Evidence of Linezolid Resistance and Virulence Factors in Enterococcus spp. Isolates from Wild and Domestic Ruminants, Italy
Abstract
:1. Introduction
2. Results
2.1. Bacterial Isolation and Antibiotic Susceptibility Test
2.2. Antibiotic Resistance Genes and Virulence Determinants
3. Discussion
4. Materials and Methods
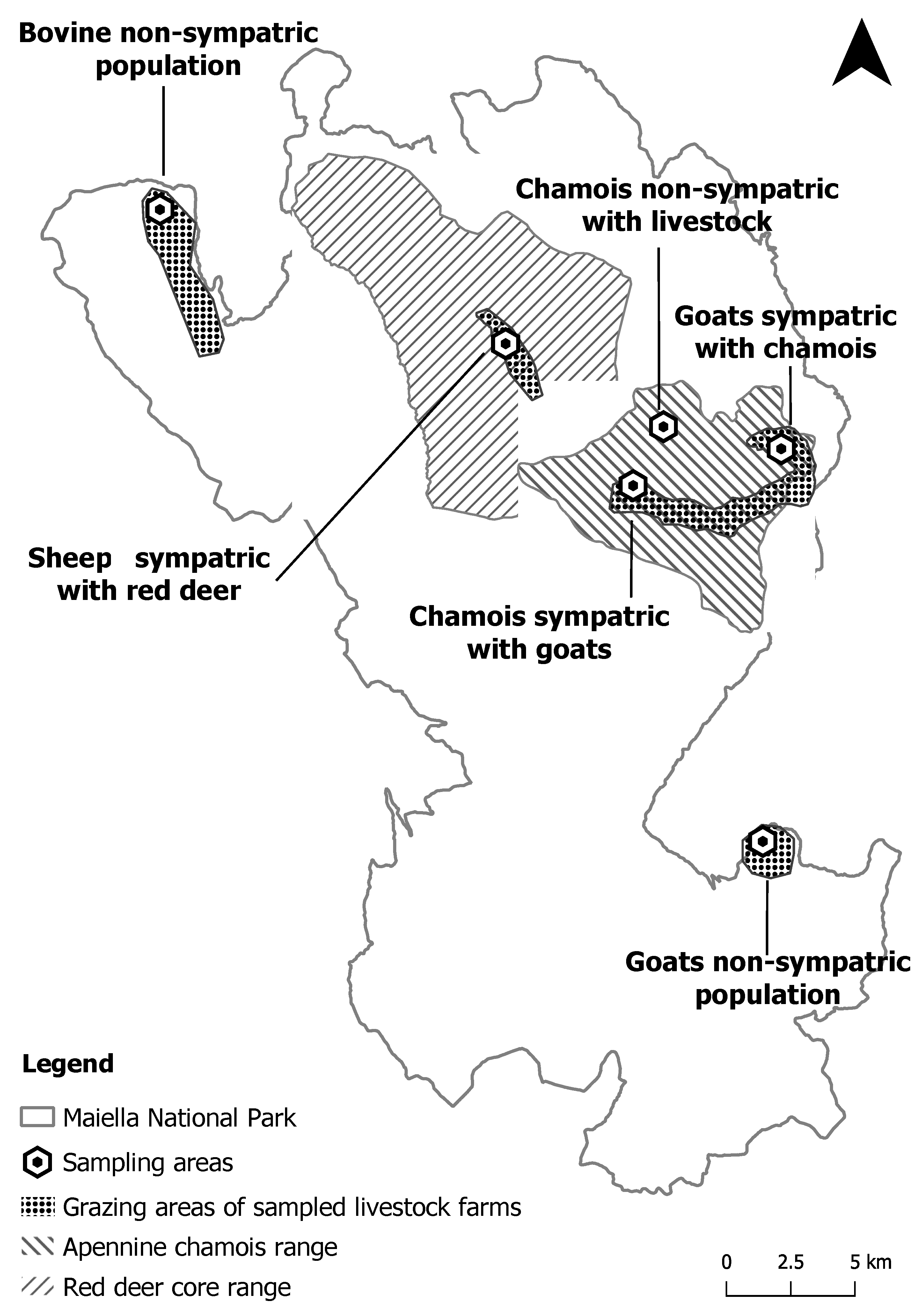
4.1. Study Area and Sampling Design
4.2. Bacteria Isolation and Antibioticssusceptibility Test
4.3. Detection of Antibiotic Resistance and Virulence Genes
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Prieto, A.M.G.; van Schaik, W.; Rogers, M.R.C.; Coque, T.M.; Ebaquero, F.; Ecorander, J.; Willems, R. Global Emergence and Dissemination of Enterococci as Nosocomial Pathogens: Attack of the Clones? Front. Microbiol. 2016, 7, 788. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Lv, Y.; Cai, J.; Schwarz, S.; Cui, L.; Hu, Z.; Zhang, R.; Li, J.; Zhao, Q.; Dacheng, W.; et al. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalisand Enterococcus faeciumof human and animal origin. J. Antimicrob. Chemother. 2015, 70, 2182–2190. [Google Scholar] [CrossRef] [Green Version]
- Torres, R.T.; Carvalho, J.; Cunha, M.V.; Serrano, E.; Palmeira, J.D.; Fonseca, C. Temporal and geographical research trends of antimicrobial resistance in wildlife—A bibliometric analysis. One Health 2020, 11, 100198. [Google Scholar] [CrossRef]
- Chajecka-Wierzchowska, W.; Zadernowska, A.; Łaniewska-Trokenheim, Ł. Virulence factors of Enterococcus spp. presented in food. LWT 2017, 75, 670–676. [Google Scholar] [CrossRef]
- Zaheer, R.; Cook, S.R.; Barbieri, R.; Goji, N.; Cameron, A.; Petkau, A.; Polo, R.O.; Tymensen, L.; Stamm, C.; Song, J.; et al. Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a One-Health continuum. Sci. Rep. 2020, 10, 3937, Corrected in Sci. Rep. 2020, 10, 13401. [Google Scholar] [CrossRef] [PubMed]
- Brenciani, A.; Fioriti, S.; Morroni, G.; Cucco, L.; Morelli, A.; Pezzotti, G.; Paniccià, M.; Antonelli, A.; Magistrali, C.F.; Rossolini, G.M.; et al. Detection in Italy of a porcine Enterococcus faecium isolate carrying the novel phenicol-oxazolidinone-tetracycline resistance gene poxtA. J. Antimicrob. Chemother. 2018, 74, 817–818. [Google Scholar] [CrossRef] [PubMed]
- Elghaieb, H.; Freitas, A.; Abbassi, M.S.; Novais, C.; Zouari, M.; Hassen, A.; Peixe, L. Dispersal of linezolid-resistant enterococci carrying poxtA or optrA in retail meat and food-producing animals from Tunisia. J. Antimicrob. Chemother. 2019, 74, 2865–2869. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wang, M.; Gao, Y.; Chen, L.; Wang, L. Emergence of plasmid-mediated oxazolidinone resistance gene poxtA from CC17 Enterococcus faecium of pig origin—authors’ response. J. Antimicrob. Chemother. 2020, 75, 1359–1361. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.-W.; Kang, Z.-Z.; Wu, S.-K.; Chen, Y.-P.; Kong, L.-H.; Wang, H.-N. Detection of the phenicol–oxazolidinone–tetracycline resistance gene poxtA in Enterococcus faecium and Enterococcus faecalis of food-producing animal origin in China. J. Antimicrob. Chemother. 2019, 74, 2459–2461. [Google Scholar] [CrossRef] [PubMed]
- Na, S.-H.; Moon, D.-C.; Kim, M.-H.; Kang, H.-Y.; Kim, S.-J.; Choi, J.-H.; Mechesso, A.-F.; Yoon, S.-S.; Lim, S.-K. Detection of the Phenicol–Oxazolidinone Resistance Gene poxtA in Enterococcus faecium and Enterococcus faecalis from Food-Producing Animals during 2008–2018 in Korea. Microorganisms 2020, 8, 1839. [Google Scholar] [CrossRef]
- Ruiz-Ripa, L.; Feßler, A.T.; Hanke, D.; Eichhorn, I.; Azcona-Gutiérrez, J.M.; Pérez-Moreno, M.O.; Seral, C.; Aspiroz, C.; Alonso, C.A.; Torres, L.; et al. Mechanisms of Linezolid Resistance Among Enterococci of Clinical Origin in Spain—Detection of optrA- and cfr(D)-Carrying E. faecalis. Microorganisms 2020, 8, 1155. [Google Scholar] [CrossRef] [PubMed]
- Torres, C.; Alonso, C.A.; Ruiz-Ripa, L.; León-Sampedro, R.; Del Campo, R.; Coque, T.M. Antimicrobial Resistance in Enterococcus spp. of animal origin. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Ramey, A.M.; Ahlstrom, C.A. Antibiotic Resistant Bacteria in Wildlife: Perspectives on Trends, Acquisition and Dissemination, Data Gaps, and Future Directions. J. Wildl. Dis. 2020, 56, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Dec, M.; Stępień-Pyśniak, D.; Gnat, S.; Fratini, F.; Urban-Chmiel, R.; Cerri, D.; Winiarczyk, S.; Turchi, B. Antibiotic Susceptibility and Virulence Genes in Enterococcus Isolates from Wild Mammals Living in Tuscany, Italy. Microb. Drug Resist. 2020, 26, 505–519. [Google Scholar] [CrossRef] [PubMed]
- Jánošková, A.; Kmet, V. Vancomycin Resistance Genes in Enterococcus spp. Strains Isolated from Alpine Accentor and Chamois. Acta Vet. Brno 2004, 73, 211–214. [Google Scholar] [CrossRef] [Green Version]
- Vandžurová, A.; Hrašková, I.; Jüdová, J.; Javorskỳ, P.; Pristaš, P. Antibiotic resistance and restriction endonucleases in fecal enterococci of chamois (Rupicapra rupicapra Linnaeus, 1758). Folia Microbiol. 2012, 57, 355–358. [Google Scholar] [CrossRef]
- Poeta, P.; Costa, D.; Igrejas, G.; Rodrigues, J.; Torres, C. Phenotypic and genotypic characterization of antimicrobial resistance in faecal enterococci from wild boars (Sus scrofa). Vet. Microbiol. 2007, 125, 368–374. [Google Scholar] [CrossRef]
- Figueiredo, N.; Radhouani, H.; Gonçalves, A.; Rodrigues, J.; Carvalho, C.; Igrejas, G.; Poeta, P. Genetic characterization of vancomycin-resistant enterococci isolates from wild rabbits. J. Basic Microbiol. 2009, 49, 491–494. [Google Scholar] [CrossRef]
- Radhouani, H.; Igrejas, G.; Carvalho, C.; Pinto, L.; Gonçalves, A.; Lopez, M.; Sargo, R.; Cardoso, L.; Martinho, A.; Rego, V.; et al. Clonal Lineages, Antibiotic Resistance and Virulence Factors in Vancomycin-Resistant Enterococci Isolated from Fecal Samples of Red Foxes (Vulpes Vulpes). J. Wildl. Dis. 2011, 47, 769–773. [Google Scholar] [CrossRef]
- Radhouani, H.; Igrejas, G.; Gonçalves, A.; Pacheco, R.; Monteiro, R.; Sargo, R.; Brito, F.; Torres, C.; Poeta, P. Antimicrobial resistance and virulence genes in Escherichia coli and enterococci from red foxes (Vulpes vulpes). Anaerobe 2013, 23, 82–86. [Google Scholar] [CrossRef]
- Gonçalves, A.; Igrejas, G.; Radhouani, H.; Correia, S.; Pacheco, R.; Santos, T.; Monteiro, R.; Guerra, A.; Petrucci-Fonseca, F.; Brito, F.; et al. Antimicrobial resistance in faecal enterococci and Escherichia coli isolates recovered from Iberian wolf. Lett. Appl. Microbiol. 2013, 56, 268–274. [Google Scholar] [CrossRef]
- Navarro-Gonzalez, N.; Casas-Díaz, E.; Porrero, C.M.; Mateos, A.; Domínguez, L.; Lavín, S.; Serrano, E. Food-borne zoonotic pathogens and antimicrobial resistance of indicator bacteria in urban wild boars in Barcelona, Spain. Vet. Microbiol. 2013, 167, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Lozano, C.; González-Barrio, D.; García, J.T.; Ceballos, S.; Olea, P.P.; Ruiz-Fons, F.; Torres, C. Detection of vancomycin-resistant Enterococcus faecalis ST6-vanB2 and E. faecium ST915-vanA in faecal samples of wild Rattus rattus in Spain. Vet. Microbiol. 2015, 177, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Ramos, E.; Cordero, J.; Molina, D.; Poeta, P.; Igrejas, G.; Alonso-Calleja, C.; Capita, R. Antimicrobial resistance and virulence genes in enterococci from wild game meat in Spain. Food Microbiol. 2016, 53, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Sassi, M.; Guerin, F.; Zouari, A.; Beyrouthy, R.; Auzou, M.; Fines-Guyon, M.; Potrel, S.; Dejoies, L.; Collet, A.; Boukthir, S.; et al. Emergence of optrA-mediated linezolid resistance in enterococci from France, 2006–16. J. Antimicrob. Chemother. 2019, 74, 1469–1472. [Google Scholar] [CrossRef]
- Almeida, L.M.; Gaca, A.; Bispo, P.M.; Lebreton, F.; Saavedra, J.T.; Silva, R.A.; Basílio-Júnior, I.D.; Zorzi, F.M.; Filsner, P.H.; Moreno, A.M.; et al. Coexistence of the Oxazolidinone Resistance—Associated Genes cfr and optrA in Enterococcus faecalis From a Healthy Piglet in Brazil. Front. Public Health 2020, 8, 518. [Google Scholar] [CrossRef]
- Guerin, F.; Sassi, M.; Dejoies, L.; Zouari, A.; Schutz, S.; Potrel, S.; Auzou, M.; Collet, A.; Lecointe, D.; Auger, G.; et al. Molecular and functional analysis of the novel cfr(D) linezolid resistance gene identified in Enterococcus faecium. J. Antimicrob. Chemother. 2020, 75, 1699–1703. [Google Scholar] [CrossRef]
- Pang, S.; Boan, P.; Lee, T.; Gangatharan, S.; Tan, S.J.; Daley, D.; Lee, Y.T.; Coombs, G.W. Linezolid-resistant ST872 Enteroccocus faecium harbouring optrA and cfr (D) oxazolidinone resistance genes. Int. J. Antimicrob. Agents 2019, 55, 105831. [Google Scholar] [CrossRef]
- Hughes, D.; I Andersson, D. Environmental and genetic modulation of the phenotypic expression of antibiotic resistance. FEMS Microbiol. Rev. 2017, 41, 374–391. [Google Scholar] [CrossRef]
- Ben Yahia, H.; Chairat, S.; Hamdi, N.; Gharsa, H.; Ben Sallem, R.; Ceballos, S.; Torres, C.; Ben Slama, K. Antimicrobial resistance and genetic lineages of faecal enterococci of wild birds: Emergence of vanA and vanB2 harbouring Enterococcus faecalis. Int. J. Antimicrob. Agents 2018, 52, 936–941. [Google Scholar] [CrossRef]
- Nowakiewicz, A.; Zięba, P.; Gnat, S.; Trościańczyk, A.; Osińska, M.; Łagowski, D.; Kosior-Korzecka, U.; Puzio, I. A significant number of multi-drug resistant Enterococcus faecalis in wildlife animals; long-term consequences and new or known reservoirs of resistance? Sci. Total Environ. 2019, 705, 135830. [Google Scholar] [CrossRef] [PubMed]
- Drobni, M.; Bonnedahl, J.; Hernandez, J.; Haemig, P.; Olsen, B. Vancomycin-Resistant Enterococci, Point Barrow, Alaska, USA. Emerg. Infect. Dis. 2009, 15, 838–839. [Google Scholar] [CrossRef]
- Wayne, P.A. Clinical and Laboratory Standards Institute: Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI Supplement M100. Available online: https://clsi.org/media/3481/m100ed30_sample.pdf (accessed on 9 February 2022).
- Nomura, T.; Hashimoto, Y.; Kurushima, J.; Hirakawa, H.; Tanimoto, K.; Zheng, B.; Ruan, G.; Xue, F.; Liu, J.; Hisatsune, J.; et al. New colony multiplex PCR assays for the detection and discrimination of vancomycin-resistant enterococcal species. J. Microbiol. Methods 2018, 145, 69–72. [Google Scholar] [CrossRef]
- Bender, J.K.; Fleige, C.; Klare, I.; Werner, G. Development of a multiplex-PCR to simultaneously detect acquired linezolid resistance genes cfr, optrA and poxtA in enterococci of clinical origin. J. Microbiol. Methods 2019, 160, 101–103. [Google Scholar] [CrossRef]
- Lee, S.-M.; Huh, H.J.; Song, D.J.; Shim, H.J.; Park, K.S.; Kang, C.-I.; Ki, C.-S.; Lee, N.Y. Resistance mechanisms of linezolid-nonsusceptible enterococci in Korea: Low rate of 23S rRNA mutations in Enterococcus faecium. J. Med. Microbiol. 2017, 66, 1730–1735. [Google Scholar] [CrossRef] [PubMed]
- Shaw, T.D.; Fairley, D.J.; Schneiders, T.; Pathiraja, M.; Hill, R.L.R.; Werner, G.; Elborn, J.S.; McMullan, R. The use of high-throughput sequencing to investigate an outbreak of glycopeptide-resistant Enterococcus faecium with a novel quinupristin-dalfopristin resistance mechanism. Eur. J. Clin. Microbiol. 2018, 37, 959–967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martín-Platero, A.M.; Valdivia, E.; Maqueda, M.; Martínez-Bueno, M. Characterization and safety evaluation of enterococci isolated from Spanish goats’ milk cheeses. Int. J. Food Microbiol. 2009, 132, 24–32. [Google Scholar] [CrossRef]
| Groups | Strain | Animal Source | MIC (μg/mL) * | Genes | |||
|---|---|---|---|---|---|---|---|
| QD | VAN | LNZ | Resistance | Virulence | |||
| Sympatric populations | E. faecalis | Apennine Chamois | 4 | 1 | 2 | gelE, efa, asa1 | |
| E. faecalis | Apennine Chamois | 4 | 1 | 2 | gelE, efa, asa1 | ||
| E. faecalis | Goats | 4 | 1 | 2 | vanC1/C2, msrC | gelE, efa, asa1 | |
| E. hirae | Apennine Chamois | 1 | 4 | 2 | gelE | ||
| E. hirae | Goats | 1 | 0.5 | 2 | |||
| E. faecium | Apennine Chamois | 16 | 32 | 8 | msrC, cfrD | gelE, efa | |
| E. casselliflavus | Goats | 1 | 4 | 2 | vanC1/C2 | ||
| E. faecalis | Red deer | 4 | 1 | 2 | gelE | ||
| E. faecalis | Red deer | 4 | 1 | 2 | gelE | ||
| E. faecium | Red deer | 4 | 1 | 2 | msrC | gelE | |
| E. faecium | Red deer | 0.5 | 0.5 | 2 | |||
| E. faecium | Sheep | 1 | 0.5 | 2 | msrC | ||
| E. gallinarum | Red deer | 16 | 32 | 8 | vanC1, msrC, cfrD | gelE | |
| E. gallinarum | Sheep | 16 | 32 | 8 | vanC1/C2, cfrD | ||
| E. gallinarum | Sheep | 16 | 32 | 8 | vanC1, msrC, cfrD | gelE, esp, efa | |
| E. hirae | Sheep | 1 | 0.5 | 2 | |||
| E. casselliflavus | Sheep | 1 | 4 | 1 | |||
| Non-sympatric populations | E. gallinarum | Cattle | 1 | 2 | 0.5 | ||
| E. casseliflavus | Cattle | 1 | 1 | 2 | vanC2 | ||
| E. faecium | Cattle | 1 | 2 | 1 | msrC | ||
| E. faecium | Cattle | 1 | 2 | 0.5 | msrC | ||
| E. faecium | Cattle | 1 | 2 | 0.5 | msrC | ||
| E. faecium | Cattle | 1 | 2 | 0.5 | msrC | ||
| E. faecium | Cattle | 1 | 2 | 0.5 | msrC | ||
| E. gallinarum | Goats | 1 | 2 | 0.5 | |||
| E. gallinarum | Goats | 16 | 8 | 32 | msrC, vanC1/C2, cfrD | ||
| E. gallinarum | Goats | 16 | 8 | 32 | msrC, cfrD | ||
| E. gallinarum | Goats | 0.5 | 2 | 0.5 | msrC | ||
| E. gallinarum | Goats | 1 | 2 | 0.5 | |||
| E. gallinarum | Goats | 1 | 2 | 0.5 | vanC1/C2, msrC | asa1 | |
| E. gallinarum | Goats | 1 | 2 | 0.5 | msrC | ||
| E. gallinarum | Goats | 0.5 | 0.5 | 2 | msrC | ||
| E. gallinarum | Goats | 4 | 1 | 2 | msrC | esp, efa, asa1 | |
| E. gallinarum | Goats | 16 | 32 | 8 | vanC2, cfrD | ||
| E. gallinarum | Goats | 1 | 2 | 0.5 | |||
| E. faecium | Goats | 0.5 | 2 | 0.5 | msrC | ||
| E. faecium | Goats | 0.5 | 2 | 4 | msrC | ||
| E. faecium | Goats | 0.5 | 2 | 0.5 | |||
| E. hirae | Goats | 1 | 2 | 0.5 | asa1 | ||
| E. hirae | Goats | 1 | 2 | 0.5 | asa1 | ||
| E. faecalis | Apennine Chamois | 2 | 2 | 1 | gelE, asa1 | ||
| E. faecalis | Apennine Chamois | 2 | 2 | 1 | gelE, efa, ace | ||
| E. faecalis | Apennine Chamois | 4 | 2 | 1 | gelE, efa, ace | ||
| E. faecalis | Apennine Chamois | 4 | 2 | 1 | gelE, asa1 | ||
| E. faecalis | Apennine Chamois | 4 | 2 | 1 | gelE, efa, ace, asa1 | ||
| E. faecalis | Apennine Chamois | 4 | 2 | 1 | gelE, efa, asa1 | ||
| E. casseliflavus | Apennine Chamois | 1 | 2 | 4 | vanC2 | gelE, efa, ace, asa1 | |
| E. hirae | Apennine Chamois | 1 | 2 | 0.5 | |||
| Groups | Animals | N. Fecal Pools | N. Colonies |
|---|---|---|---|
| Sympatric populations | Apennine chamois (8) | 2 | 4 |
| Goat (12) | 3 | 3 | |
| Red deer (16) | 4 | 5 | |
| Sheep (12) | 3 | 5 | |
| Non-sympatric populations | Cattle (20) | 5 | 7 |
| Goat (32) | 8 | 16 | |
| Red deer (12) | 3 | 0 | |
| Apennine chamois (20) | 5 | 8 | |
| Total | 33 | 48 |
| Primer | Sequence 5’- 3’ | Size (bp) | References |
|---|---|---|---|
| VanD_F1 | TGGAATCACAAAATCCGGCG | 311 | [34] |
| VanD_R2 | TWCCCGCATTTTTCACAACS | ||
| VanM_F1 | GGCAGAGATTGCCAACAACA | 425 | |
| VanM _R1 | AGGTAAACGAATCTGCCGCT | ||
| VanC2_F1 | GCAAACGTTGGTACCTGATG | 523 | |
| VanC2_R4 | GGTGATTTTGGCGCTGATCA | ||
| VanB_F1 | GATGTGTCGGTAAAATCCGC | 640 | |
| VanB_R1 | CCACTTCGCCGACAATCAAA | ||
| VanA_F1 | GCAAGTCAGGTGAAGATGGA | 721 | |
| VanA_R1 | GCTAATACGATCAAGCGGTC | ||
| VanC1_5 | GTATCAAGGAAACCTCGCGA | 836 | |
| VanC1_6 | CGTAGGATAACCCGACTTCC | ||
| VanN_F1 | CCTCAAATCAGCAGCTAGTG | 941 | |
| VanN_R1 | GCTCCTGATAAGTGATACCC | ||
| Cfr_fw | TGAAGTATAAAGCAGGTTGGGAGTCAAC | 746 | [35] |
| Cfr_rev | CATATAATTGACCACAAGCAGC | ||
| optrA_fw | TACTTGATGAACCTACTAACCA | 422 | |
| optrA_rev | CCTTGAACTACTGATTCTCGG | ||
| poxtAfw | AAAGCTACCCATAAAATATC | 533 | |
| poxtArev | TCATCAAGCTGTTCGAGTTC | ||
| cfr(B) fw | TGAGCATATACGAGTAACCTCAAGA | 293 | [36] |
| cfr(B) rev | CGCAAGCAGCGTCTATATCA | ||
| cfr(D) fw | AGAAGTCGCAACAAGTGAGGA | 595 | [11] |
| cfr(D) rev | GCAACTGCATGAGTCAAAGAA | ||
| Vat D F | TCCAGCTAACATGTATGGCG | 271 | [37] |
| Vat D R | GCTCAATAGGACCAGGTGTA | ||
| vgaA F | AGTGGTGGTGAAGTAACACG | 659 | |
| vgaA R | CTTGTCTCCTCCGCGAATAC | ||
| vgaB F | TGACAATATGAGTGGTGGTG | 576 | |
| vgaB R | GCGACCATGAAATTGCTCTC | ||
| vgbB F | CAGCAGTCTAGATCAGAGTGG | 728 | [37] |
| vgbB R | CATACGGATCCATCTTTTCC | ||
| msrC F | AAGGAATCCTTCTCTCTCCG | 343 | |
| msrC R | GTAAACAAAATCGTTCCCG | ||
| vgbA F | TACAGAGTACCCACTACCGA | 569 | |
| vgbA R | TCAATTCCTGCTCCAGCAGT | ||
| ermB F | CATTTAACGACGAAACTGGC | 424 | |
| ermB R | GGAACATCTGTGGTATGGCG | ||
| vatE F | ACTATACCTGACGCAAATGC | 511 | [37] |
| vatE R | GGTTCAAATCTTGGTCCG | ||
| gelE F | TATGACAATGCTTTTTGGGAT | 213 | [38] |
| gelE R | AGATGCACCCGAAATAATATA | ||
| esp F | AGATTTCATCTTTGATTCTTGG | 510 | |
| esp R | AATTGATTCTTTAGCATCTGG | ||
| ace F | GAATTGAGCAAAAGTTCAATCG | 1008 | |
| ace R | GTCTGTCTTTTCACTTGTTTC | ||
| efa F | GCCAATTGGGACAGACCCTC | 688 | |
| efa R | CGCCTTCTGTTCCTTCTTTGGC | ||
| asa1 F | GCACGCTATTACGAACTATGA | 375 | [38] |
| asa1 R | TAAGAAAGAACATCACCACGA | ||
| hyl F | ACAGAAGAGCTGCAGGAAATG | 276 | |
| hyl R | GACTGACGTCCAAGTTTCCAA | ||
| cylA F | ACTCGGGGATTGATAGGC | 688 | |
| cylA R | GCTGCTAAAGCTGCGCTT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smoglica, C.; Vergara, A.; Angelucci, S.; Festino, A.R.; Antonucci, A.; Marsilio, F.; Di Francesco, C.E. Evidence of Linezolid Resistance and Virulence Factors in Enterococcus spp. Isolates from Wild and Domestic Ruminants, Italy. Antibiotics 2022, 11, 223. https://doi.org/10.3390/antibiotics11020223
Smoglica C, Vergara A, Angelucci S, Festino AR, Antonucci A, Marsilio F, Di Francesco CE. Evidence of Linezolid Resistance and Virulence Factors in Enterococcus spp. Isolates from Wild and Domestic Ruminants, Italy. Antibiotics. 2022; 11(2):223. https://doi.org/10.3390/antibiotics11020223
Chicago/Turabian StyleSmoglica, Camilla, Alberto Vergara, Simone Angelucci, Anna Rita Festino, Antonio Antonucci, Fulvio Marsilio, and Cristina Esmeralda Di Francesco. 2022. "Evidence of Linezolid Resistance and Virulence Factors in Enterococcus spp. Isolates from Wild and Domestic Ruminants, Italy" Antibiotics 11, no. 2: 223. https://doi.org/10.3390/antibiotics11020223
APA StyleSmoglica, C., Vergara, A., Angelucci, S., Festino, A. R., Antonucci, A., Marsilio, F., & Di Francesco, C. E. (2022). Evidence of Linezolid Resistance and Virulence Factors in Enterococcus spp. Isolates from Wild and Domestic Ruminants, Italy. Antibiotics, 11(2), 223. https://doi.org/10.3390/antibiotics11020223







