [CrossRef]
- Meekhanon, N.; Kaewmongkol, S.; Phimpraphai, W.; Okura, M.; Osaki, M.; Sekizaki, T.; Takamatsu, D. Potentially hazardous Streptococcus suis strains latent in asymptomatic pigs in a major swine production area of Thailand. J. Med. Microbiol. 2017, 66, 662–669. [Google Scholar] [CrossRef]
- Thongkamkoon, P.; Kiatyingangsulee, T.; Gottschalk, M. Serotypes of Streptococcus suis isolated from healthy pigs in Phayao Province, Thailand. BMC Res. Notes 2017, 10, 53. [Google Scholar] [CrossRef]
- Segura, M.; Fittipaldi, N.; Calzas, C.; Gottschalk, M. Critical Streptococcus suis virulence factors: Are they all really critical? Trends Microbiol. 2017, 25, 585–599. [Google Scholar] [CrossRef]
- Zheng, H.; Du, P.; Qiu, X.; Kerdsin, A.; Roy, D.; Bai, X.; Xu, J.; Vela, A.I.; Gottschalk, M. Genomic comparisons of Streptococcus suis serotype 9 strains recovered from diseased pigs in Spain and Canada. Vet. Res. 2018, 49, 1. [Google Scholar] [CrossRef]
- Coyne, L.; Arief, R.; Benigno, C.; Giang, V.N.; Huong, L.Q.; Jeamsripong, S.; Kalpravidh, W.; McGrane, J.; Padungtod, P.; Patrick, I.; et al. Characterizing antimicrobial use in the livestock sector in three South East Asian countries (Indonesia, Thailand, and Vietnam). Antibiotics 2019, 8, 33. [Google Scholar] [CrossRef]
- Tiseo, K.; Huber, L.; Gilbert, M.; Robinson, T.P.; Van Boeckel, T.P. Global trends in antimicrobial use in food animals from 2017 to 2030. Antibiotics 2020, 9, 918. [Google Scholar] [CrossRef]
- Du, F.; Lv, X.; Duan, D.; Wang, L.; Huang, J. Characterization of a linezolid- and vancomycin-resistant Streptococcus suis isolate that harbors optrA and vanG operons. Front. Microbiol. 2019, 10, 2026. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Ma, J.; Shang, K.; Hu, X.; Liang, Y.; Li, D.; Wu, Z.; Dai, L.; Chen, L.; Wang, L. Evolution and diversity of the antimicrobial resistance associated mobilome in Streptococcus suis: A probable mobile genetic elements reservoir for other streptococci. Front. Cell. Infect. Microbiol. 2016, 6, 118. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Zhao, T.; Yang, X.; Xiao, X.; Velkov, T.; Tang, S. Pharmacokinetics and relative bioavailability of an oral amoxicillin-apramycin combination in pigs. PLoS ONE 2017, 12, e0176149. [Google Scholar] [CrossRef] [PubMed]
- Tateda, K.; Matsumoto, T.; Miyazaki, S.; Yamaguchi, K. Efficacy of beta-lactam antibiotics combined with gentamicin against penicillin-resistant pneumococcal pneumonia in CBA/J mice. J. Antimicrob. Chemother. 1999, 43, 367–371. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yu, Y.; Fang, J.T.; Zheng, M.; Zhang, Q.; Walsh, T.R.; Liao, X.P.; Sun, J.; Liu, Y.H. Combination therapy strategies against multiple-resistant Streptococcus suis. Front. Pharmacol. 2018, 9, 489. [Google Scholar] [CrossRef]
- Tarradas, C.; Arenas, A.; Maldonado, A.; Luque, I.; Miranda, A.; Perea, A. Identification of Streptococcus suis isolated from swine: Proposal for biochemical parameters. J. Clin. Microbiol. 1994, 322, 578580. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement, 30th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals, 5th ed.; CLSI Supplement VET01S; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters; Version 10.0; EUCAST: Basel, Switzerland, 2020; Available online: http://www.eucast.org (accessed on 15 September 2021).
- Food and Drug Administration. Antibacterial Susceptibility Test Interpretive Criteria; FDA: Silver Spring, MD, USA, 2019. Available online: https://www.fda.gov/drugs/development-resources/antibacterial-susceptibility-test-interpretive-criteria (accessed on 15 September 2021).
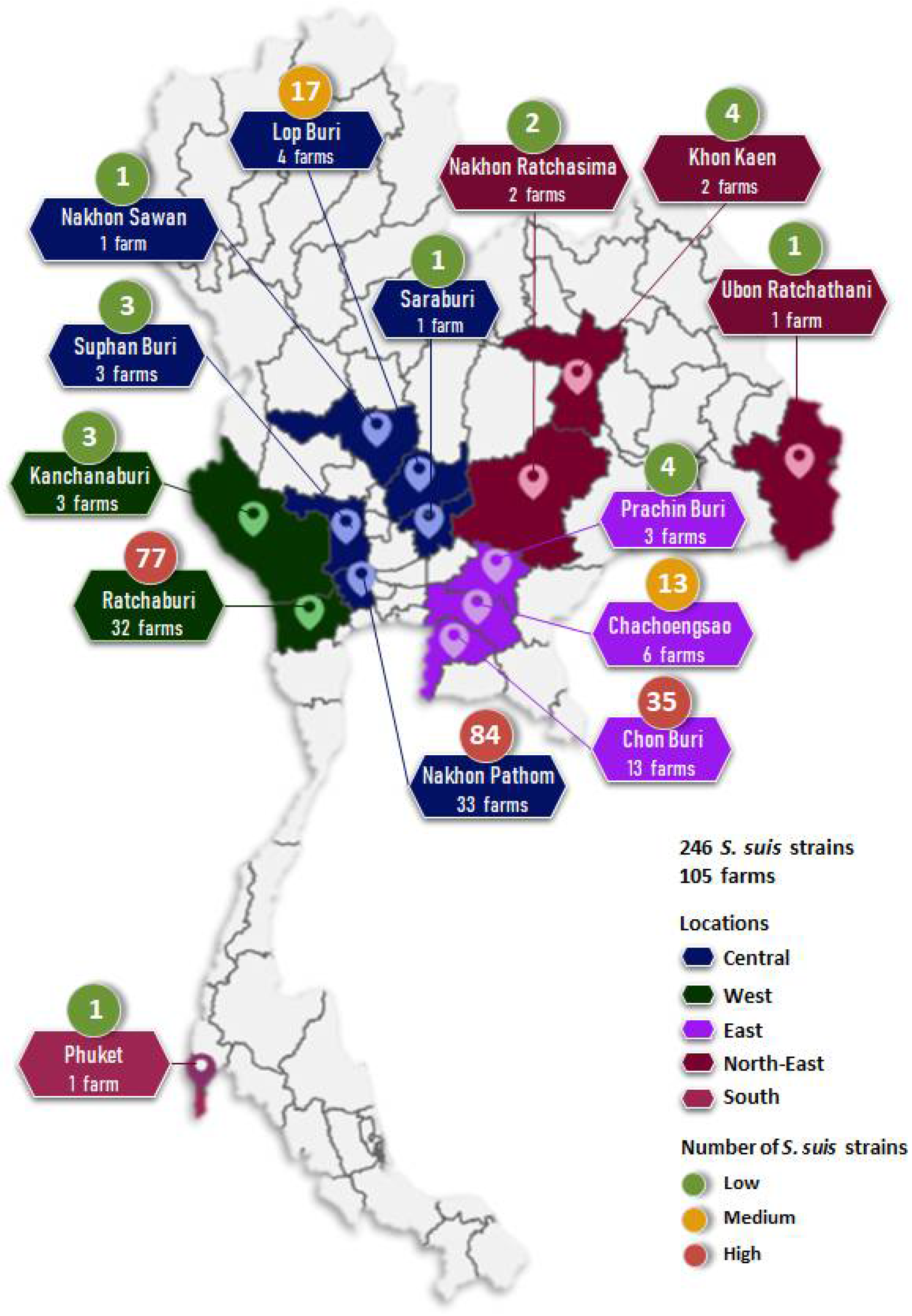
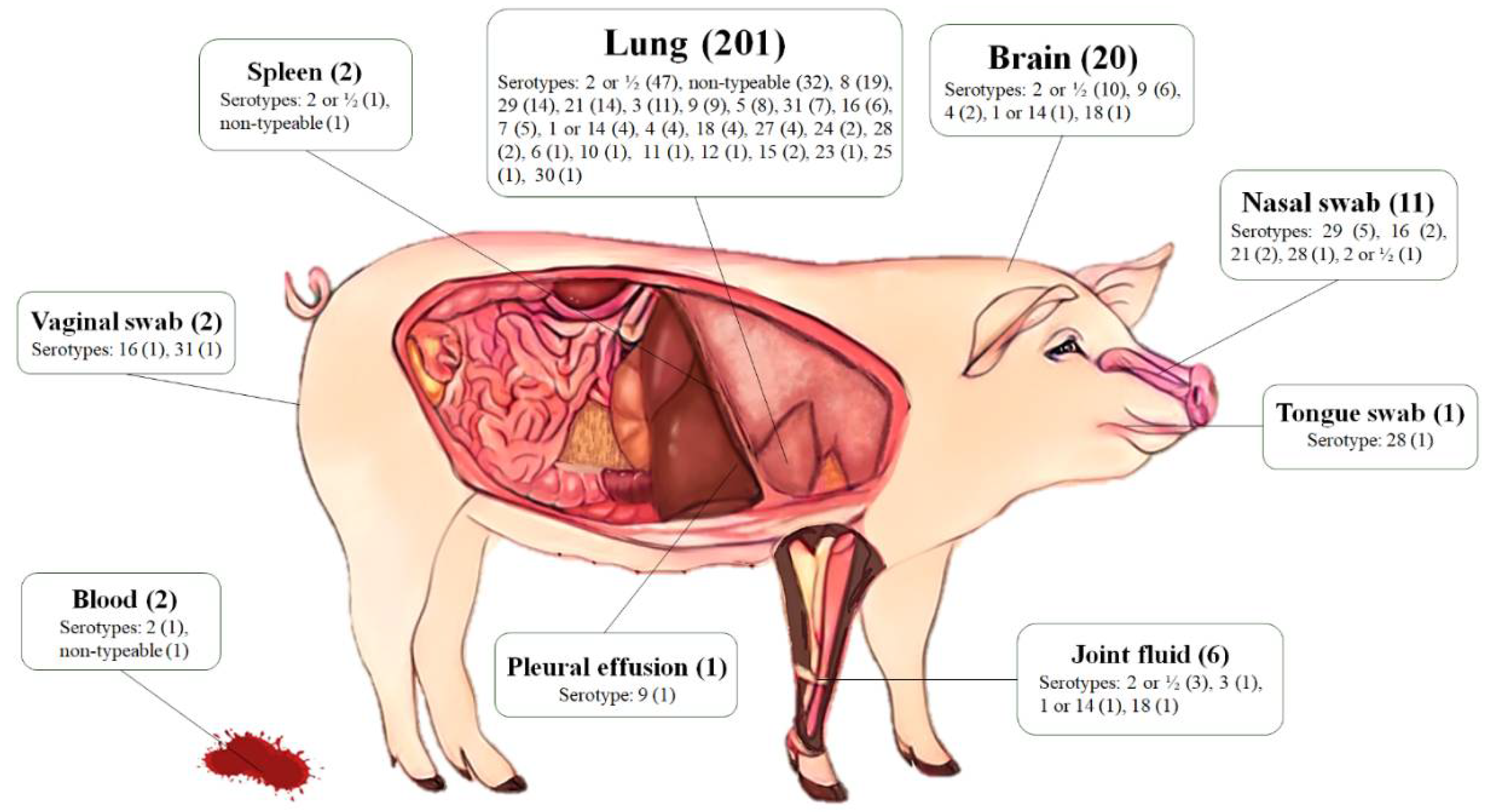
| Serotypes | Year of Isolation, n (%) | p-Values | Total | ||
|---|---|---|---|---|---|
| 2018 n = 72 | 2019 n = 97 | 2020 n = 77 | |||
| Serotype 2 or ½ | 18 (25.0) | 26 (26.8) | 19 (24.7) | 0.941 | 63 (25.6) |
| Serotype 3 | 4 (5.6) | 4 (4.1) | 4 (5.2) | 0.902 | 12 (4.9) |
| Serotype 8 | 4 (5.6) | 11 (11.3) | 4 (5.2) | 0.229 | 19 (7.7) |
| Serotype 9 | 3 (4.2) | 6 (6.2) | 7 (9.1) | 0.470 | 16 (6.5) |
| Serotype 16 | 4 (5.6) | 2 (2.1) | 3 (3.9) | 0.485 | 9 (3.7) |
| Serotype 21 | 3 (4.2) | 7 (7.2) | 6 (7.8) | 0.626 | 16 (6.5) |
| Serotype 29 | 4 (5.6) | 7 (7.2) | 8 (10.4) | 0.528 | 19 (7.7) |
| Other serotypes a | 21 (29.2) | 21 (21.6) | 16 (20.8) | 0.410 | 58 (23.6) |
| Non-typeable | 11 (15.3) | 13 (13.4) | 10 (13.0) | 0.910 | 34 (13.8) |
| Antibiotic Drugs | MIC Breakpoints (µg/mL) | MIC Values (µg/mL) a | MIC50 | MIC90 | S (%) | I (%) | R (%) | MIC Ranges | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | I | R | 0.008 | 0.016 | 0.03 | 0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | |||||||
| Amoxicillin/Clavulanic acid | ≤8/4 | 16/8 | ≥32/16 | 194 | 13 | 27 | 8 | 4 | ≤2 | 8 | 95.1 | 3.3 | 1.6 | ≤2–>16 | ||||||||||||
| Ampicillin | ≤0.5 | 1 | ≥2 | 150 | 11 | 6 | 15 | 13 | 7 | 13 | 31 | ≤0.25 | >16 | 65.4 | 2.4 | 32.1 | ≤0.25–>16 | |||||||||
| Cefepime | ≤2 | 4 | ≥8 | 168 | 26 | 18 | 18 | 10 | 6 | ≤0.5 | 4 | 68.3 | ND | 31.7 | ≤0.5–>8 | |||||||||||
| Cefotaxime | ≤0.5 | - | ≥1 | 27 | 20 | 52 | 64 | 28 | 12 | 43 | 1 | >4 | 40.2 | ND | 59.8 | ≤0.12–>4 | ||||||||||
| Ceftiofur | ≤0.5 | - | ≥1 | 142 | 24 | 29 | 15 | 5 | 15 | 16 | ≤0.25 | 8 | 85.4 | 2.0 | 12.6 | ≤0.25–>8 | ||||||||||
| Ceftriaxone | ≤0.5 | - | ≥1 | 30 | 16 | 46 | 68 | 22 | 64 | 1 | >2 | 37.4 | ND | 62.6 | ≤0.12–>2 | |||||||||||
| Cefuroxime | ≤0.5 | - | ≥1 | 79 | 76 | 38 | 7 | 46 | 1 | >4 | 32.1 | ND | 67.9 | ≤0.5–>4 | ||||||||||||
| Daptomycin | ≤1 | - | ≥2 | 22 | 98 | 107 | 6 | 6 | 1 | 6 | 0.25 | 0.25 | 97.2 | ND | 2.8 | ≤0.06–>2 | ||||||||||
| Ertapenem | ≤0.5 | - | ≥1 | 240 | 3 | 2 | 1 | ≤0.5 | ≤0.5 | 97.6 | ND | 2.4 | ≤0.5–4 | |||||||||||||
| Meropenem | ≤2 | - | ≥4 | 241 | 4 | 1 | ≤0.25 | ≤0.25 | 100.0 | ND | 0.0 | ≤0.25–1 | ||||||||||||||
| Penicillin | ≤0.25 | 0.5 | ≥1 | 15 | 10 | 20 | 28 | 41 | 19 | 16 | 27 | 16 | 54 | 1 | >8 | 29.7 | 16.7 | 53.7 | ≤0.03–>8 | |||||||
| Vancomycin | ≤1 | - | ≥2 | 237 | 2 | 3 | 1 | 3 | ≤0.5 | ≤0.5 | 97.2 | ND | 2.8 | ≤0.5–>4 | ||||||||||||
| Azithromycin | ≤0.5 | 1 | ≥2 | 7 | 1 | 1 | 2 | 235 | >2 | >2 | 3.3 | 0.4 | 96.3 | ≤0.25–>2 | ||||||||||||
| Chloramphenicol | ≤4 | 8 | ≥16 | 5 | 64 | 72 | 41 | 38 | 26 | 8 | >32 | 28.0 | 29.3 | 42.7 | 2–>32 | |||||||||||
| Chlortetracycline | ≤2 | 4 | ≥8 | 4 | 4 | 3 | 17 | 218 | >8 | >8 | 3.3 | 1.2 | 95.5 | ≤0.5–>8 | ||||||||||||
| Clindamycin | ≤0.5 | 1–2 | ≥4 | 1 | 2 | 2 | 3 | 238 | >16 | >16 | 0.4 | 0.0 | 99.6 | 0.25–>16 | ||||||||||||
| Erythromycin | ≤0.25 | 0.5 | ≥1 | 6 | 1 | 3 | 6 | 230 | >2 | >2 | 2.4 | 0.4 | 97.2 | ≤0.25–>2 | ||||||||||||
| Florfenicol | ≤2 | 4 | ≥8 | 6 | 58 | 58 | 14 | 110 | 4 | >8 | 26.0 | 23.6 | 50.4 | 1–>8 | ||||||||||||
| Gentamicin | ≤4 | 8 | ≥16 | 24 | 35 | 47 | 16 | 11 | 113 | 8 | >16 | 43.1 | 6.5 | 50.4 | ≤1–>16 | |||||||||||
| Linezolid | ≤2 | - | ≥4 | 2 | 25 | 98 | 56 | 52 | 13 | 1 | 4 | 73.6 | ND | 26.4 | ≤0.25–>4 | |||||||||||
| Neomycin | ≤16 | - | ≥32 | 26 | 70 | 51 | 33 | 66 | 16 | >32 | 59.8 | ND | 40.2 | ≤4–>32 | ||||||||||||
| Oxytetracycline | ≤4 | - | ≥8 | 3 | 2 | 4 | 6 | 231 | >8 | >8 | 3.7 | ND | 96.3 | ≤0.5–>8 | ||||||||||||
| Spectinomycin | ≤64 | - | ≥128 | 17 | 46 | 40 | 7 | 136 | >64 | >64 | 44.7 | ND | 55.3 | ≤8–>64 | ||||||||||||
| Tetracycline | ≤0.5 | 1 | ≥2 | 2 | 3 | 2 | 239 | >8 | >8 | 0.0 | 0.8 | 99.2 | ≤1–>8 | |||||||||||||
| Tiamulin | ≤16 | - | ≥32 | 9 | 8 | 12 | 2 | 7 | 13 | 7 | 188 | >32 | >32 | 20.7 | ND | 79.3 | ≤0.5–>32 | |||||||||
| Tigecycline | ≤0.25 | - | ≥0.5 | 2 | 19 | 53 | 68 | 104 | 0.12 | >0.12 | ND | ND | ND | ≤0.02–>0.12 | ||||||||||||
| Tilmicosin | ≤16 | - | ≥32 | 3 | 2 | 1 | 240 | >64 | >64 | 2.0 | ND | 98.0 | ≤4–>64 | |||||||||||||
| Tulathromycin | ND | ND | ND | 1 | 3 | 2 | 2 | 2 | 4 | 232 | >64 | >64 | ND | ND | ND | ≤1–>64 | ||||||||||
| Tylosin tartrate | ≤4 | - | ≥8 | 4 | 1 | 1 | 240 | >32 | >32 | 2.0 | ND | 98.0 | 1–>32 | |||||||||||||
| Danofloxaci | ND | ND | ND | 2 | 8 | 51 | 53 | 132 | >1 | >1 | ND | ND | ND | ≤0.12–>1 | ||||||||||||
| Enrofloxacin | ≤0.5 | 1 | ≥2 | 1 | 20 | 70 | 20 | 12 | 123 | 2 | >2 | 37.0 | 8.1 | 54.9 | ≤0.12–>2 | |||||||||||
| Levofloxacin | ≤0.01 | 0.03–2 | ≥4 | 83 | 32 | 8 | 12 | 111 | 2 | >4 | 0.0 | 50.0 | 50.0 | ≤0.5–>4 | ||||||||||||
| Moxifloxacin | ≤0.5 | - | ≥1 | 206 | 27 | 12 | 1 | ≤1 | 2 | ND | ND | ND | ≤1–8 | |||||||||||||
| Sulphadimethoxine | ND | ND | ND | 13 | 233 | >256 | >256 | ND | ND | ND | ≤256–>256 | |||||||||||||||
| Trimethoprim/sulfamethoxazole | ≤0.5/9.5 | 1/19–2/38 | ≥4/76 | 79 | 5 | 3 | 26 | 133 | >2 | >4 | 32.1 | 3.3 | 64.6 | ≤0.5–>2 | ||||||||||||
| Antibiotic Drugs | Antimicrobial Susceptibility, n (%) | p-Values | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2018 n = 72 | 2019 n = 97 | 2020 n = 77 | ||||||||
| S | I | R | S | I | R | S | I | R | ||
| Amoxicillin/Clavulanic acid | 67 (93.1) | 1 (1.4) | 4 (5.6) | 94 (96.9) | 3 (3.1) | 0 (0.0) | 73 (94.8) | 4 (5.2) | 0 (0.0) | 0.022 * |
| Ampicillin | 56 (77.8) | 1 (1.4) | 15 (20.8) | 61 (62.9) | 1 (1.0) | 35 (36.1) | 44 (57.1) | 4 (5.2) | 29 (37.7) | 0.038 * |
| Cefepime | 53 (73.6) | ND | 19 (26.4) | 69 (71.1) | ND | 28 (28.9) | 46 (59.7) | ND | 31 (40.3) | 0.142 |
| Cefotaxime | 28 (38.9) | ND | 44 (61.1) | 41 (42.3) | ND | 56 (57.7) | 30 (39.0) | ND | 47 (61.0) | 0.872 |
| Ceftiofur | 65 (90.3) | 0 (0.0) | 7 (9.7) | 83 (85.6) | 3 (3.1) | 11 (11.3) | 62 (80.5) | 2 (2.6) | 13 (16.9) | 0.373 |
| Ceftriaxone | 26 (36.1) | ND | 46 (63.9) | 40 (41.2) | ND | 57 (58.8) | 26 (33.8) | ND | 51 (66.2) | 0.578 |
| Cefuroxime | 23 (31.9) | ND | 49 (68.1) | 37 (38.1) | ND | 60 (61.9) | 19 (24.7) | ND | 58 (75.3) | 0.168 |
| Daptomycin | 69 (95.8) | ND | 3 (4.2) | 95 (97.9) | ND | 2 (2.1) | 75 (97.4) | ND | 2 (2.6) | 0.709 |
| Ertapenem | 70 (97.2) | ND | 2 (2.8) | 95 (97.9) | ND | 2 (2.1) | 75 (97.4) | ND | 2 (2.6) | 0.951 |
| Meropenem | 72 (100.0) | ND | 0 (0.0) | 97 (100.0) | ND | 0 (0.0) | 77 (100.0) | ND | 0 (0.0) | 1.000 |
| Penicillin | 28 (38.9) | 10 (13.9) | 34 (47.2) | 24 (24.7) | 23 (23.7) | 50 (51.5) | 21 (27.3) | 8 (10.4) | 48 (62.3) | 0.045 * |
| Vancomycin | 71 (98.6) | ND | 1 (1.4) | 93 (95.9) | ND | 4 (4.1) | 75 (97.4) | ND | 2 (2.6) | 0.565 |
| Azithromycin | 2 (2.8) | 0 (0.0) | 70 (97.2) | 3 (3.1) | 1 (1.0) | 93 (95.9) | 3 (3.9) | 0 (0.0) | 74 (96.1) | 0.791 |
| Chloramphenicol | 16 (22.2) | 28 (38.9) | 28 (38.9) | 34 (35.1) | 21 (21.6) | 42 (43.3) | 19 (24.7) | 23 (29.9) | 35 (45.5) | 0.113 |
| Chlortetracycline | 2 (2.8) | 2 (2.8) | 68 (94.4) | 3 (3.1) | 1 (1.0) | 93 (95.9) | 2 (2.6) | 0 (0.0) | 75 (97.4) | 0.588 |
| Clindamycin | 1 (1.4) | 0 (0.0) | 71 (98.6) | 0 (0.0) | 0 (0.0) | 97 (100.0) | 0 (0.0) | 0 (0.0) | 77 (100.0) | 0.297 |
| Erythromycin | 2 (2.8) | 0 (0.0) | 70 (97.2) | 2 (2.1) | 1 (1.0) | 94 (96.9) | 2 (2.6) | 0 (0.0) | 75 (97.4) | 0.802 |
| Florfenicol | 20 (27.8) | 20 (27.8) | 32 (44.4) | 29 (29.9) | 18 (18.6) | 50 (51.5) | 15 (19.5) | 20 (26.0) | 42 (54.5) | 0.346 |
| Gentamicin | 35 (48.6) | 3 (4.2) | 34 (47.2) | 38 (39.2) | 7 (7.2) | 52 (53.6) | 33 (42.9) | 6 (7.8) | 38 (49.4) | 0.719 |
| Linezolid | 55 (76.4) | ND | 17 (23.6) | 72 (74.2) | ND | 25 (25.8) | 54 (70.1) | ND | 23 (29.9) | 0.676 |
| Neomycin | 38 (52.8) | ND | 34 (47.2) | 63 (64.9) | ND | 34 (35.1) | 46 (59.7) | ND | 31 (40.3) | 0.280 |
| Oxytetracycline | 2 (2.8) | ND | 70 (97.2) | 5 (5.2) | ND | 92 (94.8) | 2 (2.6) | ND | 75 (97.4) | 0.600 |
| Spectinomycin | 37 (51.4) | ND | 35 (48.6) | 45 (46.4) | ND | 52 (53.6) | 28 (36.4) | ND | 49 (63.6) | 0.167 |
| Tetracycline | 0 (0.0) | 0 (0.0) | 72 (100.0) | 0 (0.0) | 2 (2.1) | 95 (97.9) | 0 (0.0) | 0 (0.0) | 77 (100.0) | 0.213 |
| Tiamulin | 19 (26.4) | ND | 53 (73.6) | 18 (18.6) | ND | 79 (81.4) | 14 (18.2) | ND | 63 (81.8) | 0.370 |
| Tilmicosin | 2 (2.8) | ND | 70 (97.2) | 1 (1.0) | ND | 96 (99.0) | 2 (2.6) | ND | 75 (97.4) | 0.666 |
| Tylosin tartrate | 2 (2.8) | ND | 70 (97.2) | 2 (2.1) | ND | 95 (97.9) | 1 (1.3) | ND | 76 (98.7) | 0.815 |
| Enrofloxacin | 23 (31.9) | 7 (9.7) | 42 (58.3) | 38 (39.2) | 9 (9.3) | 50 (51.5) | 30 (39.0) | 4 (5.2) | 43 (55.8) | 0.687 |
| Levofloxacin | 0 (0.0) | 39 (54.2) | 33 (45.8) | 0 (0.0) | 47 (48.5) | 50 (51.5) | 0 (0.0) | 37 (48.1) | 40 (51.9) | 0.701 |
| Trimethoprim/sulfamethoxazole | 26 (36.1) | 3 (4.2) | 43 (59.7) | 35 (36.1) | 3 (3.1) | 59 (60.8) | 19 (24.7) | 2 (2.6) | 56 (72.7) | 0.495 |
| Antibiotic Drugs | Antimicrobial Susceptible, n (%) | p-Values | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Serotype 2 or ½ | Serotype 3 | Serotype 8 | Serotype 9 | Serotype 16 | Serotype 21 | Serotype 29 | Other Serotypes a | Non- Typeable | ||
| n = 63 | n = 12 | n = 19 | n = 16 | n = 9 | n = 16 | n = 19 | n = 58 | n = 34 | ||
| Amoxicillin/Clavulanic Acid | 62 (98.4) | 12 (100.0) | 19 (100.0) | 16 (100.0) | 9 (100.0) | 15 (93.8) | 17 (89.5) | 55 (94.8) | 29 (85.3) | 0.566 |
| Ampicillin | 50 (79.4) | 12 (100.0) | 17 (89.5) | 12 (75.0) | 4 (44.4) | 10 (62.5) | 8 (42.1) | 38 (65.5) | 10 (29.4) | <0.001 * |
| Cefepime | 52 (82.5) | 11 (91.7) | 17 (89.5) | 12 (75.0) | 3 (33.3) | 11 (68.8) | 13 (68.4) | 36 (62.1) | 13 (38.2) | <0.001 * |
| Cefotaxime | 24 (38.1) | 6 (50.0) | 16 (84.2) | 8 (50.0) | 4 (44.4) | 6 (37.5) | 6 (31.6) | 24 (41.4) | 5 (14.7) | 0.001 * |
| Ceftiofur | 59 (93.7) | 12 (100.0) | 18 (94.7) | 16 (100.0) | 6 (66.7) | 12 (75.0) | 16 (84.2) | 49 (84.5) | 22 (64.7) | 0.013 * |
| Ceftriaxone | 23 (36.5) | 6 (50.0) | 16 (84.2) | 6 (37.5) | 3 (33.3) | 6 (37.5) | 6 (31.6) | 19 (32.8) | 7 (20.6) | 0.003 * |
| Cefuroxime | 19 (30.2) | 5 (41.7) | 15 (78.9) | 3 (18.8) | 4 (44.4) | 5 (31.3) | 3 (15.8) | 19 (32.8) | 6 (17.6) | 0.001 * |
| Daptomycin | 61 (96.8) | 12 (100.0) | 18 (94.7) | 16 (100.0) | 9 (100.0) | 14 (87.5) | 19 (100.0) | 57 (98.3) | 33 (97.1) | 0.461 |
| Ertapenem | 62 (98.4) | 12 (100.0) | 19 (100.0) | 16 (100.0) | 9 (100.0) | 15 (93.8) | 18 (94.7) | 58 (100.0) | 31 (91.2) | 0.233 |
| Meropenem | 63 (100.0) | 12 (100.0) | 19 (100.0) | 16 (100.0) | 9 (100.0) | 16 (100.0) | 19 (100.0) | 58 (100.0) | 34 (100.0) | ND |
| Penicillin | 30 (47.6) | 9 (75.0) | 8 (42.1) | 5 (31.3) | 3 (33.3) | 3 (18.8) | 0 (0.0) | 13 (22.4) | 2 (5.9) | <0.001 * |
| Vancomycin | 61 (96.8) | 12 (100.0) | 19 (100.0) | 16 (100.0) | 9 (100.0) | 14 (87.5) | 19 (100.0) | 57 (98.3) | 32 (94.1) | 0.341 |
| Azithromycin | 1 (0.02) | 0 (0.0) | 1 (5.3) | 0 (0.0) | 2 (22.2) | 2 (12.5) | 2 (10.5) | 0 (0.0) | 0 (0.0) | 0.025* |
| Chloramphenicol | 30 (47.6) | 1 (8.3) | 2 (10.5) | 7 (43.8) | 1 (11.1) | 7 (43.8) | 8 (42.1) | 9 (15.5) | 4 (11.8) | <0.001 * |
| Chlortetracycline | 1 (1.6) | 1 (8.3) | 0 (0.0) | 1 (6.3) | 0 (0.0) | 0 (0.0) | 1 (5.3) | 4 (6.9) | 0 (0.0) | 0.256 |
| Clindamycin | 0 (0.0) | 0 (0.0) | 1 (5.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0.151 |
| Erythromycin | 1 (1.6) | 0 (0.0) | 1 (5.3) | 0 (0.0) | 1 (11.1) | 1 (6.3) | 2 (10.5) | 0 (0.0) | 0 (0.0) | 0.264 |
| Florfenicol | 19 (30.2) | 2 (16.7) | 3 (15.8) | 7 (43.8) | 1 (11.1) | 6 (37.5) | 8 (42.1) | 11 (19.0) | 7 (20.6) | 0.012 * |
| Gentamicin | 19 (30.2) | 5 (41.7) | 9 (47.4) | 7 (43.8) | 2 (22.2) | 6 (37.5) | 12 (63.2) | 33 (56.9) | 13 (38.2) | 0.078 |
| Linezolid | 47 (74.6) | 8 (66.7) | 12 (63.2) | 15 (93.8) | 6 (66.7) | 9 (56.3) | 15 (78.9) | 47 (81.0) | 22 (64.7) | 0.216 |
| Neomycin | 27 (42.9) | 7 (58.3) | 11 (57.9) | 8 (50.0) | 4 (44.4) | 7 (43.8) | 16 (84.2) | 41 (70.7) | 26 (76.5) | 0.004 * |
| Oxytetracycline | 1 (1.6) | 1 (8.3) | 1 (5.3) | 1 (6.3) | 0 (0.0) | 0 (0.0) | 1 (5.3) | 4 (6.9) | 0 (0.0) | 0.641 |
| Spectinomycin | 23 (36.5) | 7 (58.3) | 14 (73.7) | 9 (56.3) | 3 (33.3) | 5 (31.3) | 10 (52.6) | 28 (48.3) | 11 (32.4) | 0.071 |
| Tetracycline | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0.978 |
| Tiamulin | 11 (17.5) | 5 (41.7) | 4 (21.1) | 2 (12.5) | 0 (0.0) | 4 (25.0) | 6 (31.9) | 15 (25.9) | 4 (11.8) | 0.216 |
| Tilmicosin | 1 (1.6) | 0 (0.0) | 1 (5.3) | 0 (0.0) | 0 (0.0) | 1 (6.3) | 2 (10.5) | 0 (0.0) | 0 (0.0) | 0.149 |
| Tylosin tartrate | 1 (1.6) | 0 (0.0) | 1 (5.3) | 1 (6.3) | 0 (0.0) | 0 (0.0) | 2 (10.5) | 0 (0.0) | 0 (0.0) | 0.149 |
| Enrofloxacin | 27 (42.9) | 9 (75.0) | 9 (47.4) | 11 (68.8) | 1 (11.1) | 2 (12.5) | 6 (31.6) | 21 (36.2) | 5 (14.7) | <0.001 * |
| Levofloxacin | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | <0.001 * |
| Trimethoprim/sulfamethoxazole | 28 (44.4) | 10 (83.3) | 12 (63.2) | 6 (37.5) | 0 (0.0) | 3 (18.8) | 1 (5.3) | 13 (22.4) | 6 (17.6) | <0.001 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lunha, K.; Chumpol, W.; Samngamnim, S.; Jiemsup, S.; Assavacheep, P.; Yongkiettrakul, S. Antimicrobial Susceptibility of Streptococcus suis Isolated from Diseased Pigs in Thailand, 2018–2020. Antibiotics 2022, 11, 410. https://doi.org/10.3390/antibiotics11030410
Lunha K, Chumpol W, Samngamnim S, Jiemsup S, Assavacheep P, Yongkiettrakul S. Antimicrobial Susceptibility of Streptococcus suis Isolated from Diseased Pigs in Thailand, 2018–2020. Antibiotics. 2022; 11(3):410. https://doi.org/10.3390/antibiotics11030410
Chicago/Turabian StyleLunha, Kamonwan, Wiyada Chumpol, Sukuma Samngamnim, Surasak Jiemsup, Pornchalit Assavacheep, and Suganya Yongkiettrakul. 2022. "Antimicrobial Susceptibility of Streptococcus suis Isolated from Diseased Pigs in Thailand, 2018–2020" Antibiotics 11, no. 3: 410. https://doi.org/10.3390/antibiotics11030410
APA StyleLunha, K., Chumpol, W., Samngamnim, S., Jiemsup, S., Assavacheep, P., & Yongkiettrakul, S. (2022). Antimicrobial Susceptibility of Streptococcus suis Isolated from Diseased Pigs in Thailand, 2018–2020. Antibiotics, 11(3), 410. https://doi.org/10.3390/antibiotics11030410






