Novel Antimicrobial Peptides Designed Using a Recurrent Neural Network Reduce Mortality in Experimental Sepsis
Abstract
1. Introduction
2. Results
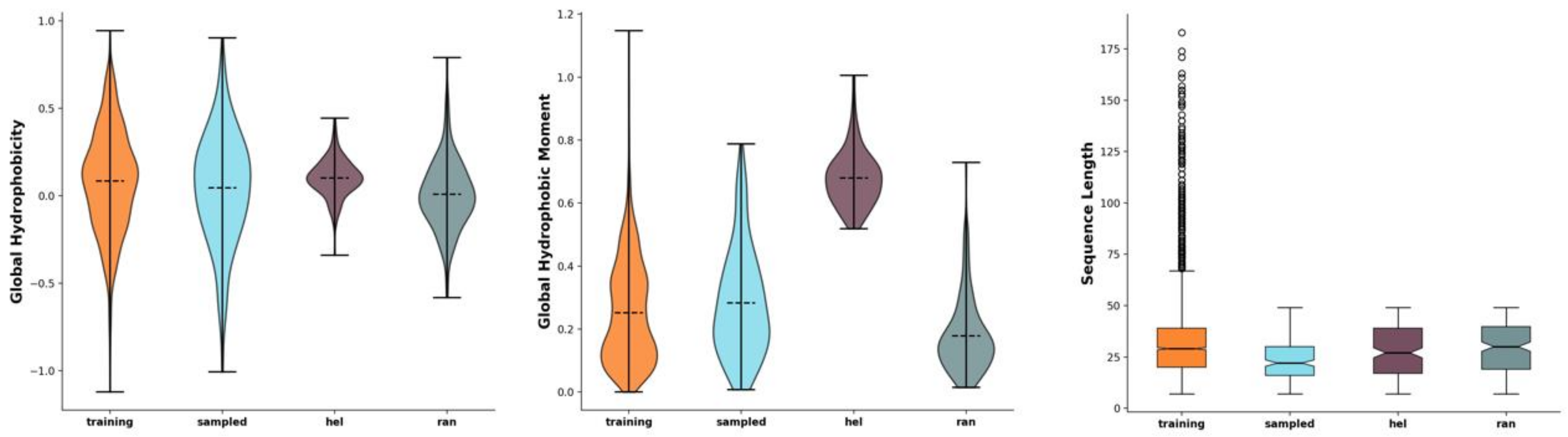
2.1. Development of Novel Peptides with Potential Antimicrobial Effect
2.2. PEP-38 and PEP-137 Are Active against Carbapenem-Resistant Gram-Negative Bacteria In Vitro
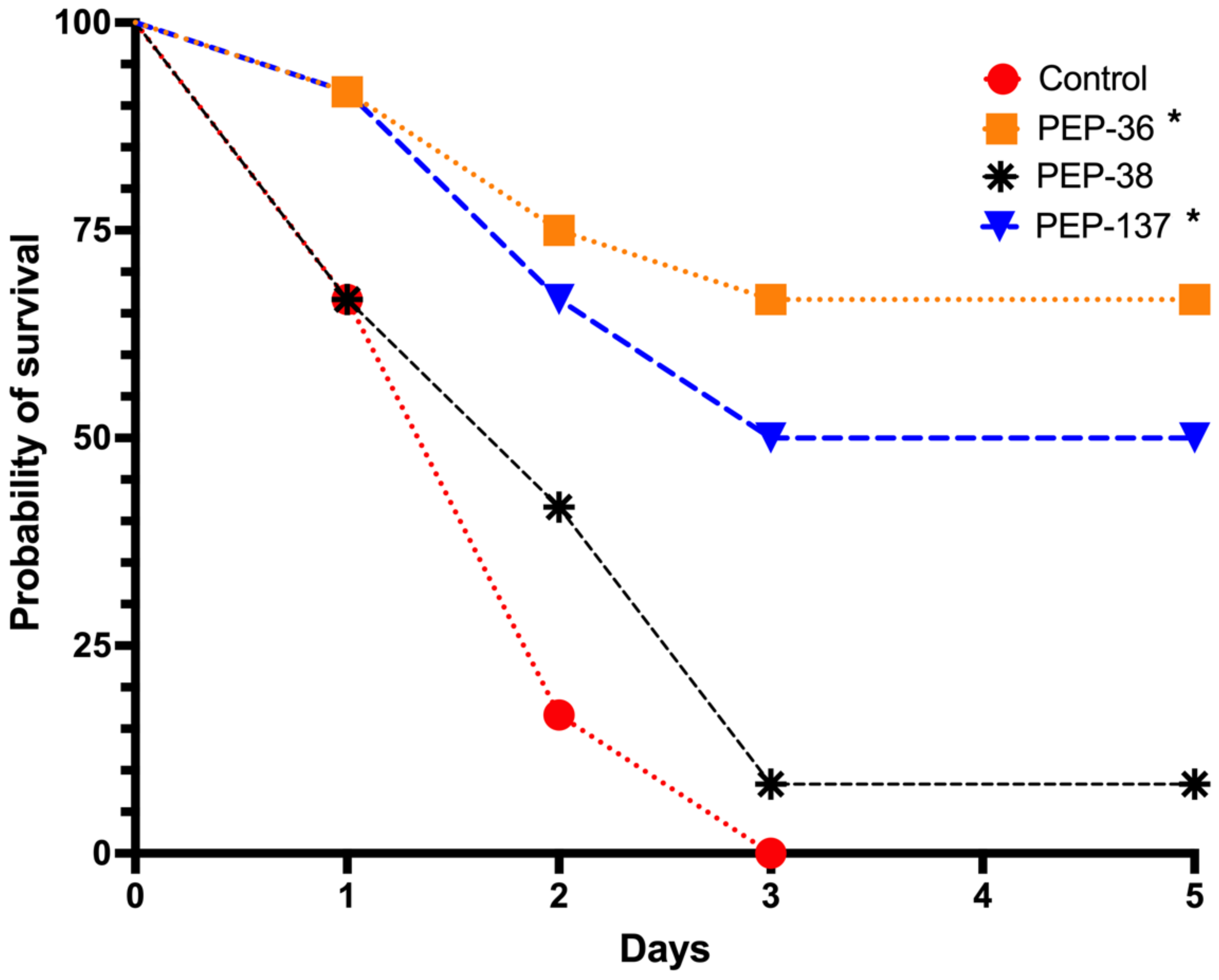
2.3. PEP-36 and PEP-137 Peptides Reduce Mortality in Experimental Sepsis
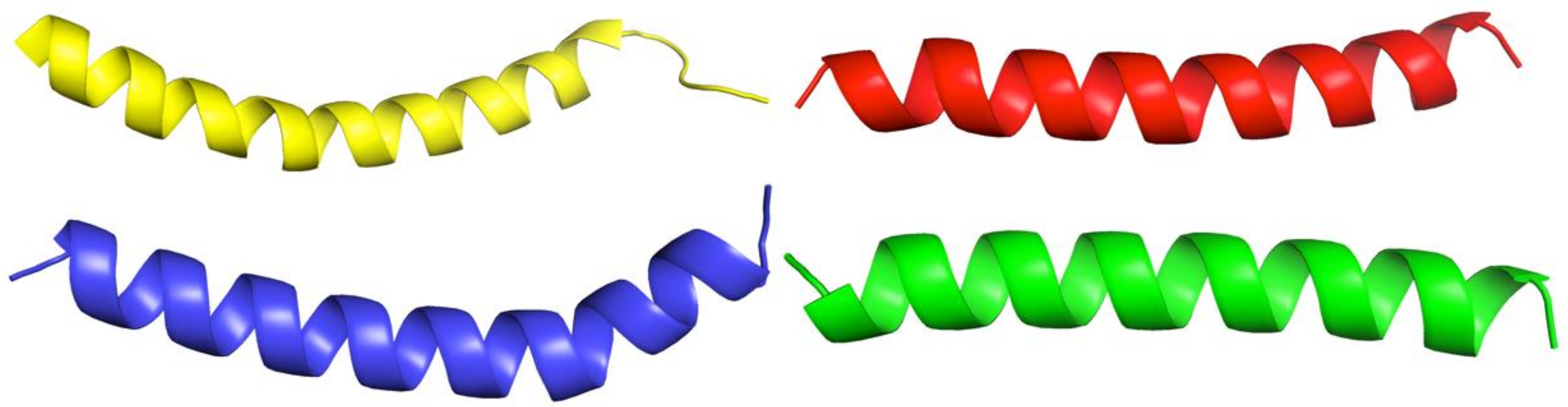
2.4. PEP-36, PEP-38 and PEP-137 Peptides Have Similarity in Their Spatial Structure
2.5. PEP-36 and PEP-38 Are Potentially Less Toxic to Red Blood Cells
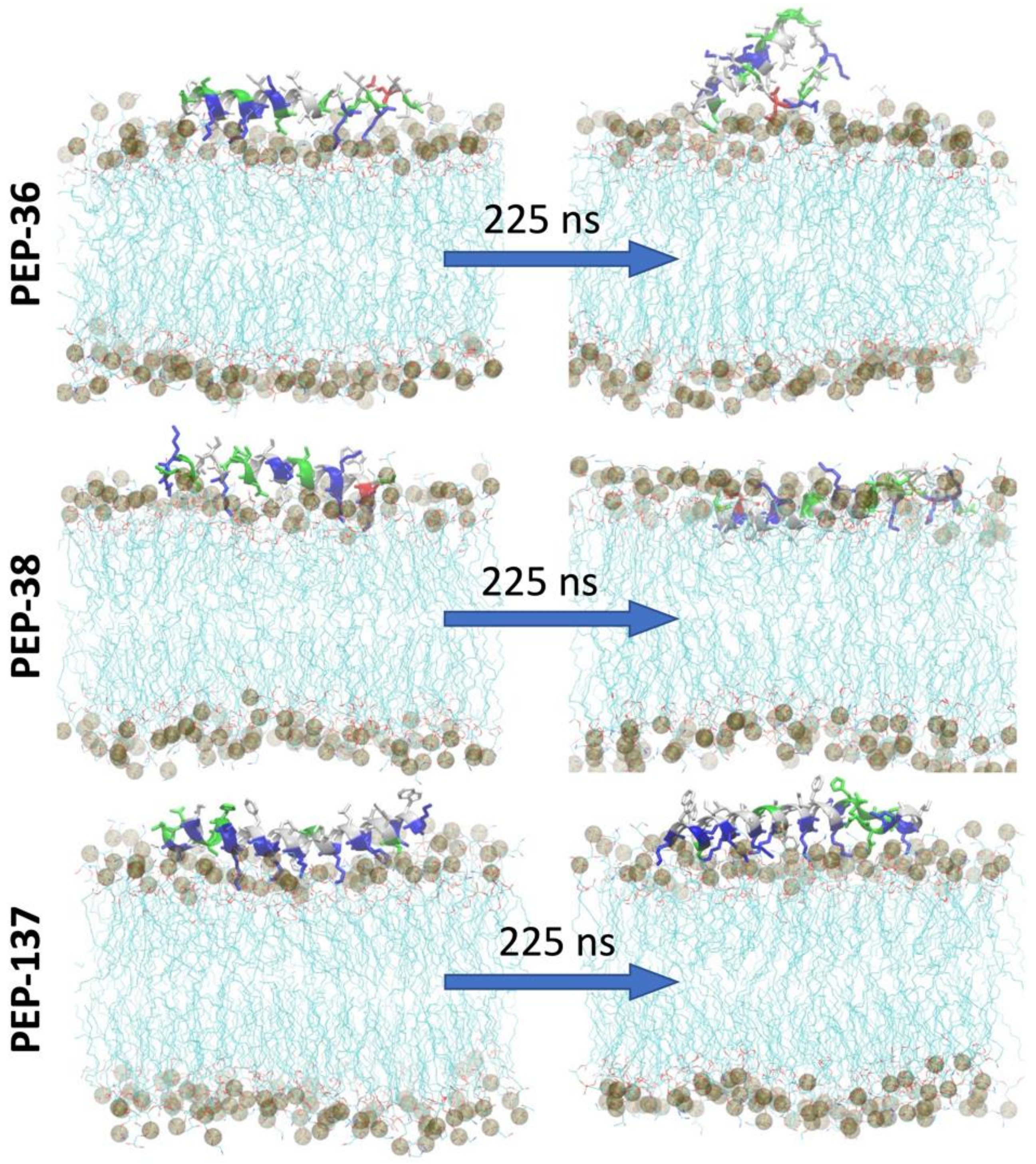
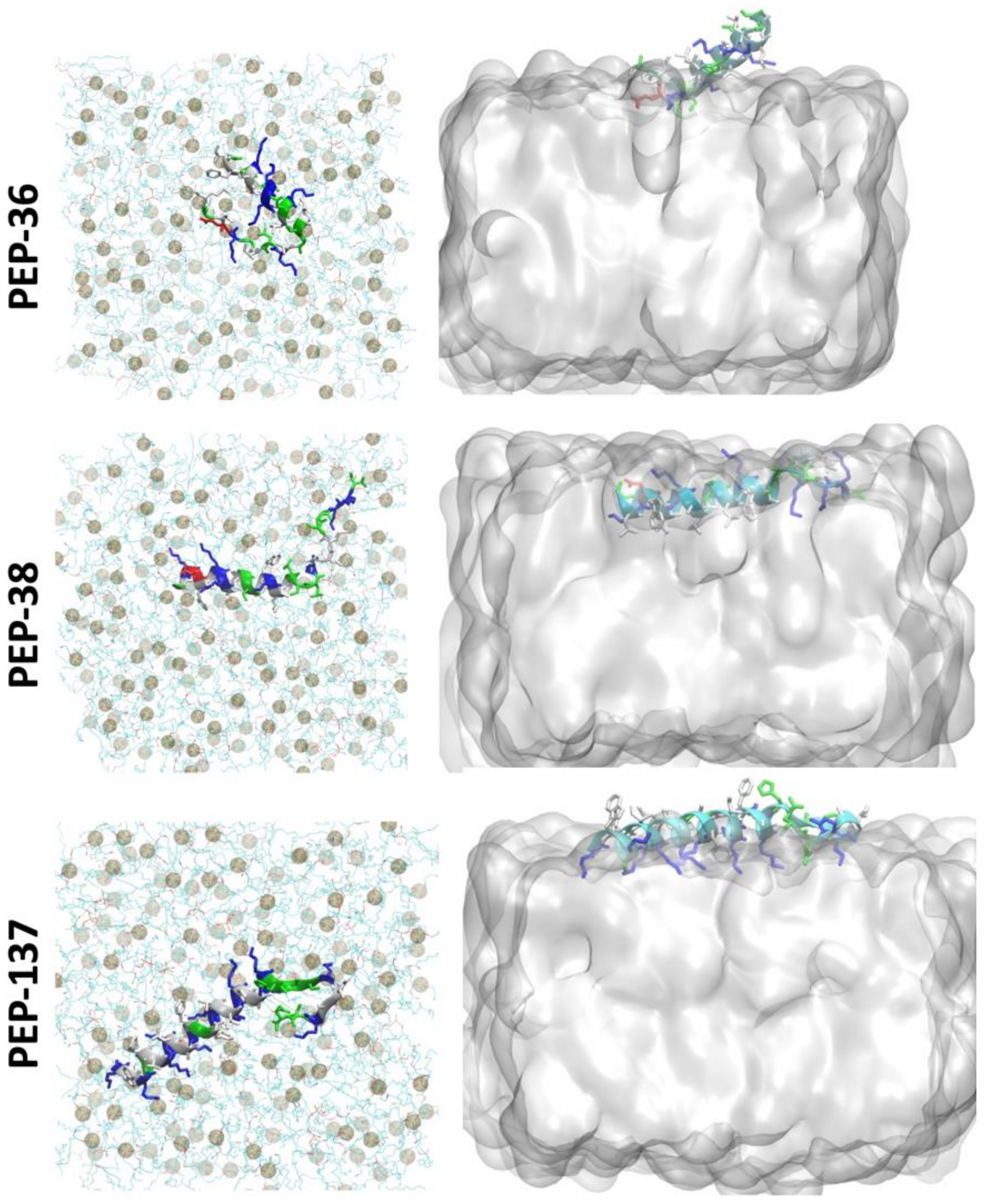
2.6. Molecular Dynamics Simulation of PEP-36, PEP-38 and PEP-137 Interaction with Bacterial Membrane
3. Discussion
4. Materials and Methods
4.1. Peptides
4.2. Bacteria
4.3. In Vitro Study of Antibacterial Activity
4.4. Murine Experimental Model of Sepsis
4.5. Modeling the Structure of Novel Peptides
4.6. In Silico Predicting of Hemolytic Potential of Novel Peptides
4.7. Statistical Analysis
4.8. Molecular Dynamics Modeling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roberts, S.C.; Zembower, T.R. Global Increases in Antibiotic Consumption: A Concerning Trend for WHO Targets. Lancet Infect. Dis. 2021, 21, 10–11. [Google Scholar] [CrossRef]
- Baturin, V.; Shchetinin, E.; Demidenko, I.; Kunitsina, E.; Korableva, O.; Baturina, M.; Kharitonova, Y. Evaluation of Klebsiella Spp. and Acinetobacter Spp. Antibiotic Resistance in Hospital Environment (Stavropol, Russia). Med. News North Cauc. 2014, 9. [Google Scholar] [CrossRef]
- Minaev, S.V.; Filipeva, N.V.; Leskin, V.V.; Shchetinin, E.V.; Golubeva, M.V.; Rakitina, E.N.; Shamadaev, E.Z.; Zhdanova, T.V. Microbiological Spectrum of Pyoinflammatory Diseases Causative Agents in Children at a Multi-Speciality Hospital. Med. News North Cauc. 2018, 13, 112–114. [Google Scholar] [CrossRef]
- Strathdee, S.A.; Davies, S.C.; Marcelin, J.R. Confronting Antimicrobial Resistance beyond the COVID-19 Pandemic and the 2020 US Election. Lancet 2020, 396, 1050–1053. [Google Scholar] [CrossRef]
- Langford, B.J.; So, M.; Raybardhan, S.; Leung, V.; Westwood, D.; MacFadden, D.R.; Soucy, J.-P.R.; Daneman, N. Bacterial Co-Infection and Secondary Infection in Patients with COVID-19: A Living Rapid Review and Meta-Analysis. Clin. Microbiol. Infect. 2020, 26, 1622–1629. [Google Scholar] [CrossRef]
- Browne, K.; Chakraborty, S.; Chen, R.; Willcox, M.D.; Black, D.S.; Walsh, W.R.; Kumar, N. A New Era of Antibiotics: The Clinical Potential of Antimicrobial Peptides. Int. J. Mol. Sci. 2020, 21, 7047. [Google Scholar] [CrossRef]
- Plackett, B. Why Big Pharma Has Abandoned Antibiotics. Nature 2020, 586, S50–S52. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 0, 2559. [Google Scholar] [CrossRef]
- Pachón-Ibáñez, M.E.; Smani, Y.; Pachón, J.; Sánchez-Céspedes, J. Perspectives for Clinical Use of Engineered Human Host Defense Antimicrobial Peptides. FEMS Microbiol. Rev. 2017, 41, 323–342. [Google Scholar] [CrossRef]
- Lee, A.C.-L.; Harris, J.L.; Khanna, K.K.; Hong, J.-H. A Comprehensive Review on Current Advances in Peptide Drug Development and Design. Int. J. Mol. Sci. 2019, 20, 2383. [Google Scholar] [CrossRef]
- Yu, G.; Baeder, D.Y.; Regoes, R.R.; Rolff, J. Predicting Drug Resistance Evolution: Insights from Antimicrobial Peptides and Antibiotics. Proc. R. Soc. B Biol. Sci. 2018, 285. [Google Scholar] [CrossRef] [PubMed]
- Wimley, W.C.; Hristova, K.; Wimley, W.C.; Hristova, K. The Mechanism of Membrane Permeabilization by Peptides: Still an Enigma. Aust. J. Chem. 2019, 73, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Baturin, V.A.; Selimov, M.A.; Bolatchiev, A.D.; Sadovoy, V.V.; Budkevich, R.O.; Baturina, M.V. The Prognosis and Investigation of α-Defensin-1 (HNP-1) Influence on Morphological Changes of Staphylococcus Aureus Cells by the Atomic-Force Microscopy Data. Med. News North Cauc. 2017, 12, 290–294. [Google Scholar] [CrossRef][Green Version]
- Bolatchiev, A. Antibacterial Activity of Human Defensins against Staphylococcus Aureus and Escherichia Coli. PeerJ 2020, 8, e10455. [Google Scholar] [CrossRef] [PubMed]
- Bolatchiev, A.; Baturin, V.; Bazikov, I.; Maltsev, A.; Kunitsina, E. Effect of Antimicrobial Peptides HNP-1 and HBD-1 on Staphylococcus Aureus Strains in Vitro and in Vivo. Fundam. Clin. Pharmacol. 2020, 34, 102–108. [Google Scholar] [CrossRef]
- Müller, A.T.; Hiss, J.A.; Schneider, G. Recurrent Neural Network Model for Constructive Peptide Design. J. Chem. Inf. Modeling 2018, 58, 472–479. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD3: The Antimicrobial Peptide Database as a Tool for Research and Education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Waghu, F.H.; Idicula-Thomas, S. Collection of Antimicrobial Peptides Database and Its Derivatives: Applications and Beyond. Protein Sci. 2020, 29, 36–42. [Google Scholar] [CrossRef]
- Vishnepolsky, B.; Pirtskhalava, M. Prediction of Linear Cationic Antimicrobial Peptides Based on Characteristics Responsible for Their Interaction with the Membranes. J. Chem. Inf. Modeling 2014, 54, 1512–1523. [Google Scholar] [CrossRef]
- Wang, G. Structures of Human Host Defense Cathelicidin LL-37 and Its Smallest Antimicrobial Peptide KR-12 in Lipid Micelles. J. Biol. Chem. 2008, 283, 32637–32643. [Google Scholar] [CrossRef]
- Timmons, P.B.; Hewage, C.M. HAPPENN Is a Novel Tool for Hemolytic Activity Prediction for Therapeutic Peptides Which Employs Neural Networks. Sci. Rep. 2020, 10, 1–18. [Google Scholar] [CrossRef]
- Pan, C.Y.; Chen, J.C.; Sheen, J.F.; Lin, T.L.; Chen, J.Y. Epinecidin-1 Has Immunomodulatory Effects, Facilitating Its Therapeutic Use in a Mouse Model of Pseudomonas Aeruginosa Sepsis. Antimicrob. Agents Chemother. 2014, 58, 4264–4274. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Pan, C.Y.; Chen, J.Y. The Antimicrobial Peptide, Epinecidin-1, Mediates Secretion of Cytokines in the Immune Response to Bacterial Infection in Mice. Peptides 2012, 36, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Bolatchiev, A. Antimicrobial Peptides Epinecidin-1 and Beta-Defesin-3 Are Effective against a Broad Spectrum of Antibiotic-Resistant Bacterial Isolates and Increase Survival Rate in Experimental Sepsis. Antibiotics 2022, 11, 76. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 10.0. 2020. Available online: http://www.Eucast.Org (accessed on 29 January 2022).
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and Broth Dilution Methods to Determine the Minimal Inhibitory Concentration (MIC) of Antimicrobial Substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID). Determination of Minimum Inhibitory Concentrations (MICs) of Antibacterial Agents by Broth Dilution. Clin. Microbiol. Infect. 2003, 9, ix. [Google Scholar] [CrossRef]
- Brunetti, J.; Falciani, C.; Roscia, G.; Pollini, S.; Bindi, S.; Scali, S.; Arrieta, U.C.; Gomez-Vallejo, V.; Quercini, L.; Ibba, E.; et al. In Vitro and in Vivo Efficacy, Toxicity, Bio-Distribution and Resistance Selection of a Novel Antibacterial Drug Candidate. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Percie du Sert, N.; Hurst, V.; Ahluwalia, A.; Alam, S.; Avey, M.T.; Baker, M.; Browne, W.J.; Clark, A.; Cuthill, I.C.; Dirnagl, U.; et al. The ARRIVE Guidelines 2.0: Updated Guidelines for Reporting Animal Research. PLoS Biol. 2020, 18, e3000410. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Qian, C.D.; Wu, X.C.; Teng, Y.; Zhao, W.P.; Li, O.; Fang, S.G.; Huang, Z.H.; Gao, H.C. Battacin (Octapeptin B5), a New Cyclic Lipopeptide Antibiotic from Paenibacillus Tianmuensis Active against Multidrug-Resistant Gram-Negative Bacteria. Antimicrob. Agents Chemother. 2012, 56, 1458–1465. [Google Scholar] [CrossRef]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Dávila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI Membrane Builder toward Realistic Biological Membrane Simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.; Kobzev, E.; Chan, J.; de Zoysa, G.H.; Sarojini, V.; Piggot, T.J.; Allison, J.R. Molecular Dynamics Simulation of the Interaction of Two Linear Battacin Analogs with Model Gram-Positive and Gram-Negative Bacterial Cell Membranes. ACS Omega 2021, 6, 388–400. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable Molecular Dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An Improved Force Field for Folded and Intrinsically Disordered Proteins. Nat. Methods 2016, 14, 71–73. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1998, 103, 8577. [Google Scholar] [CrossRef]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H.J.C. Numerical Integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of n-Alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
| Peptide | Amino Acid Sequence | Length | Molecular Weight | Charge | Hydrophobic Residues |
|---|---|---|---|---|---|
| PEP-36 | GIFSKLAGKKIKNLLISGLKNIGKEVGM | 28 | 2958 | +5.0 | 43 |
| PEP-38 | GLKDWVKKALGSLWKLANSQKAIISGKKS | 29 | 3156 | +6.0 | 41 |
| PEP-136 | KWKLFKKIWSSVKLKS | 16 | 2007 | +6.0 | 44 |
| PEP-137 | KWKSFIKKLAKFGFKVIKKFAKKHGSKIAKNQ | 32 | 3764 | +12.1 | 41 |
| PEP-174 | GILSSFKGVLKGAGKNLLGSLKDKLKN | 27 | 2786 | +5.0 | 37 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bolatchiev, A.; Baturin, V.; Shchetinin, E.; Bolatchieva, E. Novel Antimicrobial Peptides Designed Using a Recurrent Neural Network Reduce Mortality in Experimental Sepsis. Antibiotics 2022, 11, 411. https://doi.org/10.3390/antibiotics11030411
Bolatchiev A, Baturin V, Shchetinin E, Bolatchieva E. Novel Antimicrobial Peptides Designed Using a Recurrent Neural Network Reduce Mortality in Experimental Sepsis. Antibiotics. 2022; 11(3):411. https://doi.org/10.3390/antibiotics11030411
Chicago/Turabian StyleBolatchiev, Albert, Vladimir Baturin, Evgeny Shchetinin, and Elizaveta Bolatchieva. 2022. "Novel Antimicrobial Peptides Designed Using a Recurrent Neural Network Reduce Mortality in Experimental Sepsis" Antibiotics 11, no. 3: 411. https://doi.org/10.3390/antibiotics11030411
APA StyleBolatchiev, A., Baturin, V., Shchetinin, E., & Bolatchieva, E. (2022). Novel Antimicrobial Peptides Designed Using a Recurrent Neural Network Reduce Mortality in Experimental Sepsis. Antibiotics, 11(3), 411. https://doi.org/10.3390/antibiotics11030411







