An 18-Year Dataset on the Clinical Incidence and MICs to Antibiotics of Achromobacter spp. (Labeled Biochemically or by MAL-DI-TOF MS as A. xylosoxidans), Largely in Patient Groups Other than Those with CF
Abstract
1. Introduction
2. Results
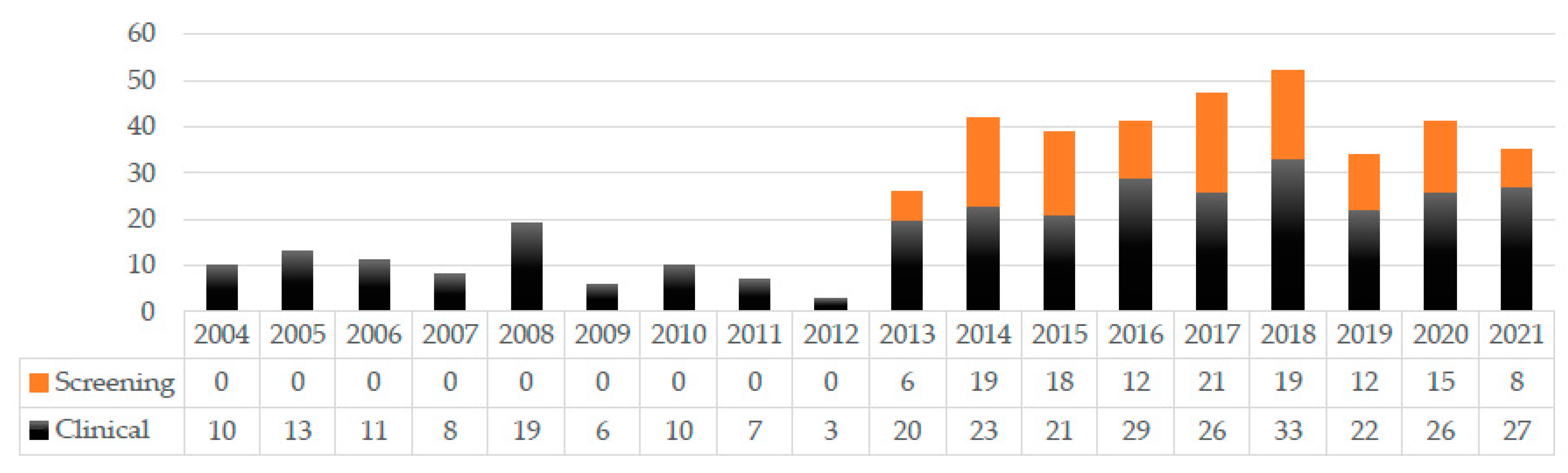
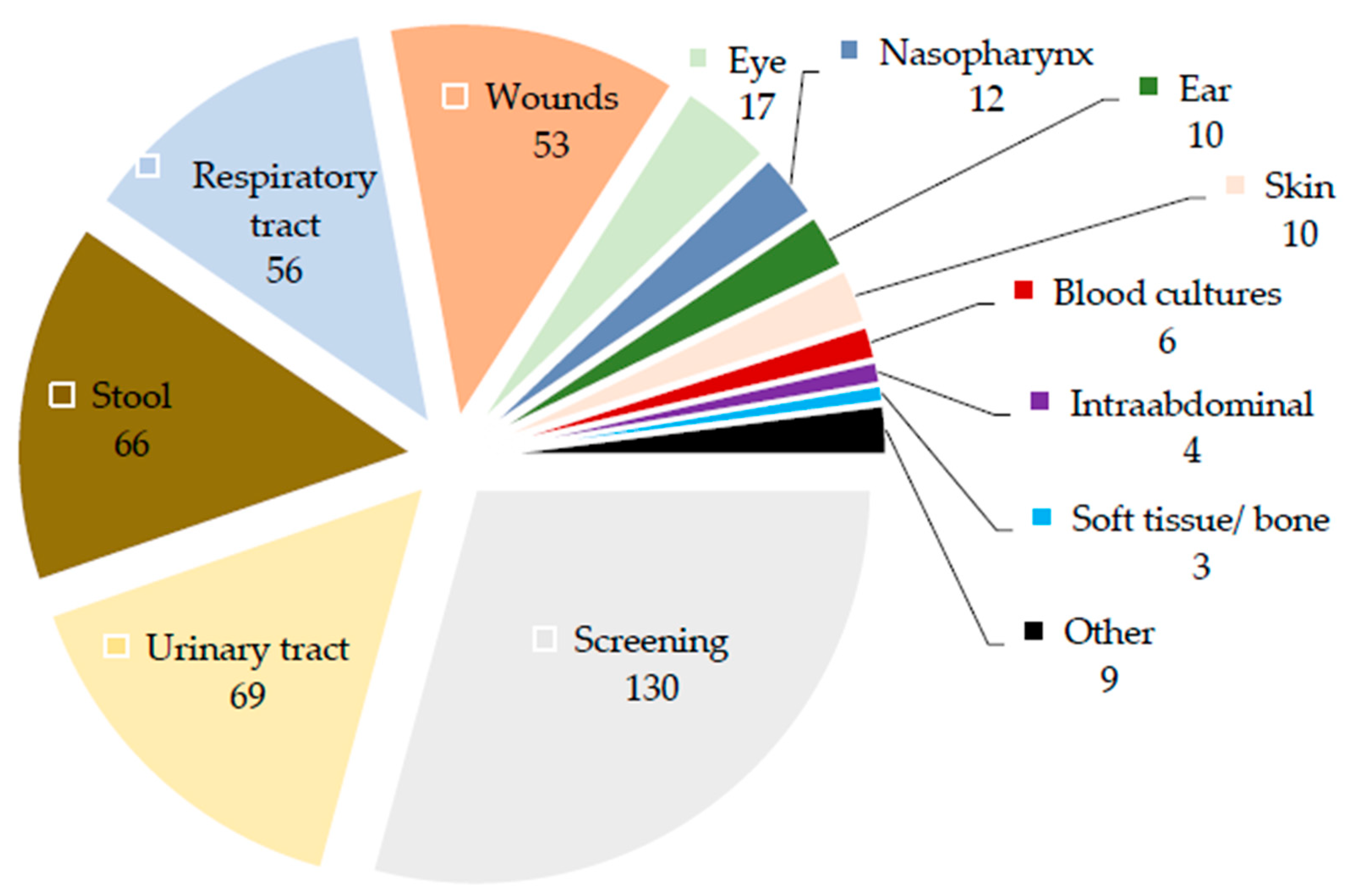
2.1. Specimens in Which Achromobacter Isolates Were Detected
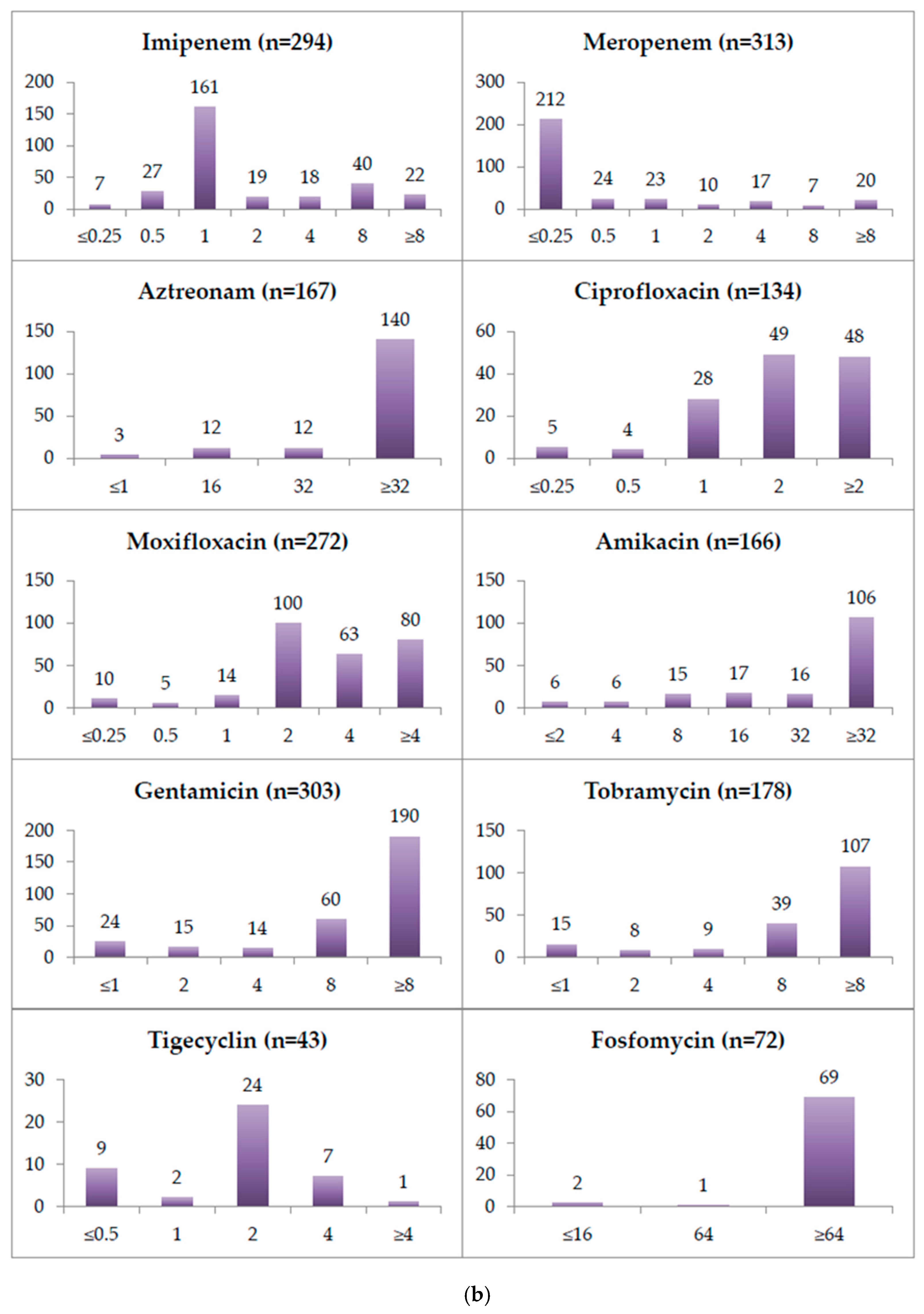
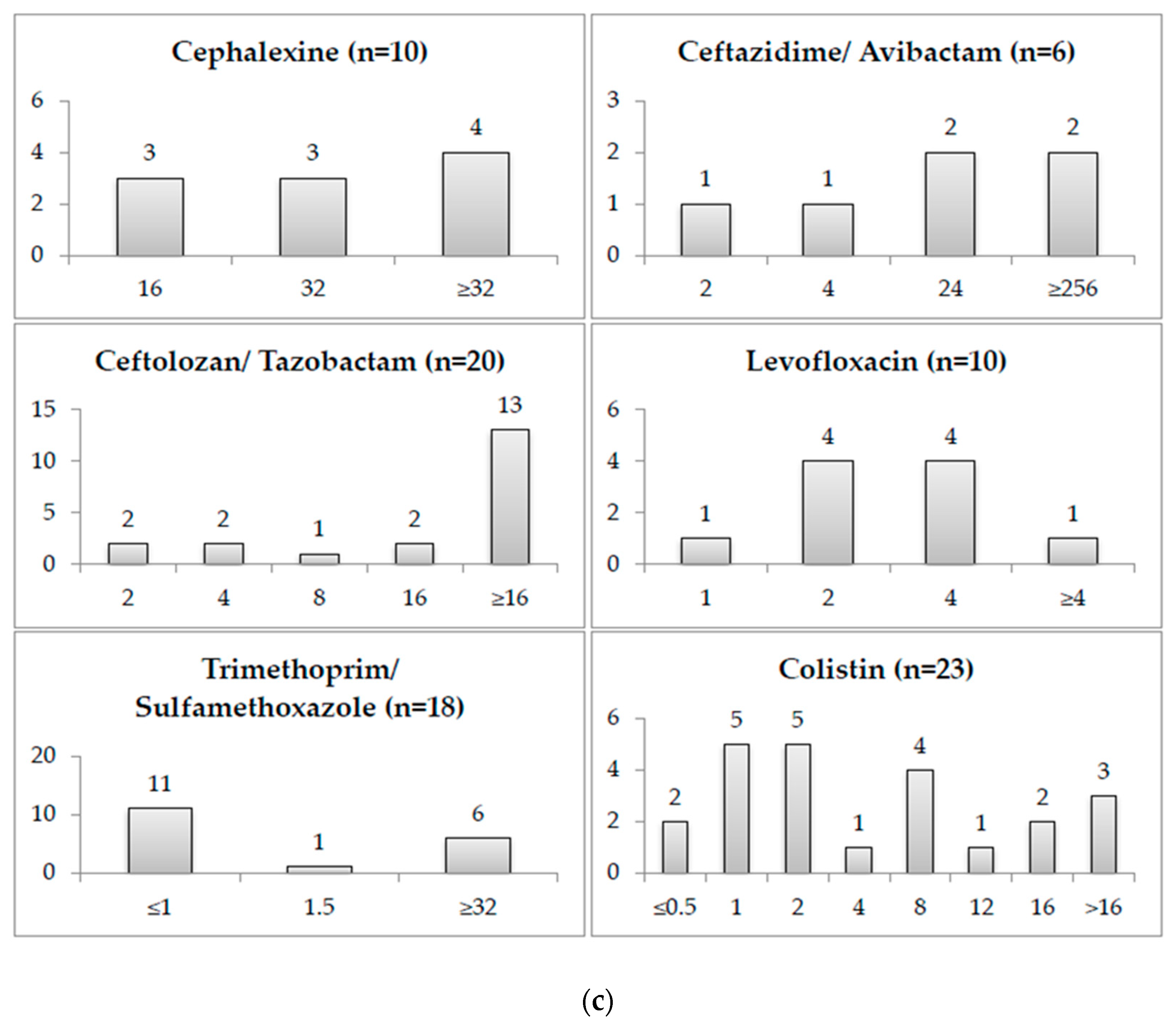
2.2. Minimum Inhibitory Concentrations
2.3. Patient Groups Detected with Achromobacter Isolates
3. Discussion
4. Materials and Methods
4.1. Data Retrieval
4.2. Ward Type and Length of Hospitalization
4.3. Minimum Inhibitory Concentrations of the Antibiotics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pressler, T.; Høiby, N.; Gormsen, M. Chronic infection with Achromobacter xylosoxidans in cystic fibrosis patients; a retrospective case control study. J. Cyst. Fibros. 2006, 5, 245–251. [Google Scholar]
- De Baets, F.; Schelstraete, P.; Van Daele, S.; Haerynck, F.; Vaneechoutte, M. Achromobacter xylosoxidans in cystic fibrosis: Prevalence and clinical relevance. J. Cyst. Fibros. 2007, 6, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Lambiase, A.; Catania, M.R.; Del Pezzo, M.; Rossano, F.; Terlizzi, V.; Sepe, A.; Raia, V. Achromobacter xylosoxidans respiratory tract infection in cystic fibrosis patients. Eur. J. Clin. Microbiol. 2011, 30, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Tena, D.; González-Praetorius, A.; Pérez-Balsalobre, M.; Sancho, O.; Bisquert, J. Urinary tract infection due to Achromobacter xylosoxidans: Report of 9 cases. Scand. J. Infect. Dis. 2008, 40, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.G.; Rays, J.; Kanegae, M.Y. Native-valve endocarditis caused by Achromobacter xylosoxidans: A case report and review of literature. Autopsy Case Rep. 2017, 7, 50. [Google Scholar] [CrossRef] [PubMed]
- Marion-Sanchez, K.; Lion, F.; Olive, C.; Cailleaux, G.; Roques, F. Mediastinitis superinfected by Achromobacter xylosoxidans. A case report. J. Infect. Chemother. 2018, 24, 987–989. [Google Scholar] [CrossRef] [PubMed]
- Duggan, J.M.; Goldstein, S.J.; Chenoweth, C.E.; Kauffman, C.A.; Bradley, S.F. Achromobacter xylosoxidans bacteremia: Report of four cases and review of the literature. Clin. Infect. Dis. 1996, 23, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Raghuraman, K.; Ahmed, N.H.; Baruah, F.K.; Grover, R.K. Achromobacter Xylosoxidans bloodstream infection in elderly patient with Hepatocellular Carcinoma: Case report and review of literature. J. Lab. Phys. 2015, 7, 124–127. [Google Scholar] [CrossRef]
- Legrand, C.; Anaissie, E. Bacteremia due to Achromobacter xylosoxidans in patients with cancer. Clin. Infect. Dis. 1992, 14, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Hansen, C.R.; Pressler, T.; Nielsen, K.G.; Jensen, P.; Bjarnsholt, T.; Høiby, N. Inflammation in Achromobacter xylosoxidans infected cystic fibrosis patients. J. Cyst. Fibros. 2010, 9, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Eshwara, V.K.; Mukhopadhyay, C.; Mohan, S.; Prakash, R.; Pai, G. Two unique presentations of Achromobacter xylosoxidans infections in clinical settings. J. Infect. Dev. Ctries. 2011, 5, 138–141. [Google Scholar] [CrossRef]
- Reverdy, M.E.; Freney, J.; Fleurette, J.; Coulet, M.; Surgot, M.; Marmet, D.; Ploton, C. Nosocomial colonization and infection by Achromobacter xylosoxidans. J. Clin. Microbiol. 1984, 19, 140–143. [Google Scholar] [CrossRef] [PubMed]
- Ridderberg, W.; Bendstrup, E.; Olesen, H.V.; Jensen-Fangel, S.; Nørskov-Lauritsen, N. Marked increase in incidence of Achromobacter xylosoxidans infections caused by sporadic acquisition from the environment. J. Cyst. Fibros. 2011, 10, 466–469. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Cerezo, J.; Suárez, I.; Ríos, J.J.; Peña, P.; García De Miguel, M.J.G.; De José, M.; Monteagudo, O.; Linares, P.; Barbado-Cano, A.; Vázquez, J.J. Achromobacter xylosoxidans bacteremia: A 10-year analysis of 54 cases. Eur. J. Clin. Microbiol. Infect. Dis. 2003, 22, 360–363. [Google Scholar] [CrossRef] [PubMed]
- Veschetti, L.; Sandri, A.; Patuzzo, C.; Melotti, P.; Malerba, G.; Lleo, M.M. Genomic characterization of Achromobacter species isolates from chronic and occasional lung infection in cystic fibrosis patients. Microb. Genom. 2021, 7, 606. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekar, P.H.; Arathoon, E.; Levine, D.P. Infections due to Achromobacter xylosoxidans. Case report and review of the literature. Infection 1986, 14, 279–282. [Google Scholar] [CrossRef] [PubMed]
- Amoureux, L.; Bador, J.; Siebor, E.; Taillefumier, N.; Fanton, A.; Neuwirth, C. Epidemiology and resistance of Achromobacter xylosoxidans from cystic fibrosis patients in Dijon, Burgundy: First French data. J. Cyst. Fibros. 2013, 12, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Garrigos, T.; Dollat, M.; Magallon, A.; Chapuis, A.; Varin, V.; Bador, J.; Makki, N.; Cremet, L.; Persyn, E.; Cardot-Martin, E.; et al. Matrix-assisted laser desorption ionization—Time of flight mass spectrometry for rapid detection of isolates belonging to the epidemic clones Achromobacter xylosoxidans ST137 and Achromobacter ruhlandii DES from cystic fibrosis patients. J. Clin. Microbiol. 2021, 59, e0094621. [Google Scholar] [CrossRef] [PubMed]
- Amoureux, L.; Bador, J.; Zouak, F.B.; Chapuis, A.; de Curraize, C.; Neuwirth, C. Distribution of the species of Achromobacter in a French cystic fibrosis centre and multilocus sequence typing analysis reveal the predominance of A. xylosoxidans and clonal relationships between some clinical and environmental isolates. J. Cyst. Fibros. 2016, 15, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Aisenberg, G.; Rolston, K.V.; Safdar, A. Bacteremia caused by Achromobacter and Alcaligenes species in 46 patients with cancer (1989–2003). Cancer 2004, 101, 2134–2140. [Google Scholar] [CrossRef]
- Turel, O.; Kavuncuoğlu, S.; Hoşaf, E.; Ozbek, S.; Aldemir, E.; Uygur, T.; Hatipoglu, N.; Siraneci, R. Bacteremia due to Achromobacter xylosoxidans in neonates: Clinical features and outcome. Braz. J. Infect. Dis. 2013, 17, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Gade, S.S.; Nørskov-Lauritsen, N.; Ridderberg, W. Prevalence and species distribution of Achromobacter sp. cultured from cystic fibrosis patients attending the Aarhus centre in Denmark. J. Med Microbiol. 2017, 66, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Edwards, B.D.; Greysson-Wong, J.; Somayaji, R.; Waddell, B.; Whelan, F.J.; Storey, D.; Rabin, H.R.; Surette, M.G.; Parkins, M.D. Prevalence and outcomes of Achromobacter species infections in adults with cystic fibrosis: A North American cohort study. J. Clin. Microbiol. 2017, 55, 2074–2085. [Google Scholar] [CrossRef] [PubMed]
- Amoureux, L.; Sauge, J.; Sarret, B.; Lhoumeau, M.; Bajard, A.; Tetu, J.; Bador, J.; Neuwirth, C.; Caillon, J.; Cardot-Martin, E.; et al. Study of 109 Achromobacter spp. isolates from 9 French CF centres reveals the circulation of a multiresistant clone of A. xylosoxidans belonging to ST 137. J. Cyst. Fibros. 2019, 18, 804–807. [Google Scholar] [CrossRef] [PubMed]
- Bizzini, A.; Durussel, C.; Bille, J.; Greub, G.; Prod’Hom, G. Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J. Clin. Microbiol. 2010, 48, 1549–1554. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Ye, L.; Zhao, Q.; Ma, Y.; Yang, J.; Luo, Y. Comparative study of MALDI-TOF MS and VITEK 2 in bacteria identification. J. Thorac. Dis. 2014, 6, 534–538. [Google Scholar]
- Febbraro, F.; Rodio, N.M.; Puggioni, G.; Antonelli, G.; Pietropaolo, V.; Trancassini, M. MALDI-TOF MS Versus VITEK® 2: Comparison of systems for the identification of microorganisms responsible for bacteremia. Curr. Microbiol. 2016, 73, 843–850. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Marko, D.C.; Saffert, R.T.; Cunningham, S.A.; Hyman, J.; Walsh, J.; Arbefeville, S.; Howard, W.; Pruessner, J.; Safwat, N.; Cockerill, F.R.; et al. Evaluation of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry systems for identification of nonfermenting Gram-negative bacilli isolated from cultures from cystic fibrosis patients. J. Clin. Microbiol. 2012, 50, 2034–2039. [Google Scholar] [CrossRef] [PubMed]
- Panagopoulos, M.I.; Jean, M.S.; Brun, D.; Guiso, N.; Bekal, S.; Ovetchkine, P.; Tapiero, B. Bordetella holmesii bacteremia in asplenic children: Report of four cases initially misidentified as Acinetobacter lwoffii. J. Clin. Microbiol. 2010, 48, 3762–3764. [Google Scholar] [CrossRef][Green Version]
- Buechler, C.; Neidhöfer, C.; Hornung, T.; Neuenhoff, M.; Parčina, M. Detection and characterization of clinical Bordetella trematum isolates from chronic wounds. Pathogens 2021, 10, 966. [Google Scholar] [CrossRef] [PubMed]
- Villers, D.; Espaze, E.; Coste-Burel, M.; Giauffret, F.; Ninin, E.; Nicolas, F.; Richet, H. Nosocomial Acinetobacter baumannii infections: Microbiological and clinical epidemiology. Ann. Intern. Med. 1998, 129, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Driscoll, J.A.; Brody, S.L.; Kollef, M.H. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 2007, 67, 351–368. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhu, Y.; Ma, Y.; Liu, F.; Lu, N.; Yang, X.; Luan, C.; Yi, Y.; Zhu, B. Genomic insights into intrinsic and acquired drug resistance mechanisms in Achromobacter xylosoxidans. Antimicrob. Agents Chemother. 2014, 59, 1152–1161. [Google Scholar] [CrossRef] [PubMed]
- Bador, J.; Amoureux, L.; Duez, J.-M.; Drabowicz, A.; Siebor, E.; Llanes, C.; Neuwirth, C. First description of an RND-type multidrug efflux pump in Achromobacter xylosoxidans, AxyABM. Antimicrob. Agents Chemother. 2011, 55, 4912–4914. [Google Scholar] [CrossRef] [PubMed]
- Isler, B.; Kidd, T.J.; Stewart, A.G.; Harris, P.; Paterson, D.L. Achromobacter infections and treatment options. Antimicrob. Agents Chemother. 2020, 64, e01025-20. [Google Scholar] [CrossRef] [PubMed]
- Bador, J.; Amoureux, L.; Blanc, E.; Neuwirth, C. Innate aminoglycoside resistance of Achromobacter xylosoxidans is due to AxyXY-OprZ, an RND-type multidrug efflux pump. Antimicrob. Agents Chemother. 2013, 57, 603–605. [Google Scholar] [CrossRef]
- Bador, J.; Neuwirth, C.; Grangier, N.; Muniz, M.; Germé, L.; Bonnet, J.; Pillay, V.-G.; Llanes, C.; de Curraize, C.; Amoureux, L. Role of AxyZ transcriptional regulator in overproduction of AxyXY-OprZ multidrug efflux system in Achromobacter species mutants selected by tobramycin. Antimicrob. Agents Chemother. 2017, 61, e00290-17. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Pan, F.; Guo, J.; Yan, W.; Jin, Y.; Liu, C.; Qin, L.; Fang, X. Hospital acquired pneumonia due to Achromobacter spp. in a geriatric ward in China: Clinical characteristic, genome variability, biofilm production, antibiotic resistance and integron in isolated strains. Front. Microbiol. 2016, 7, 621. [Google Scholar] [CrossRef] [PubMed]
- Traglia, G.M.; Almuzara, M.; Merkier, A.K.; Adams, C.; Galanternik, L.; Vay, C.; Centrón, D.; Ramírez, M.S. Achromobacter xylosoxidans: An emerging pathogen carrying different elements involved in horizontal genetic transfer. Curr. Microbiol. 2012, 65, 673–678. [Google Scholar] [CrossRef][Green Version]
- Filipic, B.; Malesevic, M.; Vasiljevic, Z.; Lukic, J.; Novovic, K.; Kojic, M.; Jovcic, B. Uncovering differences in virulence markers associated with Achromobacter species of CF and non-CF origin. Front. Cell. Infect. Microbiol. 2017, 7, 224. [Google Scholar] [CrossRef]
- Vali, P.; Shahcheraghi, F.; Seyfipour, M.; Zamani, M.A.; Allahyar, M.R.; Feizabadi, M.M. Phenotypic and genetic characterization of carbapenemase and ESBLs producing gram-negative bacteria (GNB) isolated from patients with cystic fibrosis (CF) in Tehran hospitals. J. Clin. Diagn. Res. 2014, 8, 26. [Google Scholar] [PubMed]
- Neuwirth, C.; Freby, C.; Ogier-Desserrey, A.; Perez-Martin, S.; Houzel, A.; Péchinot, A.; Duez, J.-M.; Huet, F.; Siebor, E. VEB-1 in Achromobacter xylosoxidans from cystic fibrosis patient, France. Emerg. Infecti. Dis. 2006, 12, 1737. [Google Scholar] [CrossRef] [PubMed]
- Shibata, N.; Doi, Y.; Yamane, K.; Yagi, T.; Kurokawa, H.; Shibayama, K.; Kato, H.; Kai, K.; Arakawa, Y. PCR typing of genetic determinants for metallo-β-lactamases and integrases carried by gram-negative bacteria isolated in Japan, with focus on the class 3 integron. J. Clin. Microbiol. 2003, 41, 5407–5413. [Google Scholar] [CrossRef] [PubMed]
- Senda, K.; Arakawa, Y.; Ichiyama, S.; Nakashima, K.; Ito, H.; Ohsuka, S.; Shimokata, K.; Kato, N.; Ohta, M. PCR detection of metallo-beta-lactamase gene (blaIMP) in gram-negative rods resistant to broad-spectrum beta-lactams. J. Clin. Microbiol. 1996, 34, 2909–2913. [Google Scholar] [CrossRef] [PubMed]
- Riccio, M.L.; Pallecchi, L.; Fontana, R.; Rossolini, G.M. In70 of plasmid pAX22, a bla VIM-1-containing integron carrying a new aminoglycoside phosphotransferase gene cassette. Antimicrob. Agents Chemother. 2001, 45, 1249–1253. [Google Scholar] [CrossRef]
- Potron, A.; Fournier, D.; Emeraud, C.; Triponney, P.; Plésiat, P.; Naas, T.; Dortet, L. Evaluation of the immunochromatographic NG-Test Carba 5 for rapid identification of carbapenemase in nonfermenters. Antimicrob. Agents Chemother. 2019, 63, e00968-19. [Google Scholar] [CrossRef]
- Sofianou, D.; Markogiannakis, A.; Metzidie, E.; Pournaras, S.; Tsakris, A. VIM-2 metallo-β-lactamase in Achromobacter xylosoxidans in Europe. Eur. J. Clin. Microbiol. 2005, 24, 854–855. [Google Scholar] [CrossRef] [PubMed]
- Shin, K.S.; Han, K.; Lee, J.; Hong, S.B.; Son, B.R.; Youn, S.J.; Kim, J.; Shin, H.S. Imipenem-resistant Achromobacter xylosoxidans carrying blaVIM-2-containing class 1 integron. Diagn. Microbiol. Infect. Dis. 2005, 53, 215–220. [Google Scholar] [CrossRef]
- El Salabi, A.; Borra, P.S.; Toleman, M.A.; Samuelsen, Ø.; Walsh, T.R. Genetic and biochemical characterization of a novel metallo-β-lactamase, TMB-1, from an Achromobacter xylosoxidans strain isolated in Tripoli, Libya. Antimicrob. Agents Chemother. 2012, 56, 2241–2245. [Google Scholar] [CrossRef]
- Amoureux, L.; Bador, J.; Verrier, T.; Mjahed, H.; De Curraize, C.; Neuwirth, C. Achromobacter xylosoxidans is the predominant Achromobacter species isolated from diverse non-respiratory samples. Epidemiol. Infect. 2016, 144, 3527–3530. [Google Scholar] [CrossRef]
- Amoureux, L.; Bador, J.; Fardeheb, S.; Mabille, C.; Couchot, C.; Massip, C.; Salignon, A.-L.; Berlie, G.; Varin, V.; Neuwirth, C. Detection of Achromobacter xylosoxidans in hospital, domestic, and outdoor environmental samples and comparison with human clinical isolates. Appl. Environ. Microbiol. 2013, 79, 7142–7149. [Google Scholar] [CrossRef] [PubMed]
- Recio, R.; Brañas, P.; Martinez, M.T.; Chaves, F.; Orellana, M.A. Effect of respiratory Achromobacter spp. infection on pulmonary function in patients with cystic fibrosis. J. Med. Microbiol. 2018, 67, 952–956. [Google Scholar] [CrossRef] [PubMed]
- Katchanov, J.; Asar, L.; Klupp, E.-M.; Both, A.; Rothe, C.; König, C.; Rohde, H.; Kluge, S.; Maurer, F.P. Carbapenem-resistant Gram-negative pathogens in a German university medical center: Prevalence, clinical implications and the role of novel β-lactam/β-lactamase inhibitor combinations. PLoS ONE 2018, 13, e0195757. [Google Scholar] [CrossRef] [PubMed]
- Aira, A.; Fehér, C.; Rubio, E.; Soriano, A. The intestinal microbiota as a reservoir and a therapeutic target to fight multi-drug-resistant bacteria: A narrative review of the literature. Infect. Dis. Ther. 2019, 8, 469–482. [Google Scholar] [CrossRef] [PubMed]
- Neidhöfer, C.; Buechler, C.; Neidhöfer, G.; Bierbaum, G.; Hannet, I.; Hoerauf, A.; Parčina, M. Global distribution patterns of carbapenemase-encoding bacteria in a new light: Clues on a role for ethnicity. Front. Cell. Infect. Microbiol. 2021, 11, 659753. [Google Scholar] [CrossRef] [PubMed]
- Gargiullo, L.; Del Chierico, F.; D’Argenio, P.; Putignani, L. Gut microbiota modulation for multidrug-resistant organism decolonization: Present and future perspectives. Front. Microbiol. 2019, 10, 1704. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Tariq, R.; Tosh, P.K.; Pardi, D.S.; Khanna, S. Faecal microbiota transplantation for eradicating carriage of multidrug-resistant organisms: A systematic review. Clin. Microbiol. Infect. 2019, 25, 958–963. [Google Scholar] [CrossRef] [PubMed]
- Crum-Cianflone, N.F.; Sullivan, E.; Ballon-Landa, G. Fecal microbiota transplantation and successful resolution of multidrug-resistant-organism colonization. J. Clin. Microbiol. 2015, 53, 1986–1989. [Google Scholar] [CrossRef] [PubMed]


| Specimen Type | Amount of Isolates Resistant to | |
|---|---|---|
| Piperacillin–tazobactam | Meropenem | |
| Blood | 1/7 | 0/7 |
| Urine | 15.69% (8/51) | 4.08% (2/49) |
| Stool | 37.21% (16/43) | 16.28% (7/43) |
| Screening | 17.09% (20/117) | 7.76% (9/116) |
| All specimens | 17.98% (57/317) | 8.63% (27/313) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neidhöfer, C.; Berens, C.; Parčina, M. An 18-Year Dataset on the Clinical Incidence and MICs to Antibiotics of Achromobacter spp. (Labeled Biochemically or by MAL-DI-TOF MS as A. xylosoxidans), Largely in Patient Groups Other than Those with CF. Antibiotics 2022, 11, 311. https://doi.org/10.3390/antibiotics11030311
Neidhöfer C, Berens C, Parčina M. An 18-Year Dataset on the Clinical Incidence and MICs to Antibiotics of Achromobacter spp. (Labeled Biochemically or by MAL-DI-TOF MS as A. xylosoxidans), Largely in Patient Groups Other than Those with CF. Antibiotics. 2022; 11(3):311. https://doi.org/10.3390/antibiotics11030311
Chicago/Turabian StyleNeidhöfer, Claudio, Christina Berens, and Marijo Parčina. 2022. "An 18-Year Dataset on the Clinical Incidence and MICs to Antibiotics of Achromobacter spp. (Labeled Biochemically or by MAL-DI-TOF MS as A. xylosoxidans), Largely in Patient Groups Other than Those with CF" Antibiotics 11, no. 3: 311. https://doi.org/10.3390/antibiotics11030311
APA StyleNeidhöfer, C., Berens, C., & Parčina, M. (2022). An 18-Year Dataset on the Clinical Incidence and MICs to Antibiotics of Achromobacter spp. (Labeled Biochemically or by MAL-DI-TOF MS as A. xylosoxidans), Largely in Patient Groups Other than Those with CF. Antibiotics, 11(3), 311. https://doi.org/10.3390/antibiotics11030311






