Abstract
Fungal natural products play a prominent role in the development of pharmaceuticalagents. Two new cyclic tetrapeptides (CTPs), westertide A (1) and B (2), with eight known compounds (3–10) were isolated from the fungus Aspergillus westerdijkiae guided by OSMAC (one strain-many compounds) and molecular networking strategies. The structures of new compounds were unambiguously determined by a combination of NMR and mass data analysis, and chemical methods. All of the isolates were evaluated for antimicrobial effects, synergistic antifungal activity, cytotoxic activity, and HDAC inhibitory activity. Compounds 1–2 showed synergistic antifungal activity against Candida albicans SC5314 with the presence of rapamycin and weak HDAC (histone deacetylase) inhibitory activity. These results indicate that molecular networking is an efficient approach for dereplication and identification of new CTPs. CTPs might be a good starting point for the development of synergistic antifungal agents.
1. Introduction
Fungal natural products play a prominent role in the development of pharmaceutical agents [1,2]. Cyclic tetrapeptides (CTPs) are a type of important bioactive natural product that were found to have a broad range of pharmacological properties, including antimicrobial [3,4], cytotoxic [5,6,7], and HDAC (histone deacetylase) inhibitory properties [8]. Most of the naturally occurring CTPs are obtained from fungi, such as HC toxin with cytotoxic and antimitogenic activities from Cochliobolus carbonum [9], apicidin with antiprotozoan activities from Fusarium strains [10], and microsporins A-B with antitumor activity from Microsporum cf. gypseum [11]. In recent years, some naturally occurring CTPs have been found to inhibit HDAC and regulate gene expression, which are very useful as cancer therapeutics. In addition to use as antineoplastic drugs, HDAC inhibitors (HDACis) also have anti-interstitial fibrosis [12], anti-inflammatory [13], immunomodulatory [14], and metabolic regulation activities [15].
Naturally occurring CTPs are usually produced in low yields, which limits the discovery of new CTPs. MS/MS-based molecular networking paves the way to solving this problem. As a promising strategy, molecular networking can provide guidance and improve efficiency for the discovery of new bioactive analogues with a specific skeleton from complex mixtures. In the field of bioactive peptides discovery, neoantimycin L with excellent cytotoxicity from Streptomyces conglobatus RJ8 [16] and thermoactinoamide A with moderate antiproliferative activity from Thermoactinomyces vulgaris DSM 43016 [17] were obtained based on molecular networking.
Aspergillus westerdijkiae is an important ochratoxin A (OTA)-producing fungus, whose genome harbors 17 non-ribosomal peptide synthetase (NRPS) genes [18]. However, most NRPS genes are unexpressed under standard laboratory conditions. In this study, we used the one strain-many compounds (OSMAC) method to activate silenced genes and MS/MS-based molecular networking to search for novel and bioactive peptides from A. westerdijkiae L1295. As a result, two new cyclic tetrapeptides, westertides A (1) and B (2), and eight known compounds (3–10) were obtained (Figure 1). This work describes the details of the isolation, structure elucidation, and biological activities of secondary metabolites from A. westerdijkiae L1295.
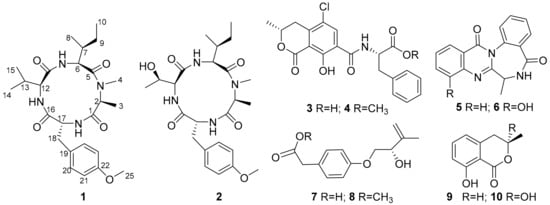
Figure 1.
Chemical structures of 1–10.
2. Results
In this study, a molecular networking-OSMAC strategy was applied to accelerate the discovery of cyclic tetrapeptides. First, the fungus A. westerdijkiae L1295 was fermented in different culture media and conditions using the OSMAC method (Table S1). Then, the ethyl acetate extracts were further investigated by UPLC-HRMS/MS. The LC-MS/MS data were used to generate a visualized molecular networking that was further annotated by Cytoscape 3.8.2. From the full molecular network, several independent families of molecules were obviously visualized in the crude extracts of A. westerdijkiae L1295 fermented on rice, which were different from the other crude extracts (Figure 2 and Figure S1). Further analysis of the molecular network found that a cluster with 19 nodes represented a peptide family, showing MS/MS patterns containing the dipeptide [Ala-Phe] fragment (m/z 219.1), which has been widely found in the peptide family [19,20,21] (Figure 2 and Figure S2). Guided by MS/MS and molecular networking, two new cyclic tetrapeptides, westertides A (1) and B (2), with eight known compounds ochratoxin A (3) [22], ochratoxin A methyl ester (4) [23], circumdatin F (5) [24], circumdatin G (6) [25], stachyline B (7) [26], westerdijkin A (8) [27], mellein (9) [28], and 3-hydroxymellein (10) [25] were obtained from the solid culture on rice medium and their structure identifications are described below.
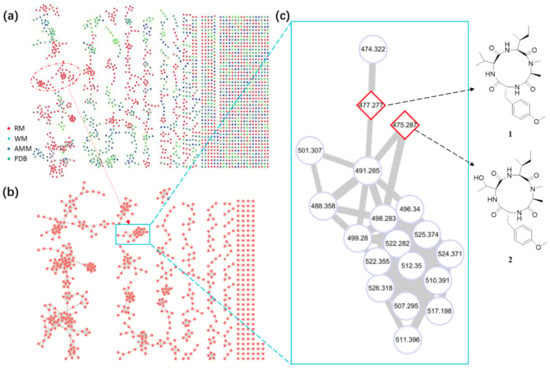
Figure 2.
Metabolic analysis of crude extracts from A. westerdijkiae L1295. (a) Tandem MS/MS-based full molecular networking cluster analysis of different culture extracts of A. westerdijkiae L1295. (RM: Rice medium; WM: wheat medium; AMM: Aspergillus Minimal Medium; PDB: Potato-Dextrose Broth) of the fungus. (b) Molecular networking of A. westerdijkiae L1295 fermented on rice. (c) The specific subnetwork predicted to contain CTPs in the MS/MS-based molecular networking. The full GNPS network and subnetwork are presented in Figures S1 and S3 in the Supplementary Materials.
Compound 1 was isolated as a pale amorphous solid, which possessed a molecular formula of C25H38N4O5 (9 degrees of unsaturation) on the basis of HRESIMS and NMR data (Table 1). The 1H, 13C NMR, and HSQC spectra of 1 revealed the presence of 7 methyl groups including 1 N-methyl [δH/δC 3.32 (3H, s)/30.9] and 1 O-methyl [δH/δC 3.78 (3H, s)/55.8], 2 methylene groups [δH/δC 1.32 (1H, m), 1.82 (1H, m)/25.8; 3.66 (1H, m), 3.91 (1H, m)/35.6], 1 para-disubstituted benzene [δH/δC 7.05 (2H, d, J = 6.9 Hz)/114.8; 7.29 (2H, d, J = 6.9 Hz)/132.0; δC 132.8 and 159.5], 6 methines including 4 characteristic α-methine signals [δH/δC 4.29 (1H, d, J =5.8 Hz)/65.3; 4.41 (1H, m)/55.8; 4.69 (1H, m)/54.7; 5.17 (1H, dd, J =7.0, 10.0 Hz)/55.3], 3 amide N-H protons (δH 7.38, 8.83, and 9.88), and 4 amide carbonyls (δC 171.2, 173.0, 174.0, and 174.1), suggesting that 1 comprised 4 amino acid residues. HMBC correlations from H3-3 (δH 1.43) to C-2 (δC 54.7) and C-1 (δC 174.0) and from H3-4 (δH 3.32) to C-2 and C-5 (δC 171.2) together with the 1H-1H COSY correlations of H-2-H3-3 led to the identification of the N-Me-Ala residue. The 1H-1H COSY correlations of H3-10-H2-9-H-7-H-6 and H3-8-H-7 together with the HMBC correlations were detected from H-6 (δH 5.17) to C-5 (δC 171.2), C-7 (δC 37.7), C-8 (δC 17.6), C-9 (δC 25.8), and C-11 (δC 173.0); from H-7 (δH 2.34) to C-6 (δC 55.3); from H3-8 (δH 1.14) to C-6, C-7, and C-9; and from H3-10 (δH 0.98) to C-7 and C-9, which confirmed the presence of the Ile moiety. Similarly, two other amino acid units Val and O-Me-Tyr were completely assigned.

Table 1.
1H (500 MHz) and 13C (125 MHz) NMR data of compound 1 in Pyridine-d5.
The amino acid sequence of 1 was deduced from the observed key HMBC correlations, NOESY correlations, and MS data. The HMBC correlations from N-CH3 (δH 3.32) of Ala to the Ile carbonyl group C-5 (δC 171.2), from H-6 (δH 5.17) of Ile to the Val carbonyl group C-11 (δC 173.0), and from H-12 (δH 4.31) of Val to the O-Me-Tyr carbonyl group C-16 (δC 174.1) suggested a partial sequence of N-Me-Ala-Ile-Val-O-Me-Tyr (Figure 3). The H3-4 (δH 1.43) of N-Me-Ala showed an NOESY correlation with H-20/24 (δH 7.29), indicating that 1 was a cyclic peptide, and this conclusion was also confirmed by the 9 degrees of unsaturation and the molecular formula. Additionally, the ESI-MS/MS experimental results (Figure 4 and Figure S4) also confirmed the connections of these residues as cyclo-[N-Me-Ala-Ile-Val-O-Me-Tyr].
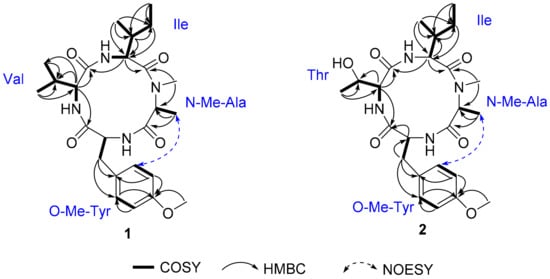
Figure 3.
Key 1H-1H COSY, HMBC, and NOESY correlations of 1 and 2.
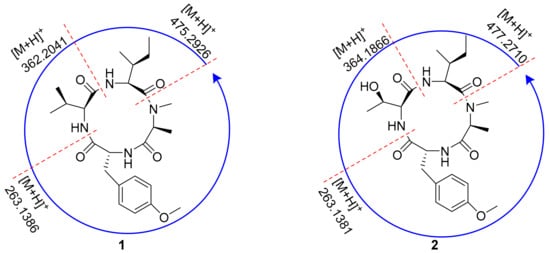
Figure 4.
ESI-MS/MS analysis of 1 and 2.
The absolute configuration of the amino acids from compound 1 was established by the advanced Marfey’s method [29]. The mixture obtained after hydrolyzing compound 1 and further derivatization with l-FDAA was analyzed by HPLC-DAD. HPLC analyses of the mixture of hydrolysates and appropriate amino acid standards confirmed the d configurations for O-Me-Tyr and the l configurations for Tyr, N-Me-Ala, and Ile in 1 (Figure 5). Consequently, the structure of 1 was elucidated as cyclo-[l-N-Me-Ala-l-Ile-l-Val-d-O-Me-Tyr] and named westertide A.
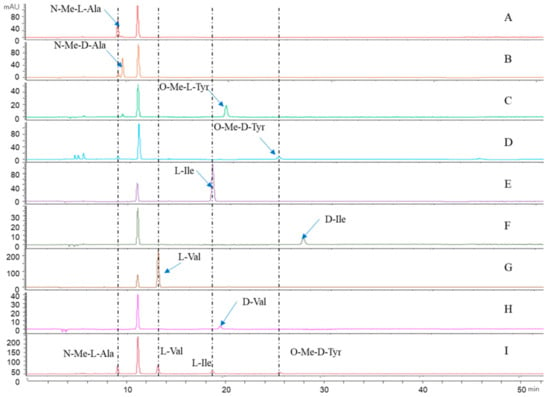
Figure 5.
Advanced Marfey’s analysis of compound 1. (A–H): The retention times for the FDAA derivatives of N-Me-l-Ala, N-Me-d-Ala, O-Me-l-Tyr, O-Me-d-Tyr, l-Ile, d-Ile, l-Val, and d-Val, respectively. (I): The FDAA derivatives of the hydrolysate of 1. The derivatives of the acid hydrolysate and the standard amino acids were subjected to RP HPLC analysis (Kromasil C18 column; 5 μm, 4.6 mm× 250 mm; 1.0 mL/min; UV detection at 340 nm) with a linear gradient of acetonitrile (35–45%) in water (TFA, 0.01%) over 40 min.
Compound 2 was isolated as a white amorphous powder. It was assigned a molecular formula of C24H36N4O6 (9 degrees of unsaturation) based on its HRESIMS and NMR data (Table 2). The 1D NMR spectroscopic data showed that compound 2 was a cyclic tetrapeptide similar to 1 but bearing a threonine (Thr) residue with signals at δH/δC 1.38 (3H, d, J = 6.9 Hz)/30.9 (CH3), δH/δC 4.42 (1H, overlapped)/65.4 (CH), δH/δC 4.81 (1H, m)/68.1 (CH), and δC 173.5 (C), instead of the valine residue. A comprehensive analysis of its relevant 1H-1H COSY, HMQC, HMBC, and NOESY correlations (Figure 3), and the ESI-MS/MS experimental results (Figure 4 and Figure S5) confirmed that 2 has the same planar structure as that of violaceomide A [20]. However, the optical rotation data of 2 ([α = +249.5, c = 1.0, MeOH) were opposite to that of violaceomide A ([α = −230.0, c = 0.6, MeOH), implying that they are optical isomers. The HPLC analysis of the acid hydrolysate of 2 after derivatization with l-FDAA revealed that l-N-Me-Ala, l-Ile, l-Thr, and d-O-Me-Tyr were present in 2 (Figure S6). This result shows that the main difference between compound 2 and violaceomide A is the substitution of l-O-Me-Tyr with d-O-Me-Tyr. Thus, compound 2 was assigned as cyclo-[l-N-Me-Ala-l-Ile-l-Thr-d-O-Me-Tyr] and named westertide B.

Table 2.
1H (500 MHz) and 13C (125 MHz) NMR data of compound 2 in Pyridine-d5.
Compounds 1–10 showed no significant bioactivity in the antibacterial, antifungal, and cytotoxicity assays at the dose of 100 μM. In our previous work, we found that peptide-like compounds showed a synergistic antifungal effect with rapamycin [30]. So, we tested whether the new cyclic tetrapeptide compounds could also cause synergistic antifungal activity with rapamycin against Candida albicans SC5314. When checkerboard assays were used to obtain the MICs (minimum inhibitory concentrations) with rapamycin for achieving 90% growth inhibition, only 0.008μM of rapamycin was required together with a very low amount (6.25 μM) of compounds 1 and 2. Based on the fractional inhibitory concentration index (FICI), westertides 1 and 2 showed effective synergism with rapamycin, and the FICI was 0.078 for both compounds 1 and 2 (Table 3). Our results showed that compounds 1 and 2 had strong synergistic antifungal activity with rapamycin. Furthermore, the effects of compounds 1 and 2 on histone deacetylation (HDAC) at the cell level were also evaluated, and compound 1 showed weak HDAC inhibitory activity, with IC50 of about 70 μM.

Table 3.
MIC values of compounds 1–2 with rapamycin against C. abicans SC5314.
3. Discussion
With a low molecular weight, low hydrophobicity, and the presence of a hydrogen-bond acceptor and donor, CTPs have been demonstrated to possess diverse pharmacological activities, including antimicrobial [4], cytotoxic [5,6,7], and HDAC inhibitory bioactivities. In the last decade or so, more than 40 cyclic peptides have been approved by the FDA and EMA, such as vorinostat and romidepsin [31,32,33].
However, it is relatively difficult to discover CTPs due to their narrow distribution and low yield. As the main natural sources of CTPs, fungi have an abundance of NRPS biosynthetic gene clusters, whereas some of these genes are not expressed under normal experimental conditions. These silent gene clusters outnumber the constitutively expressed clusters by a factor of 5–10 [34]. Hence, strategies that rationally activate silent gene clusters will dramatically enhance our reservoir of potentially therapeutic small molecules [35]. In order to efficiently discover novel cyclic peptides, the molecular network and OSMAC strategy are used in combination with gene mining techniques [36]. Molecular networking can efficiently dereplicate known natural products, thus aiding the discovery of new analogues with a specific skeleton from complex mixtures [37]. The OSMAC strategy can activate some silent genes of target strains to produce more secondary metabolites and obtain novel secondary metabolites [38]. Genome mining is a powerful approach to direct the production of novel and interesting CTPs, which become relevant in the future to search for unculturable microorganisms as a new source of novel bioactive CTPs [39]. In this work, the discovery of two new cyclopeptides from A. westerdijkiae using the OSMAC strategy and the MS/MS molecular networking further expanded the structural diversity of the CTPs and the source of CTPs producers.
An estimated 1.2 billion people worldwide suffer from a fungal disease, of which 1.5 to 2 million people die of a fungal infection each year, surpassing those killed by either malaria or tuberculosis [40,41,42]. About 30% of serious infections are caused by Candida albicans, with a mortality rate of up to 40% [43]. Unfortunately, resistance to existing classes of drugs is on the rise due to the limited class of antifungal drugs available and the decline in new drug development. As the process of de novo antifungal discovery fails to meet clinical needs, the approach of repurposing approved drugs has drawn much attention.
Rapamycin, also called sirolimus, is characterized primarily by its antifungal activity against several human fungal pathogens, such as Candida albicans [44], Cryptococcus neoformans [45], and Fusarium oxysporum [46], and potent immunosuppressive activity [47]. The dual effects of rapamycin on antifungus and immunosuppression seem to effectively solve the threat of Candida infection when patients are treated with immunosuppressive drugs. However, rapamycin showed weak antifungal activity at the dose used to suppress the immune response in patients. The identification of synergistic actions on rapamycin against fungi can possibly solve this problem. In an early report, Tong et al. showed that some commercial or natural peptide-like compounds synergistically increased the antifungal effect of rapamycin, by targeting the Rbp1 protein (homologue of the FKBP12 protein in mammals) of C. albicans to increase the binding of rapamycin-Rbp1 complex with Tor1C protein [30]. In this work, we found two new natural peptide compounds, westertides A and B, showing strong synergistic antifungal activity with rapamycin from A. westerdijkiae. The mechanism of their synergistic antifungal effect with rapamycin may be similar to the known peptide compounds, but this requires deep investigation because they showed no antifungal effects alone.
4. Materials and Methods
4.1. General
UV data and optical rotation were recorded on a Thermo Genesys-10S UV-Vis spectrophotometer and Anton Paar MCP 200 Automatic Polarimeter, respectively. High-resolution electrospray ionization mass spectrometry (HRESIMS) data were obtained on an Agilent Accurate-Mass-Q-TOF LC/MS 6520 instrument. NMR spectral data were obtained with a Bruker AVANCE-500 spectrometer (δC/δH: Pyridine-d5, 150.4, 135.9, 123.9/8.74, 7.58, 7.22; DMSO, 39.5/2.50). Silica gel (Qingdao Haiyang Chemical Co., Ltd., Qingdao, China, 200–300 mesh), Sephadex LH-20 (GE Healthcare, Uppsala, Sweden), and ODS (50 μm, YMC CO., LTD, YMC Pack, Kyoto, Japan) were used for column chromatography. Semi-preparative HPLC was performed on an Agilent 1200 HPLC system equipped with an Agilent DAD UV−vis spectrometric detector, using a reversed-phase column (C18, 5 μm 9.4 mm × 250 mm, YMC Pack, Kyoto, Japan) with a flow rate of 2.0 mL/min. Biological reagents, chemicals, and media were purchased from standard commercial sources unless stated.
4.2. Fungal Material
A. westerdijkiae was isolated from the mildewed wheat, China, in September 2017. The fungus was identified mainly based on the morphological observation, molecular multilocus phylogeny analysis, and morphological features [48] (Figure S7). The fungus was deposited in China General Microbiological Culture Collection (CGMCC No. 19033).
4.3. Fermentation and Extraction
A. westerdijkiae was cultured on a slant of PDA at 25 °C for 5 days. To prepare inoculum, the spores of the strain on the plate were collected and adjusted to 1 × 106 CFU/mL. A large-scale fermentation was carried out in 40 × 500 mL Fernbach culture flasks, with each flask containing 80 g of rice and 60 mL of distilled water (each with 0.5 mL of spore suspension), incubated at 25 °C for 3 weeks. The fermented rice substrate was extracted repeatedly with ethyl acetate by exhaustive maceration (3 × 4 L), and the organic solvent was evaporated to dryness under vacuum to afford the crude extract (20.1 g).
4.4. LC-MS/MS and Molecular Networking Analysis
LC-MS-MS was performed on an Agilent series 1290 Infinity HPLC instrument, coupled with a Q-TOF Mass spectrometer (Agilent Technologies Inc., Santa Clara, CA, USA), with a YMC C18 column [(YMC Co., Ltd. Kyoto, Japan) YMC-Park, ODS-A, 250 mm × 2.1 mm, S-5 μm, 12 nm, 0.5 mL/min]. The total extracts (0.5 mg/mL, 10 μL) were analyzed by LC-MS with a gradient program of MeCN−H2O (0.01% TFA) [0–25 min 5–80%, 25–32 min 80–100%, 32–38 min 100%; 0.5 mL/min; MS scan 150–2000 Da] and then with an automated full-dependent MS-MS scan. Mass spectral networks were assembled as described in the reference. Differentiation of the protonated molecules, adducts, and fragment ions was done by identification of the [M+H]+ ion. The All MS/MS data files were converted to “.mzML” format files using MSConver software and uploaded on the GNPS Web platform (http://gnps.ucsd.edu, accessed on 6 August 2021) for MN analysis using Classic mode. For the network creation, a parent mass tolerance of 0.02 Da and a fragment ion tolerance of 0.05 Da were applied. The generated molecular network was visualized in Cytoscape 3.8.2 (www.cytoscape.org, accessed on 6 August 2021) and guided the isolation of 1–8. The MS/MS molecular network can be browsed and downloaded from the GNPS Web site with the following links: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=6794bab0d59245bf875b14c6ebb84ff4 and https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=84b42a96c887412db918a18f20491b8b (accessed on 6 August 2021).
4.5. Isolation and Characterization Data
The EtOAc fraction was subjected to silica gel column chromatography (CC) using petroleum/ethyl acetate (P/E) in a gradient elution (v/v, 100:0, 100:1, 100:2, 100:4, 100:10) and dichloromethane/acetone (v/v, 100:0, 100:2, 100:4, 100:8, 100:12, 100:20, 0:100) to give 16 fractions (AW.1–AW.16).
Fraction AW.6 (0.85 g) was further separated on a silica gel column by elution with increasing concentrations of ethyl acetate in petroleum to give 15 fractions (AW.6-1–AW.6-15). Compound 9 (4.5 mg) was obtained from subfractions AW.6-8 (45 mg) by sephadex LH-20 chromatography eluted with dichloromethane/methanol (v/v, 1:1). AW.6-10 (75 mg) was purified finally by RP-HPLC with acetonitrile/water (50:50) to give 10 (13.5 mg, tR 22.3 min).
Fraction AW.13 (4.3 g) eluted with CH2Cl2-Acetone (v/v 20:1) was first separated by ODS using a gradient of increasing methanol (30%, 45%, 60%, 75%, and 100%) in water to afford 25 subfractions (AW.13-1–AW.13-25). Compound 6 (30.5 mg, tR 15.1 min) was obtained from AW.13-9 (152 mg) by RP-HPLC using 21% acetonitrile in acidic water (0.005% TFA). Subfractions AW.13-11 (170 mg) were followed by RP-HPLC using 32% acetonitrile in water to afford a mixture of 7 (9.1 mg, tR 22.1 min), 5 (5.6 mg, tR 31.1 min), and 8 (6.5 mg, tR 33.5 min). Compounds 1 (2.0 mg, tR 40.5 min) and 2 (8.0 mg, tR 32.5 min) were obtained from AW.13-15 (55 mg) by RP-HPLC using 45% acetonitrile in acidic water (0.005% TFA). Compound 3 (325.0 mg) was obtained from AW.13-21 by recrystallization in acetonitrile. Compound 4 (20.0 mg) was obtained from subfractions and AW.13-22 by sephadex LH-20 chromatography eluted with methanol, respectively.
Westertide A (1): pale amorphous solid; [α +235.57 (c 0.5, MeOH); UV (MeOH) λmax (log ε) 222 (4.03), 275 (1.30); Positive HRESIMS: m/z 475.2926 [M+H]+ (calcd for C25H38N4O5, 475. 2920). NMR data, see Table 1 and Figures S8–S13.
Westertide B (2): pale amorphous solid, [α +249.48 (c 1.0, MeOH); UV (MeOH) λmax (log ε) 220 (2.78), 275 (1.43) nm; Positive HRESIMS: m/z m/z 477.2710 [M+H]+ (calcd.for C24H37N4O6, 477.2713). NMR data, see Table 2 and Figures S14–S19.
4.6. Absolute Configuration of Amino Acids
Compound (1.0 mg) was dissolved in 6 N HCl (2.0 mL) and heated at 110 °C for 24 h. The solutions were then evaporated to dryness and placed in a 4 mL reaction vial and treated with a 1 g/100 mL solution of FDAA (200 μL) in acetone, followed by 1.0 M NaHCO3 (40 μL). The reaction mixtures were heated at 45 °C for 90 min, and the reactions were quenched by the addition of HCl (1 M, 40 µL). In a similar fashion, standard N-Me-l-Ala, N-Me-d-Ala, O-Me-l-Tyr, O-Me-d-Tyr, l-Ile, d-Ile, l-Val, d-Val, l-Thr, and d-Thr, were derivatized separately. The derivatives of the acid hydrolysate and the standard amino acids were subjected to RP HPLC analysis (Kromasil C18 column; 5 μm, 4.6 mm × 250 mm; 1.0 mL/min; UV detection at 340 nm) with a linear gradient of acetonitrile (35–45%) in water (TFA, 0.01%) over 40 min. The retention times for the FDAA derivatives of N-Me-l-Ala, N-Me-d-Ala, O-Me-l-Tyr, O-Me-d-Tyr, l-Ile, d-Ile, l-Val, d-Val, l-Thr, and d-Thr were 9.1, 9.4, 19.0, 25.0, 18.1, 25.8, 12.9, 18.4, 5.2, and 6.1 min, respectively, whereas those for the FDAA derivatives of N-Me-Ala, O-Me-Tyr, Ile, and Val in the hydrolysate of 1 were 9.1 (N-Me-l-Ala), 25.0 (O-Me-d-Tyr), 18.1 (l-Ile), and 12.9 (l-Val) min, and N-Me-Ala, O-Me-Tyr, Ile, and Thr in the hydrolysate of 2 were 9.1 (N-Me-l-Ala), 25.0 (O-Me-d-Tyr), 18.1 (l-Ile), and 5.2 (l-Thr) min, respectively.
4.7. Evaluation of Biological Activities
4.7.1. Antifungal and Synergistic Antifungal Assay
Candida albicans SC5314 was used as a test strain for the antifungal and synergistic antifungal bioassay. Checkerboard assays were carried out as described previously [29,30]. Overnight cultures were chosen to prepare the strain suspension with RPMI 1640 medium. RPMI 1640 was purchased from Invitrogen, and was used according to the manufacturer’s protocol, by supplementing 2% glucose, 3.45% MOS, then adjusting the pH to 7.0. Compounds were dissolved in DMSO. Final concentrations ranged from 0.002 to 2 μg/mL for rapamycin and 0.39 to 25 μg/mL for peptide-like compounds, respectively. Rapamycin was 2-fold diluted from 1 to 11 (column), while selected compounds were 2-fold diluted from A to G (row) of the 96-well microtiter plate. The fractional inhibitory concentration index (FICI) is defined as the sum of the MIC of each drug when used in combination divided by the MIC of the drug used alone. Synergism and antagonism were defined by FICI of ≤0.5 and >4, respectively.
4.7.2. Cytotoxicity Assay
Cytotoxicity tests against A549, HepG2, and K562 cell lines were carried out as previously described [49]. Taxol, 5-Flourouracil, and Cisplatin were used as the positive control.
4.7.3. HDAC Activity Assay
The HDAC activity of the compounds was measured using an HDAC8 Deacetylase Fluorometric (Human) Assay Kit (Cat KA4444, Abnova, Taipei, Taiwan) according to the manufacturer’s instructions. Fluorescence signal was detected with excitation at 360 nm and emission at 460 nm using a fluorescence microplate reader (Perkin-Elmer, Waltham, MA, USA). Experiments were performed in triplicate and data were analyzed using GraphPad Prism (version 6.0), Kd values were calculated by nonlinear curve fitting using a 1-site binding (hyperbola) model (Y = Bmax*X/(Kd + X).
5. Conclusions
Uncovered by OSMAC and molecular networking strategies, 2 new cyclic tetrapeptides (1–2), together with 7 known compounds (3–10) were isolated from A. westerdijkiae. All of the isolates were evaluated for an antifungal effect, synergistic antifungal activity, cytotoxic activity, and HDAC inhibitory activity. As a result, 1–10 showed no significant bioactivity in the antifungal assays and cytotoxicity assays at the dose of 100 μM. However, compounds 1–2 showed strong synergistic antifungal activity against C. albicans with rapamycin. In addtion, compound 1 showed weak HDAC inhibitory activity.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics11020166/s1, Table S1: Culture media with different compositions and conditions for A. westerdijkiae; Figure S1: The molecular network obtained by combining the LC-MS/MS analyses of rice fermentation extract extracts from A. westerdijkiae L1295; Figure S2: Cyclictetrapeptides-cluster and the MS/MS spectrum of each node; Figure S3: The cluster corresponding to compounds observed in the molecular networking; Figure S4: The ESI-MS/MS spectrum of 1; Figure S5: The ESI-MS/MS spectrum of 2; Figure S6: Advanced Marfey’s analysis of compound 2. Figure S7: Phylogenetic analysis and morphological characters of A. westerdijkiae L1295; Figure S8: 1H NMR spectrum of westertide A (1) in pyridine-d5 (500 MHz); Figure S9: 13C NMR spectrum of westertide A (1) in pyridine-d5 (125 MHz; Figure S10: 1H-1H COSY spectrum of westertide A (1) in pyridine-d5; Figure S11: HSQC spectrum of westertide A (1) in pyridine-d5; Figure S12: HMBC spectrum of westertide A (1) in pyridine-d5; Figure S13: ROESY spectrum of westertide A (1) in pyridine-d5; Figure S14: 1H NMR spectrum of westertide B (2) in pyridine-d5 (500 MHz); Figure S15: 13C NMR spectrum of westertide B (2) in pyridine-d5 (125 MHz); Figure S16: 1H-1H COSY spectrum of westertide B (2) in pyridine-d5; Figure S17: HSQC spectrum of westertide B (2) in pyridine-d5; Figure S18: HMBC spectrum of westertide B (2) in pyridine-d5; Figure S19: ROESY spectrum of westertide B (2) in pyridine-d5.
Author Contributions
Conceptualization, J.H. and H.W.; methodology, J.H. and B.C.; validation and data curation, R.Z., Z.L. and H.D.; formal analysis, R.Z., W.W. and J.H.; investigation, J.H. and H.L.; resources, T.W. and J.S.; writing—original draft preparation, H.W., R.Z. and J.H.; writing—review and editing, Z.L. and H.L.; supervision and project administration, J.H. and H.L.; funding acquisition, E.L., F.S., H.L. and J.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Special Project for Key Science and Technology of Food Safety (grant No. 2017YFC1601302), and the National Natural Science Foundation (Grant Nos. 22177131, 82073723 and 81872771).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors thank the National Special Project and the National Natural Science Foundation for funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Al-Fakih, A.A.; Almaqtri, W.Q.A. Overview on antibacterial metabolites from terrestrial Aspergillus spp. Mycology 2019, 10, 191–209. [Google Scholar] [CrossRef]
- Chin, J.M.W.; Puchooa, D.; Bahorun, T.; Jeewon, R. Antimicrobial properties of marine fungi from sponges and brown algae of Mauritius. Mycology 2021, 12, 231–244. [Google Scholar] [CrossRef]
- Fazal, A.; Webb, M.E.; Seipke, R.F. The Desotamide Family of Antibiotics. Antibiotics 2020, 9, 452. [Google Scholar] [CrossRef]
- Chakraborty, S.; Tai, D.F.; Lin, Y.C.; Chiou, T.W. Antitumor and antimicrobial activity of some cyclic tetrapeptides and tripeptides derived from marine bacteria. Mar. Drugs 2015, 13, 3029–3045. [Google Scholar] [CrossRef]
- He, F.; Bao, J.; Zhang, X.Y.; Tu, Z.C.; Shi, Y.M.; Qi, S.H. Asperterrestide A, a cytotoxic cyclic tetrapeptide from the marine-derived fungus Aspergillus terreus SCSGAF0162. J. Nat. Prod. 2013, 76, 1182–1186. [Google Scholar] [CrossRef]
- Sun, Y.; Tian, L.; Huang, Y.F.; Sha, Y.; Pei, Y.H. A new cyclotetrapeptide from marine fungus Trichoderma reesei. Pharmazie 2006, 61, 809–810. [Google Scholar]
- Gao, C.H.; Chen, Y.N.; Pan, L.X.; Lei, F.; Long, B.; Hu, L.Q.; Zhang, R.C.; Ke, K.; Huang, R.M. Two new cyclic tetrapeptides from deep-sea bacterium Bacillus amyloliquefaciens GAS 00152. J. Antibiot. 2014, 67, 541–543. [Google Scholar] [CrossRef]
- Abdalla, M.A. Medicinal significance of naturally occurring cyclotetrapeptides. J. Nat. Med. 2016, 70, 708–720. [Google Scholar] [CrossRef]
- Walton, J.D. HC-toxin. Phytochemistry 2006, 67, 1406–1413. [Google Scholar] [CrossRef]
- von Bargen, K.W.; Niehaus, E.M.; Bergander, K.; Brun, R.; Tudzynski, B.; Humpf, H.U. Structure Elucidation and Antimalarial Activity of Apicidin F: An Apicidin-like Compound Produced by Fusarium fujikuroi. J. Nat. Prod. 2013, 76, 2136–2140. [Google Scholar] [CrossRef]
- Gu, W.; Cueto, M.; Jensen, P.R.; Fenical, W.; Silverman, R.B. Microsporins A and B: New histone deacetylase inhibitors from the marine-derived fungus Microsporum cf. gypseum and the solid-phase synthesis of microsporin A. Tetrahedron 2007, 63, 6535–6541. [Google Scholar] [CrossRef]
- Davies, E.R.; Haitchi, H.M.; Thatcher, T.H.; Sime, P.J.; Kottmann, R.M.; Ganesan, A.; Packham, G.; O’Reilly, K.M.; Davies, D.E. Spiruchostatin A inhibits proliferation and differentiation of fibroblasts from patients with pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2012, 46, 687–694. [Google Scholar] [CrossRef]
- Leoni, F.; Zaliani, A.; Bertolini, G.; Porro, G.; Pagani, P.; Pozzi, P.; Dona, G.; Fossati, G.; Sozzani, S.; Azam, T.; et al. The antitumor histone deacetylase inhibitor suberoylanilide hydroxamic acid exhibits anti-inflammatory properties via suppression of cytokines. Proc. Natl. Acad. Sci. USA 2002, 99, 2995–3000. [Google Scholar] [CrossRef]
- Roger, T.; Lugrin, J.; Le Roy, D.; Goy, G.; Mombelli, M.; Koessler, T.; Ding, X.C.; Chanson, A.-L.; Reymond, M.K.; Miconnet, I.; et al. Histone deacetylase inhibitors impair innate immune responses to Toll-like receptor agonists and to infection. Blood 2011, 117, 1205–1217. [Google Scholar] [CrossRef]
- Lenoir, O.; Flosseau, K.; Ma, F.X.; Blondeau, B.; Mai, A.; Bassel-Duby, R.; Ravassard, P.; Olson, E.N.; Haumaitre, C.; Scharfmann, R. Specific control of pancreatic endocrine β- and δ-cell mass by class IIa histone deacetylases HDAC4, HDAC5, and HDAC9. Diabetes 2011, 60, 2861–2871. [Google Scholar] [CrossRef]
- Lin, X.; Chai, L.; Zhu, H.R.; Zhou, Y.; Lin., H.W. Applying molecular networking for targeted isolation of depsipeptides. RSC Adv. 2021, 11, 2774. [Google Scholar] [CrossRef]
- Sala, G.D.; Mangoni, A.; Costantino, V.; Teta, R. Identification of the biosynthetic gene cluster of thermoactinoamides and discovery of new congeners by integrated genome mining and MS-Based molecular networking. Front. Chem. 2020, 8, 397. [Google Scholar] [CrossRef]
- Han, X.L.; Chakrabortti, A.; Zhu, J.D.; Liang, Z.X.; Li, J.M. Sequencing and functional annotation of the whole genome of the filamentous fungus Aspergillus westerdijkiae. BMC Genom. 2016, 17, 633. [Google Scholar] [CrossRef]
- Hou, X.M.; Li, Y.Y.; Shi, Y.W.; Fang, Y.W.; Chao, R.; Gu, Y.C.; Wang, C.Y.; Shao, C.L. Integrating molecular networking and H NMR to target the isolation of chrysogeamides from a library of marine-derived Penicillium fungi. J. Org. Chem. 2019, 84, 1228–1237. [Google Scholar] [CrossRef]
- Liu, J.T.; Gu, B.B.; Yang, L.J.; Yang, F.; Lin, H.W. New Anti-inflammatory cyclopeptides from a sponge-derived fungus Aspergillus violaceofuscus. Front. Chem. 2018, 6, 226. [Google Scholar] [CrossRef]
- Yang, X.; Yang, Y.; Peng, T.; Yang, F.; Zhou, H.; Zhao, L.; Xu, L.; Ding, Z. A New cyclopeptide from endophytic Streptomyces Sp. Yim 64018. Nat. Prod. Commun. 2013, 8, 1753–1754. [Google Scholar] [CrossRef]
- Jesus, A.D.; Steyn, P.S.; Vleggaar, R.; Wessels, P.L. Carbon-13 nuclear magnetic resonance assignments and biosynthesis of the mycotoxin ochratoxin A. J. Chem. Soc. Perk. Trans. 1980, 52–54. [Google Scholar] [CrossRef]
- Xu, X.y.; He, F.; Zhang, X.Y.; Bao, J.; Qi, S.H. New mycotoxins from marine-derived fungus Aspergillus sp. SCSGAF0093. Food Chem. Toxicol. 2013, 53, 46–51. [Google Scholar] [CrossRef]
- Joshi, B.K.; Gloer, J.B.; Wicklow, D.T.; Dowd, P.F. Sclerotigenin: A new antiinsectan benzodiazepine from the sclerotia of Penicillium sclerotigenum. J. Nat. Prod. 1999, 62, 650–652. [Google Scholar] [CrossRef]
- Dai, J.R.; Carté, B.K.; Sidebottom, P.J.; Sek Yew, A.L.; Ng, S.B.; Huang, Y.C.; Butler, M.S. Circumdatin G, a New alkaloid from the fungus Aspergillus ochraceus. J. Nat. Prod. 2001, 64, 125–126. [Google Scholar] [CrossRef]
- Almeida, C.; Part, N.; Bouhired, S.; Kehraus, S.; König, G.M. Stachylines AD from the sponge-derived fungus Stachylidium sp. J. Nat. Prod. 2011, 74, 21–25. [Google Scholar] [CrossRef]
- Fredimoses, M.; Zhou, X.F.; Ai, W.; Tian, X.P.; Yang, B.; Lin, X.P.; Xian, J.Y.; Liu, Y.H. Westerdijkin A, a new hydroxyphenylacetic acid derivative from deep sea fungus Aspergillus westerdijkiae SCSIO 05233. Nat. Prod. Res. 2015, 29, 158–162. [Google Scholar] [CrossRef]
- Djoukeng, J.D.; Polli, S.; Larignon, P.; Abou-Mansour, E. Identification of phytotoxins from Botryosphaeria obtusa, a pathogen of black dead arm disease of grapevine. Eur. J. Plant Pathol. 2009, 124, 303–308. [Google Scholar] [CrossRef]
- Wu, W.; Dai, H.Q.; Bao, L.; Ren, B.; Lu, J.C.; Luo, Y.M.; Guo, L.D.; Zhang, L.X.; Liu, H.W. Isolation and structural elucidation of proline-containing cyclopentapeptides from an endolichenic Xylaria sp. J. Nat. Prod. 2011, 74, 1303–1308. [Google Scholar] [CrossRef]
- Tong, Y.; Zhang, J.; Wang, L.; Wang, Q.; Huang, H.; Chen, X.; Zhang, Q.; Li, H.; Sun, N.; Liu, G.; et al. Hyper-synergistic antifungal activity of rapamycin and peptide-like compounds against Candida albicans orthogonally via tor1 kinase. ACS Infect Dis. 2021, 7, 2826–2835. [Google Scholar] [CrossRef]
- Sarojini, V.; Cameron, A.J.; Varnava, K.G.; Denny, W.A.; Sanjayan, G. Cyclic Tetrapeptides from nature and design: A review of synthetic methodologies, structure, and function. Chem. Rev. 2019, 119, 10318–10359. [Google Scholar] [CrossRef]
- Campas-Moya, C. Romidepsin for the treatment of cutaneous T-cell lymphoma. Drugs Today 2009, 45, 787–795. [Google Scholar] [CrossRef]
- Marks, P.A.; Breslow, R. Dimethyl sulfoxide to vorinostat: Development of this histone deacetylase inhibitor as an anticancer drug. Nat. Biotech. 2007, 25, 84–90. [Google Scholar] [CrossRef]
- Baltz, R.H. Gifted microbes for genome mining and natural product discovery. J. Ind. Microbiol. Biotechnol. 2017, 44, 573–588. [Google Scholar] [CrossRef]
- Rutledge, P.J.; Challis, G.L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 2015, 13, 509–523. [Google Scholar] [CrossRef]
- Paulo, B.S.; Sigrist, R.; Angolini, C.F.F.; De Oliveira, L.G. New cyclodepsipeptide derivatives revealed by genome mining and molecular networking. ChemistrySelect 2019, 4, 7785–7790. [Google Scholar] [CrossRef]
- Yang, J.Y.; Sanchez, L.M.; Rath, M.; Liu, X.T.; Boudreau, P.D.; Bruns, N.; Glukhov, E.; Wodtke, A.; de Felicio, R.; Fenner, A.; et al. Molecular Networking as a Dereplication Strategy. J. Nat. Prod. 2013, 76, 1686–1699. [Google Scholar] [CrossRef]
- Romano, S.; Jackson, S.; Patry, S.; Dobson, A. Extending the “One Strain Many Compounds” (OSMAC) Principle to Marine Microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef]
- Malek, Z.; Gregory, L.C. Strategies for the discovery of new natural products by genome mining. ChemBioChem 2009, 10, 625–633. [Google Scholar]
- Denning, D.W.; Bromley, M.J. How to bolster the antifungal pipeline. Science 2015, 347, 1414–1416. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Gow, N.A.R.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv13. [Google Scholar] [CrossRef]
- Kumaria, A.; Tripathia, A.H.; Gautamb, P.; Gahtoria, R.; Pandec, A.; Singhd, Y.; Madane, T.; Upadhyay, S.K. Adhesins in the virulence of opportunistic fungal pathogens of human. Mycology 2021, 12, 296–324. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of invasive candidiasis: A persistent public health problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef]
- Singh, K.; Sun, S.; Vezina, C. Rapamycin (AY-22,989), a new antifungal antibiotic. IV. Mechanism of action. J. Antibiot. 1979, 32, 630–645. [Google Scholar] [CrossRef]
- Cruz, M.C.; Cavallo, L.M.; Görlach, J.M.; Cox, G.; Perfect, J.R.; Cardenas, M.E.; Heitman, J. Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologs in Cryptococcus neoformans. Mol. Cell. Biol. 1999, 19, 4101–4112. [Google Scholar] [CrossRef]
- Wong, G.K.; Griffith, S.; Kojima, I.; Demain, A.L. Antifungal activities of rapamycin and its derivatives, prolylrapamycin, 32-desmethylrapamycin, and 32-desmethoxyrapamycin. J. Antibiot. 1998, 51, 487–491. [Google Scholar] [CrossRef][Green Version]
- Eng, C.P.; Sehgal, S.N.; Vezina, C. Activity of rapamycin (AY-22,989) against transplanted tumors. J. Antibiot. 1984, 37, 1231–1237. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Frank, J.M.; Houbraken, J.A.M.P.; Kuijpers, A.F.A.; Samson, R.A. New ochratoxin A producing species of Aspergillus section Circumdati. Study Mycol. 2004, 50, 23–43. [Google Scholar]
- Han, J.J.; Bao, L.; Tao, Q.Q.; Yao, Y.J.; Liu, X.Z.; Yin, W.B.; Liu, H.W. Gloeophyllins A−J, cytotoxic ergosteroids with various skeletons from a chinese tibet fungus Gloeophyllum abietinum. Org. Lett. 2015, 17, 2538–2541. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).