Staphylococcus aureus-Cure-Associated Antigens Elicit Type 3 Immune Memory T Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Production of S. aureus Recombinant Proteins
2.2. GM-CSF-Based DNA Vaccine
2.3. Liposome Preparation and Entrapment of Plasmid DNA
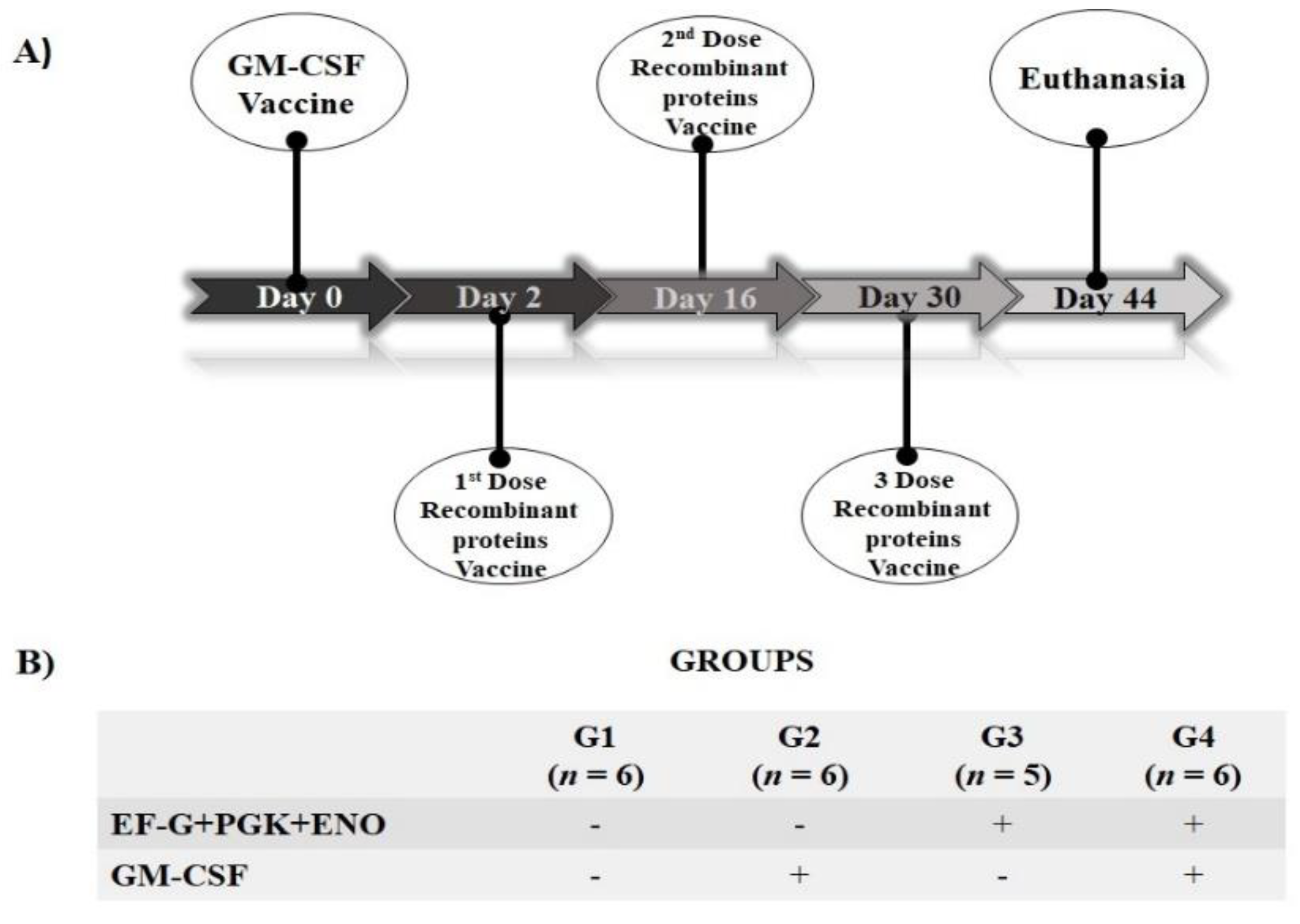
2.4. S. aureus Antigens Candidates: Immunological Evaluation in Mice
2.5. Spleen and Cell Recovery
2.6. Preparation of S. aureus Inoculum
2.7. Immunophenotyping and Lymphocyte Proliferation Evaluation of Cultured Splenic Cells
2.8. Cytokine Measurement
2.9. Statistical Analysis
3. Results
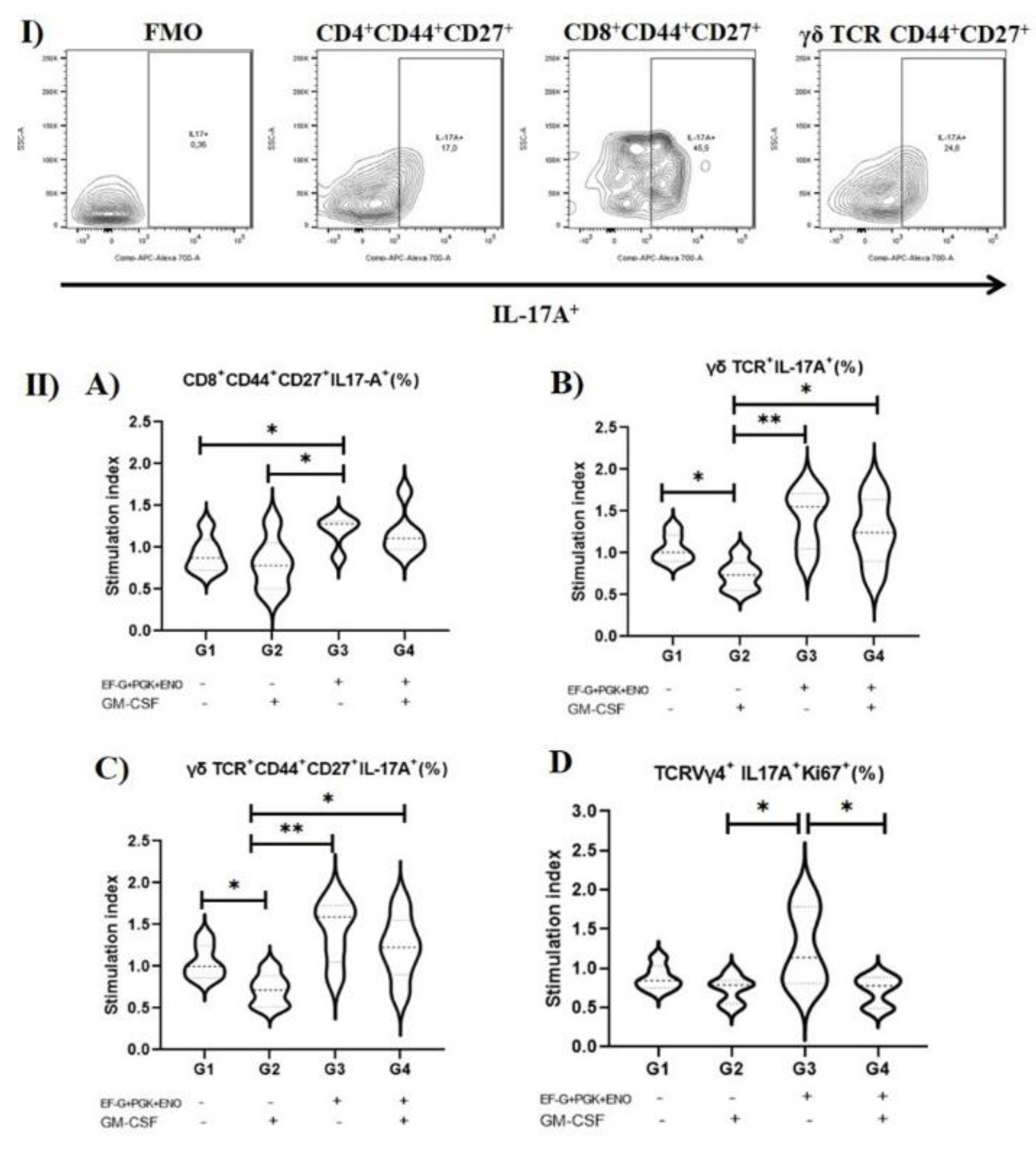
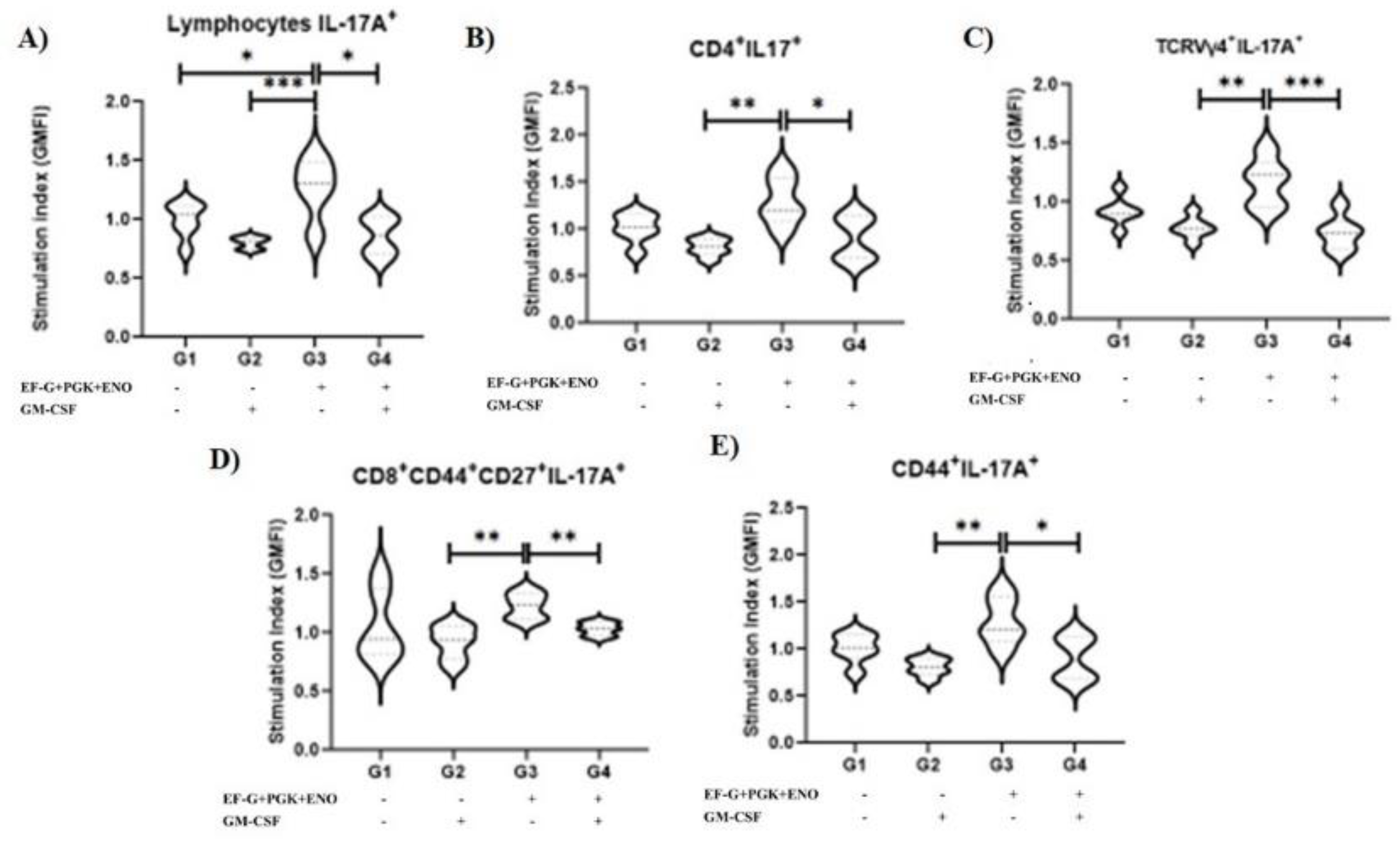
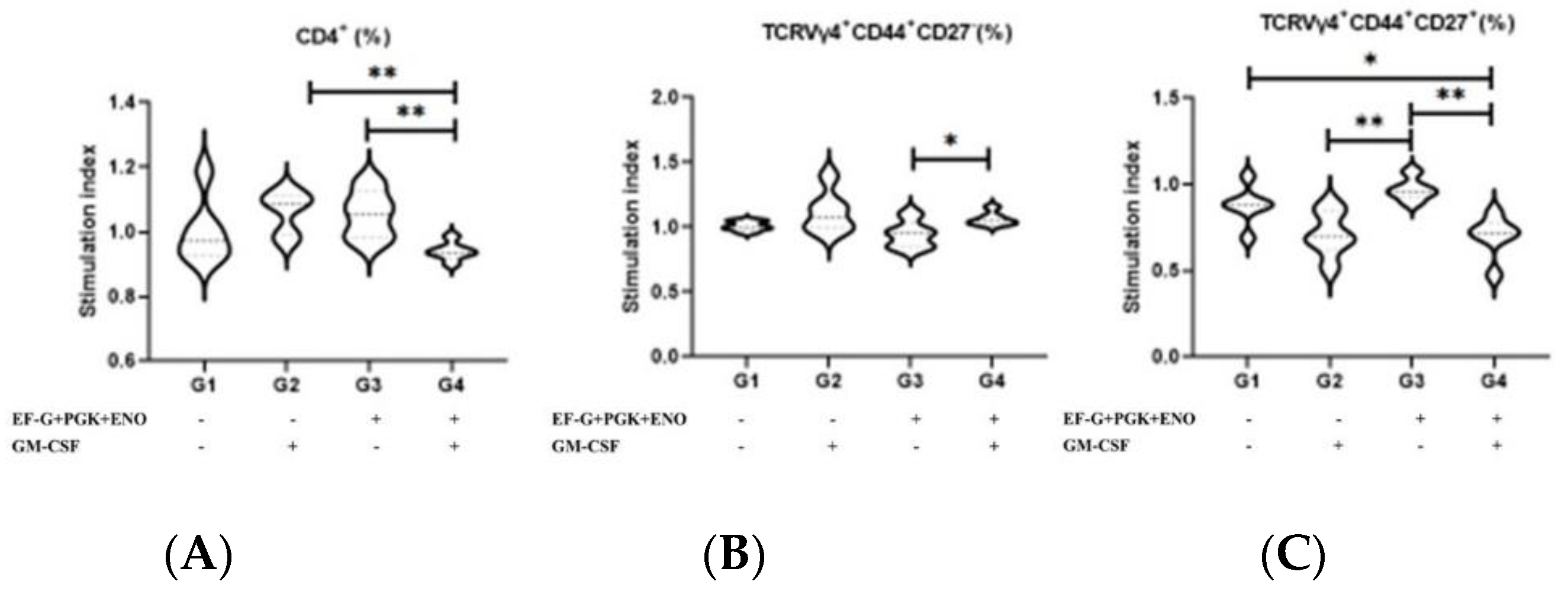
3.1. Type 3 Immunity Is Triggered by αβ and γδ T Memory Cells in Immunized Animals with the Three Recombinant Proteins
3.2. Animals Immunized with Three Recombinant Proteins in Association with the GM-CSF DNA Vaccine Was Associated an Anti-Inflammatory Cytokines Production
4. Discussion
5. Conclusions
6. Patent
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stevens, M.; Piepers, S.; De Vliegher, S. Mastitis prevention and control practices and mastitis treatment strategies associated with the comsuption of (critically important) antimicrobials o dairy herds in Flanders, Belgium. J. Dairy Sci. 2016, 99, 2896–2903. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.P.; Souza, F.N.; Oliveira, A.C.D.; de Souza Filho, A.F.; Aizawa, J.; Moreno, L.Z.; da Cunha, A.F.; Cortez, A.; Della Libera, A.M.M.P.; Heinemann, M.B.; et al. Molecular typing and antimicrobial susceptibility profile of Staphylococcus aureus isolates recovered from bovine mastitis and nasal samples. Animals 2020, 10, 2143. [Google Scholar] [CrossRef] [PubMed]
- Rainard, P.; Foucras, G.; Fitzgerald, J.R.; Watts, J.L.; Koop, G.; Middleton, J.R. Knowledge gaps and research priorities in Staphylococcus aureus mastitis control. Transbound. Emerg. Dis. 2018, 65, 149–165. [Google Scholar] [CrossRef]
- Richardson, E.J.; Bacigalupe, R.; Harrison, E.M.; Weinert, L.A.; Lycett, S.; Vrieling, M.; Robb, K.; Hoskisson, P.A.; Holden, M.T.G.; Feil, E.J.; et al. Gene exchange drives the ecological success of a multi-host bacterial pathogen. Nat. Ecol. Evol. 2018, 2, 1468–1478. [Google Scholar] [CrossRef]
- Campos, B.; Pickering, A.C.; Rocha, L.S.; Aguilar, A.P.; Fabres-Klein, M.H.; de Oliveira Mendes, T.A.; Fitzgerald, J.R.; de Oliveira Barros Ribon, A. Diversity and pathogenesis of Staphylococcus aureus from bovine mastitis: Current understanding and future perspectives. BMC Vet. Res. 2022, 18, 115. [Google Scholar] [CrossRef]
- Poolman, J.T. Expanding the role of bacterial vaccines into life-course vaccination strategies and prevention of antimicrobial-resistant infections. Npj Vaccines 2020, 5, 84. [Google Scholar] [CrossRef]
- Cunha, A.F.; Andrade, H.M.; Souza, F.N.; Fialho Júnior, L.C.; Rosa, D.L.S.O.; Ramos Sanchez, E.M.; Gidlund, M.; Goto, H.; Brito, M.A.V.P.; Guimarães, A.S.; et al. Comparison of antibody repertories against Staphylococcus aureus in healthy and infected dairy cows with a distinct mastitis history and vaccinated with a polyvalent mastitis vaccine. J. Dairy Sci. 2020, 103, 4588–4605. [Google Scholar] [CrossRef] [PubMed]
- Hemmadi, V.; Biswas, M. An overview of moonlighting proteins in Staphylococcus aureus infection. Arch. Microbiol. 2021, 203, 481–498. [Google Scholar] [CrossRef]
- Le Maréchal, C.; Jardin, J.; Briard-Bion, V.; Rault, L.; Berkova, N.; Vautor, E.; Thiéry, R.; Even, S.; Le Loir, Y. Staphylococcus aureus proteins differentially produced in ewe gangrenous mastitis or ewe milk. Vet. Microbiol. 2013, 164, 150–157. [Google Scholar] [CrossRef]
- Bergmann, S.; Rohde, M.; Chhatwal, G.S.; Hammerschmidt, S. α-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 2001, 40, 1273–1287. [Google Scholar] [CrossRef]
- Yu, X.; Zheng, L.; Yang, J.; Lei, T.; Ji, Y. Characterization of essential enolase in Staphylococcus aureus. World J. Microbiol. Biotechnol. 2011, 27, 897–905. [Google Scholar] [CrossRef]
- Carneiro, C.R.W.; Postol, E.; Nomizo, R.; Reis, L.F.L.; Brentani, R.R. Identification of enolase as a laminin-binding protein on the surface of Staphylococcus aureus. Microbes Infect. 2004, 6, 604–608. [Google Scholar] [CrossRef]
- Adamczyk-Poplawska, M.; Markowicz, S.; Jagusztyn-Krynicka, E.K. Proteomics for development of vaccine. J. Proteom. 2011, 74, 2596–2616. [Google Scholar] [CrossRef] [PubMed]
- Foulston, L.; Elsholz, A.K.W.; DeFrancesco, A.S.; Losick, R. The extracellular matrix of Staphylococcus aureus biofilms comprises cytoplasmic proteins that associate with the cell surface in response to decreasing pH. MBio 2014, 5, e01667-14. [Google Scholar] [CrossRef] [PubMed]
- Becker, P.; Hufnagle, W.; Peters, G.; Herrmann, M. Detection of Differential Gene Expression in Biofilm-Forming versus Planktonic Populations of Staphylococcus aureus Using Micro-Representational-Difference Analysis. Appl. Environ. Microbiol. 2001, 67, 2958–2965. [Google Scholar] [CrossRef] [PubMed]
- Uhlmann, J. Neutrophil Interactions with Streptococcus Pyogenes and Staphylococcus aureus. PhD. Thesis, Karolinska Institutet, Stockholm, Sweden, 2017. [Google Scholar]
- Chen, Y.; Koripella, R.K.; Sanyal, S.; Selmer, M. Staphylococcus aureus elongation factor G- Structure and analysis of a target for fusidic acid. FEBS J. 2010, 277, 3789–3803. [Google Scholar] [CrossRef]
- Guo, X.; Peisker, K.; Bäckbro, K.; Chen, Y.; Koripella, R.K.; Mandava, C.S.; Sanyal, S.; Selmer, M. Structure and function of FusB: An elongation factor G-binding fusidic acid resistance protein active in ribosomal translocation and recycling. Open Biol. 2012, 2, 120016. [Google Scholar] [CrossRef] [PubMed]
- Koripella, R.K.; Chen, Y.; Peisker, K.; Koh, C.S.; Selmer, M.; Sanyal, S. Mechanism of elongation factor-G-mediated fusidic acid resistance and fitness compensation in Staphylococcus aureus. J. Biol. Chem. 2012, 287, 30257–30267. [Google Scholar] [CrossRef] [PubMed]
- Nyfeler, B.; Hoepfner, D.; Palestrant, D.; Kirby, C.A.; Whitehead, L.; Yu, R.; Deng, G.; Caughlan, R.E.; Woods, A.L.; Jones, A.K.; et al. Identification of elongation factor g as the conserved cellular target of argyrin B. PLoS ONE 2012, 7, e42657. [Google Scholar] [CrossRef]
- Bekeredjian-Ding, I. Deciphering the significance of the T-cell response to Staphylococcus aureus. Future Microbiol. 2017, 12, 1023–1026. [Google Scholar] [CrossRef]
- Kehrli, M.E.; Cullor, J.S.; Nickerson, S.C. Immunobiology of Hematopoietic Colony-Stimulating Factors: Potential Application to Disease Prevention in the Bovine. J. Dairy Sci. 1991, 74, 4399–4412. [Google Scholar] [CrossRef]
- Shi, Y.; Liu, C.H.; Roberts, A.I.; Das, J.; Xu, G.; Ren, G.; Zhang, Y.; Zhang, L.; Zeng, R.Y.; Tan, H.S.W.; et al. Granulocyte-macrophage colony-stimulating factor (GM-CSF) and T-cell responses: What we do and don’t know. Cell Res. 2006, 16, 126–133. [Google Scholar] [CrossRef]
- Santos, K.R.; Souza, F.N.; Ramos-Sanchez, E.M.; Batista, C.F.; Reis, L.C.; Fotoran, W.F.; Heinemann, M.B.; Goto, H.; Gidlund, M.; Cunha, A.F.; et al. Staphylococcus aureus protection-related type 3 cell-mediated immune response elicited by recombinant proteins and gm-csf dna vaccine. Vaccines 2021, 9, 899. [Google Scholar] [CrossRef]
- Mahdavi, M.; Tajik, A.H.; Ebtekar, M.; Rahimi, R.; Adibzadeh, M.M.; Moozarmpour, H.R.; Beikverdi, M.S.; Olfat, S.; Hassan, Z.M.; Choopani, M.; et al. Granulocyte-macrophage colony-stimulating factor, a potent adjuvant for polarization to Th-17 pattern: An experience on HIV-1 vaccine model. Apmis 2017, 125, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, C.P.; Daniels, M.; Zhao, F.; Alegre, M.L.; Chong, A.S.; Daum, R.S. Protective immunity against recurrent Staphylococcus aureus skin infection requires antibody and interleukin-17A. Infect. Immun. 2014, 82, 2125–2134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Ledue, O.; Jun, M.; Goulart, C.; Malley, R.; Lu, Y.J. Protection against Staphylococcus aureus colonization and infection by B-and T-cell-mediated mechanisms. MBio 2018, 9, e01949-18. [Google Scholar] [CrossRef]
- Fotoran, W.L.; Santangelo, R.; de Miranda, B.N.M.; Irvine, D.J.; Wunderlich, G. DNA-Loaded Cationic Liposomes Efficiently Function as a Vaccine against Malarial Proteins. Mol. Ther. Methods Clin. Dev. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Souza, F.N.; Piepers, S.; Della Libera, A.M.M.P.; Heinemann, M.B.; Cerqueira, M.M.O.P.; De Vliegher, S. Interaction between bovine-associated coagulase-negative staphylococci species and strains and bovine mammary epithelial cells reflects differences in ecology and epidemiological behavior. J. Dairy Sci. 2016, 99, 2867–2874. [Google Scholar] [CrossRef]
- Soares, T.C.S.; Santos, K.R.; Lima, D.M.; Maia, R.B.M.; Ramos-Sanchez, E.M.; Reis, L.C.; Gidlund, M.; da Cunha, A.F.; Ordinola-Ramirez, C.M.; Cerqueira, M.M.O.P.; et al. Memory CD4+ and CD8+ T lymphocyte proliferation in vaccinated dairy cows with different histories of Staphylococcus aureus mastitis. Vet. Immunol. Immunopathol. 2022, 53, 110508. [Google Scholar] [CrossRef] [PubMed]
- Souza, F.N.; Santos, K.R.; Ferronatto, J.A.; Ramos-Sanchez, E.M.; Toledo-Silva, B.; Heinemann, M.B.; De Vliegher, S.; Della Libera, A.M.M.P. Bovine-associated staphylococci and mammaliicocci trigger T-lymphocyte proliferative response and cytokine production differently. J. Dairy Sci. 2022. accepted for publication. [Google Scholar]
- Giunchetti, R.C.; Corrêa-Oliveira, R.; Martins-Filho, O.A.; Teixeira-Carvalho, A.; Roatt, B.M.; de Oliveira Aguiar-Soares, R.D.; de Souza, J.V.; das Dores Moreira, N.; Malaquias, L.C.C.; Mota e Castro, L.L.; et al. Immunogenicity of a killed Leishmania vaccine with saponin adjuvant in dogs. Vaccine 2007, 25, 7674–7686. [Google Scholar] [CrossRef]
- Abdeladhim, M.; Ahmed, M.; Marzouki, S.; Hmida, N.B.; Boussoffara, T.; Hamida, N.B.; Salah, A.; Louzir, H. Human cellular immune response to the saliva of Phlebotomus papatasi is mediated by IL-10-producing CD8+ T cells and TH1-Polarized CD4+ lymphocytes. PLoS Negl. Trop. Dis. 2011, 5, e1345. [Google Scholar] [CrossRef]
- Duz, A.L.C.; Vieira, P.M.D.A.; Roatt, B.M.; Aguiar-Soares, R.D.O.; Cardoso, J.M.D.O.; de Oliveira, F.C.B.; Reis, L.E.S.; Tafuri, W.L.; Veloso, V.M.; Reis, A.B.; et al. The TcI and TcII Trypanosoma cruzi experimental infections induce distinct immune responses and cardiac fibrosis in dogs. Mem. Inst. Oswaldo Cruz 2014, 109, 1005–1013. [Google Scholar] [CrossRef]
- Mackroth, M.S.; Abel, A.; Steeg, C.; Schulze zur Wiesch, J.; Jacobs, T. Acute Malaria Induces PD1+CTLA4+ Effector T Cells with Cell-Extrinsic Suppressor Function. PLoS Pathog. 2016, 12, e1005909. [Google Scholar] [CrossRef]
- Mann, S.E.; Zhou, Z.; Landry, L.G.; Anderson, A.M.; Alkanani, A.K.; Fischer, J.; Peakman, M.; Mallone, R.; Campbell, K.; Michels, A.W.; et al. Multiplex T Cell Stimulation Assay Utilizing a T Cell Activation Reporter-Based Detection System. Front. Immunol. 2020, 11, 9–23. [Google Scholar] [CrossRef]
- Armentrout, E.I.; Liu, G.Y.; Martins, G.A. T cell immunity and the quest for protective vaccines against Staphylococcus aureus infection. Microorganisms 2020, 8, 1936. [Google Scholar] [CrossRef]
- Bröker, B.; Mrochen, D.; Péton, V. The T Cell Response to Staphylococcus aureus. Pathogens 2016, 5, 31. [Google Scholar] [CrossRef]
- Rainard, P.; Cunha, P.; Martins, R.P.; Gilbert, F.B.; Germon, P.; Foucras, G. Type 3 immunity: A perspective for the defense of the mammary gland against infections. Vet. Res. 2020, 51, 129. [Google Scholar] [CrossRef] [PubMed]
- Murphy, A.G.; O’Keeffe, K.M.; Lalor, S.J.; Maher, B.M.; Mills, K.H.G.; McLoughlin, R.M. Staphylococcus aureus Infection of Mice Expands a Population of Memory γδ T Cells That Are Protective against Subsequent Infection. J. Immunol. 2014, 192, 3697–3708. [Google Scholar] [CrossRef] [PubMed]
- Marchitto, M.C.; Dillen, C.A.; Liu, H.; Miller, R.J.; Archer, N.K.; Ortines, R.V.; Alphonse, M.P.; Marusina, A.I.; Merleev, A.A.; Wang, Y.; et al. Clonal Vγ6+Vδ4+ T cells promote IL-17–mediated immunity against Staphylococcus aureus skin infection. Proc. Natl. Acad. Sci. USA 2019, 166, 10917–10926. [Google Scholar] [CrossRef] [PubMed]
- Warren, T.L.; Weiner, G.J. Uses of granulocyte-macrophage colony-stimulating factor in vaccine development. Curr. Opin. Hematol. 2000, 7, 168–173. [Google Scholar] [CrossRef]
- Kumar, A.; Taghi Khani, A.; Sanchez Ortiz, A.; Swaminathan, S. A double-edged sword in cancer immunotherapy. Front. Immunol. 2022, 13, 901277. [Google Scholar] [CrossRef]
| mAbs | Fluorescent Probes | Clone | Host | Cat. n. |
|---|---|---|---|---|
| Anti-IL-17A 1 | Alexa 700 | TC11-18H10 | Rat | 554,412 |
| Anti-IFN-γ 1 | PE | XMG1.2 | Rat | 554,412 |
| Anti-TCRVγ4 1 | FITC | GL2 | Hamster | 552,143 |
| Anti- γδ TCR 1 | BV650 | GL3 | Hamster | 563,993 |
| Anti-CD4 1 | APC-Cy7 | GK1.5 | Rat | 552,051 |
| Anti-CD8 1 | BV510 | 53-6.7 | Rat | 563,068 |
| Anti-CD44 1 | BV421 | IM7 | Rat | 563,970 |
| Anti-CD27 1 | BV750 | LG.3A10 | Hamster | 747,399 |
| Anti-CD19 1 | PE-Cy7 | 1D3 | Rat | 552,854 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, K.R.; Souza, F.N.; Ramos-Sanchez, E.M.; Batista, C.F.; Reis, L.C.; Fotoran, W.L.; Heinemann, M.B.; Cunha, A.F.; Rocha, M.C.; Faria, A.R.; et al. Staphylococcus aureus-Cure-Associated Antigens Elicit Type 3 Immune Memory T Cells. Antibiotics 2022, 11, 1831. https://doi.org/10.3390/antibiotics11121831
Santos KR, Souza FN, Ramos-Sanchez EM, Batista CF, Reis LC, Fotoran WL, Heinemann MB, Cunha AF, Rocha MC, Faria AR, et al. Staphylococcus aureus-Cure-Associated Antigens Elicit Type 3 Immune Memory T Cells. Antibiotics. 2022; 11(12):1831. https://doi.org/10.3390/antibiotics11121831
Chicago/Turabian StyleSantos, Kamila R., Fernando N. Souza, Eduardo M. Ramos-Sanchez, Camila F. Batista, Luiza C. Reis, Wesley L. Fotoran, Marcos B. Heinemann, Adriano F. Cunha, Mussya C. Rocha, Angélica R. Faria, and et al. 2022. "Staphylococcus aureus-Cure-Associated Antigens Elicit Type 3 Immune Memory T Cells" Antibiotics 11, no. 12: 1831. https://doi.org/10.3390/antibiotics11121831
APA StyleSantos, K. R., Souza, F. N., Ramos-Sanchez, E. M., Batista, C. F., Reis, L. C., Fotoran, W. L., Heinemann, M. B., Cunha, A. F., Rocha, M. C., Faria, A. R., Andrade, H. M., Cerqueira, M. M. O. P., Gidlund, M., Goto, H., & Della Libera, A. M. M. P. (2022). Staphylococcus aureus-Cure-Associated Antigens Elicit Type 3 Immune Memory T Cells. Antibiotics, 11(12), 1831. https://doi.org/10.3390/antibiotics11121831










