Studies Regarding the Antimicrobial Behavior of Clotrimazole and Limonene
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Microorganisms and Culture Media
2.3. Devices
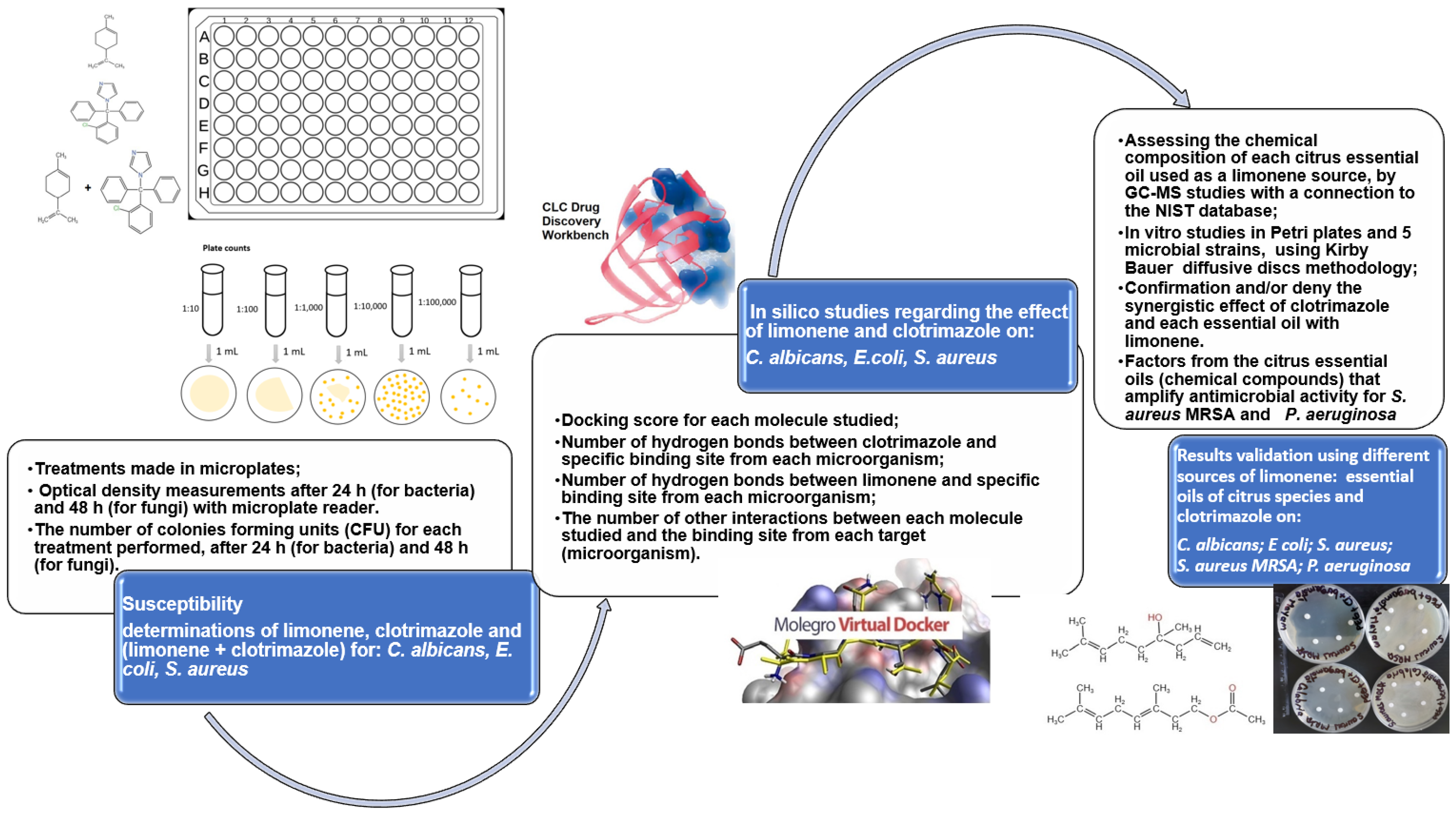
2.4. Experimental Design on Microplates
2.5. Docking Studies
- in the case of C. albicans, the follow was considered as a ligand (co-crystallized): NADPH Dihydro-Nicotinamide-Adenine-Dinucleotide Phosphate (NDP);
- in the case of S. aureus, the following was considered as a ligand: Trimethoprim (TOP);
- in the case of E. coli, the following was considered as a ligand: 8-hydroxyquinoline-5-carboxylic acid (8XQ). The co-crystallized ligands introduced in the protein fragments chosen from the PDB bank also guided the docking simulations for the studied ligands (clotrimazole; limonene), in the same binding site. In the case of the selected protein fragments, the two programs validated the docking protocol, both for each co-crystallized (the reference ligands), and the selected ligands (limonene and clotrimazole).
2.6. Results Validation
3. Results
3.1. Studies Performed on Microplates
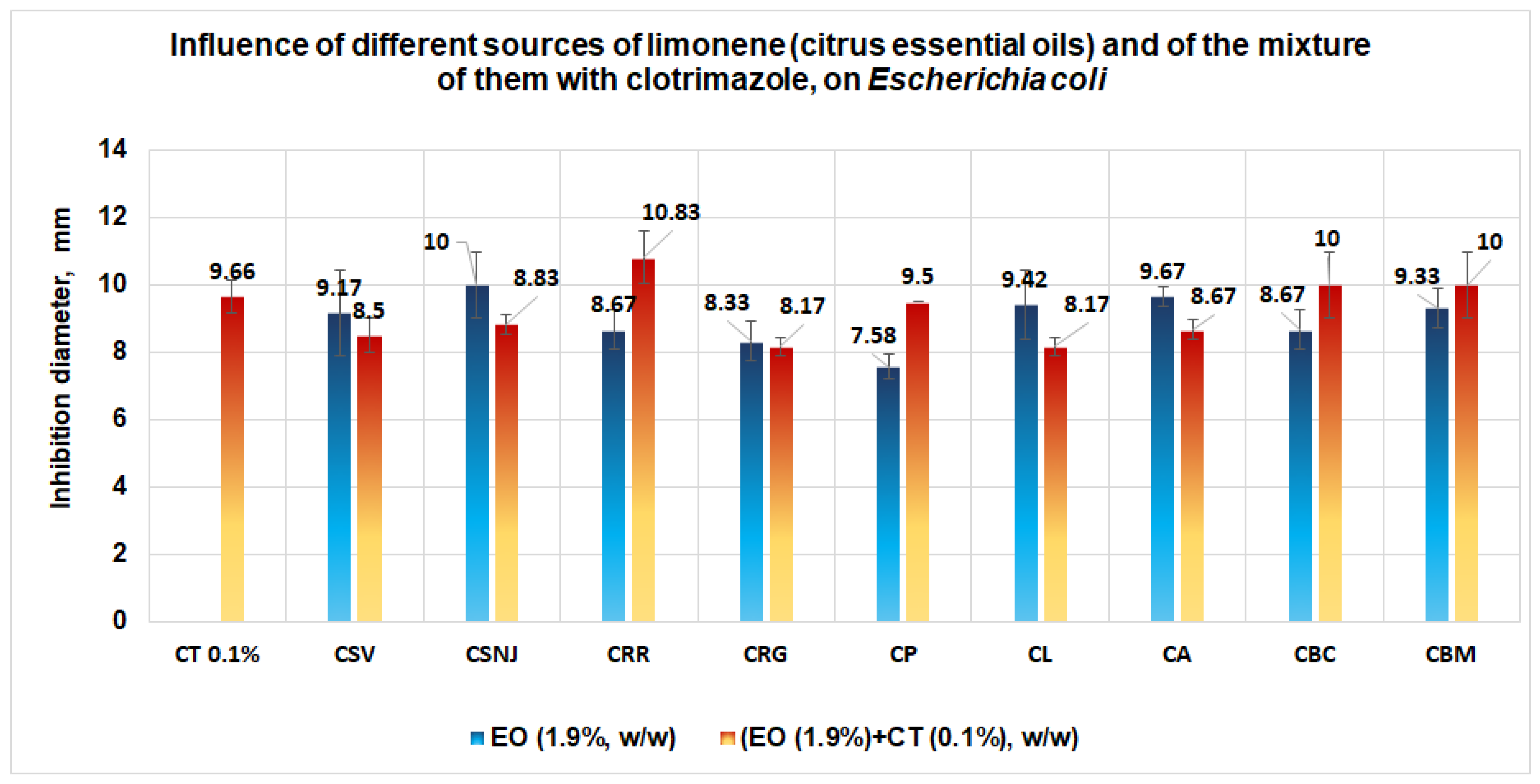
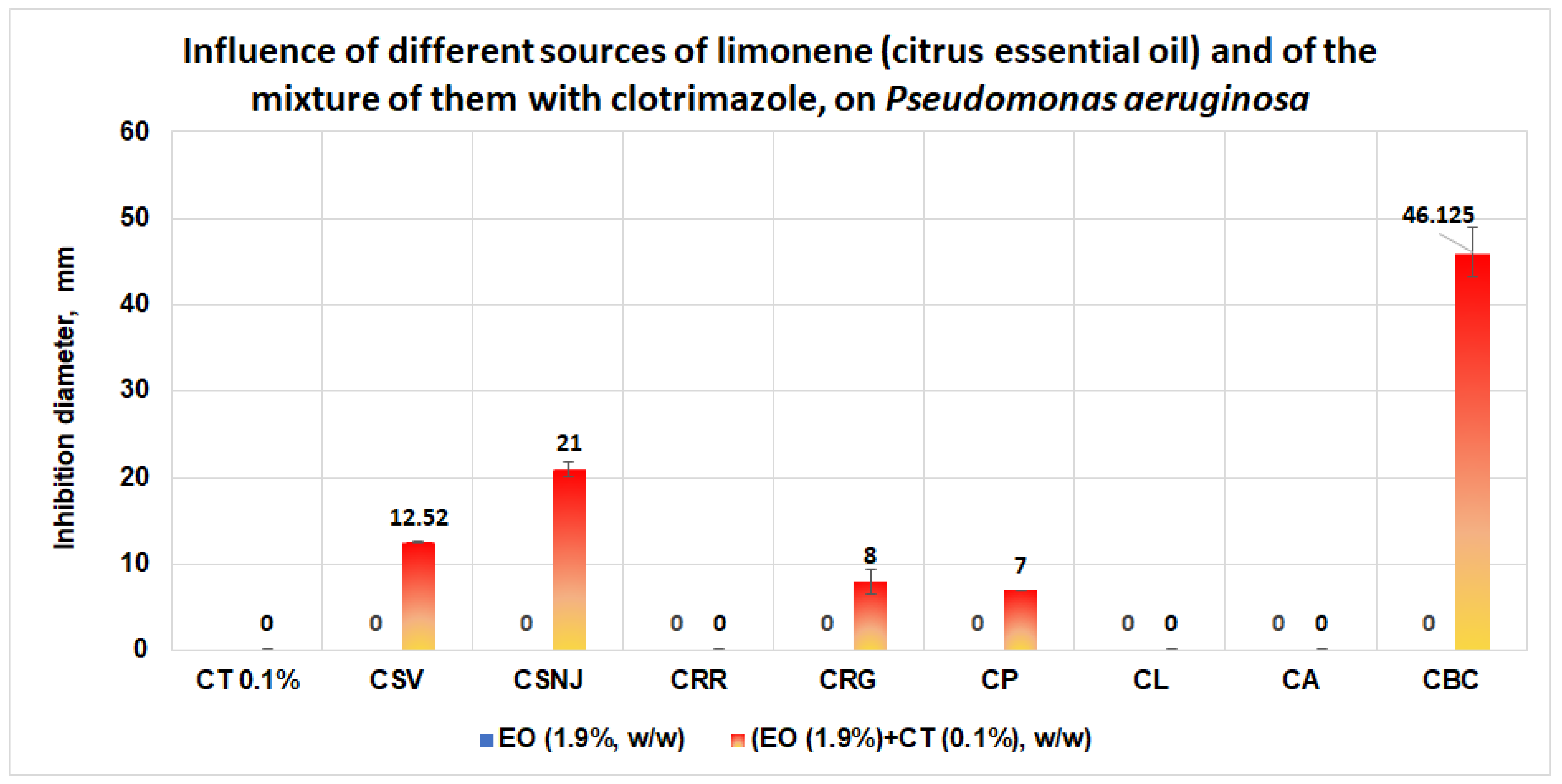
3.1.1. Studies on Microplates Performed on E. coli
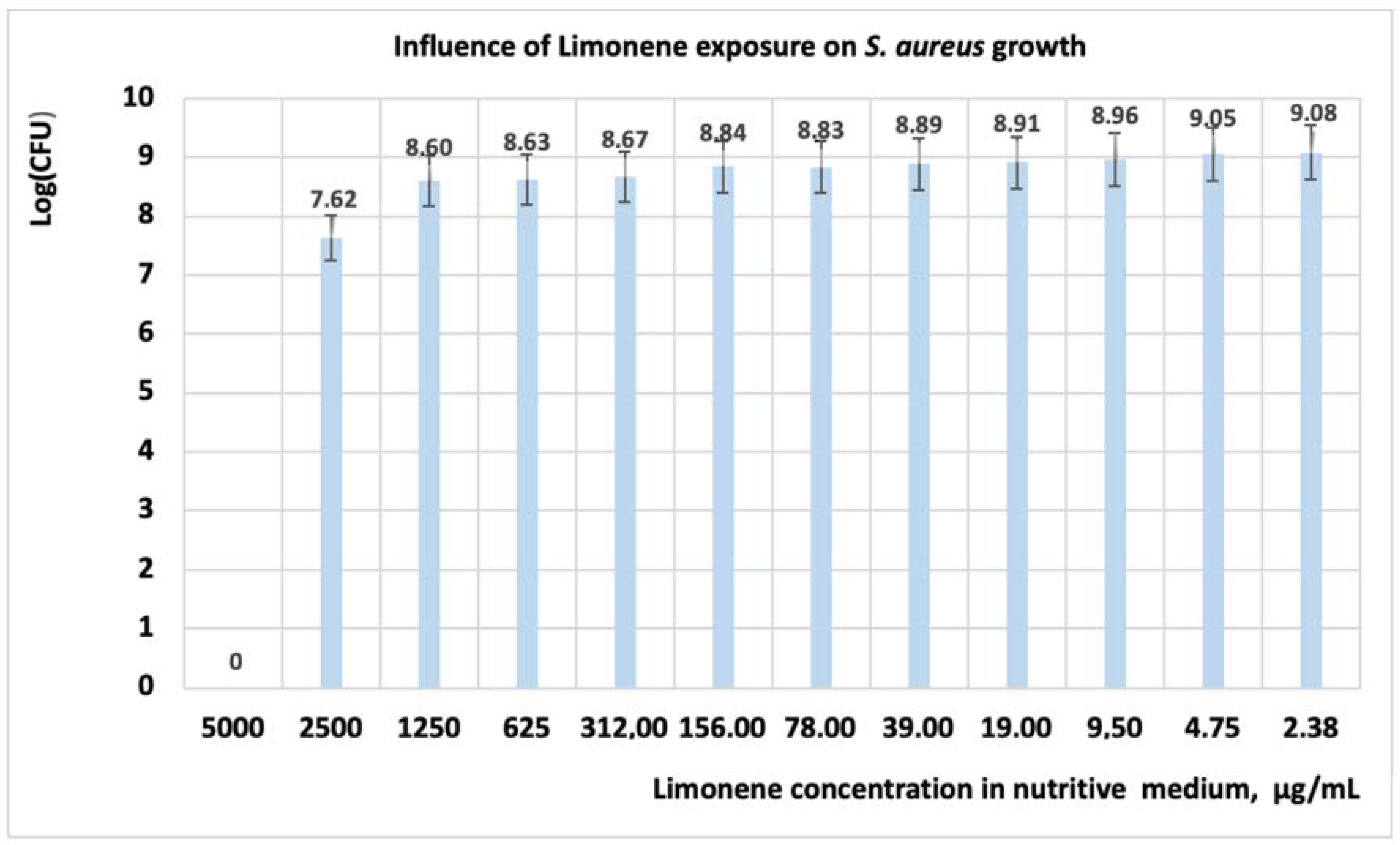
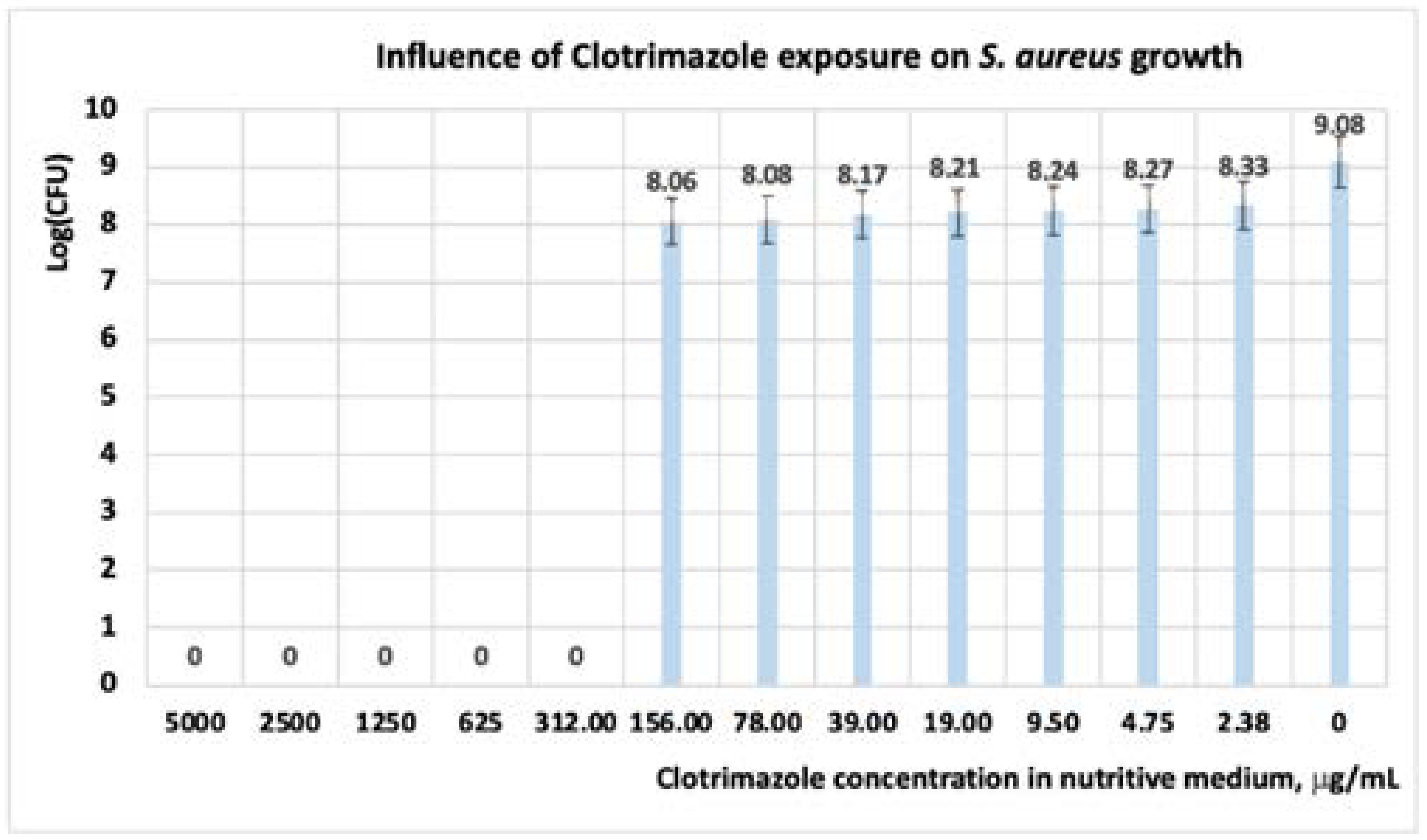
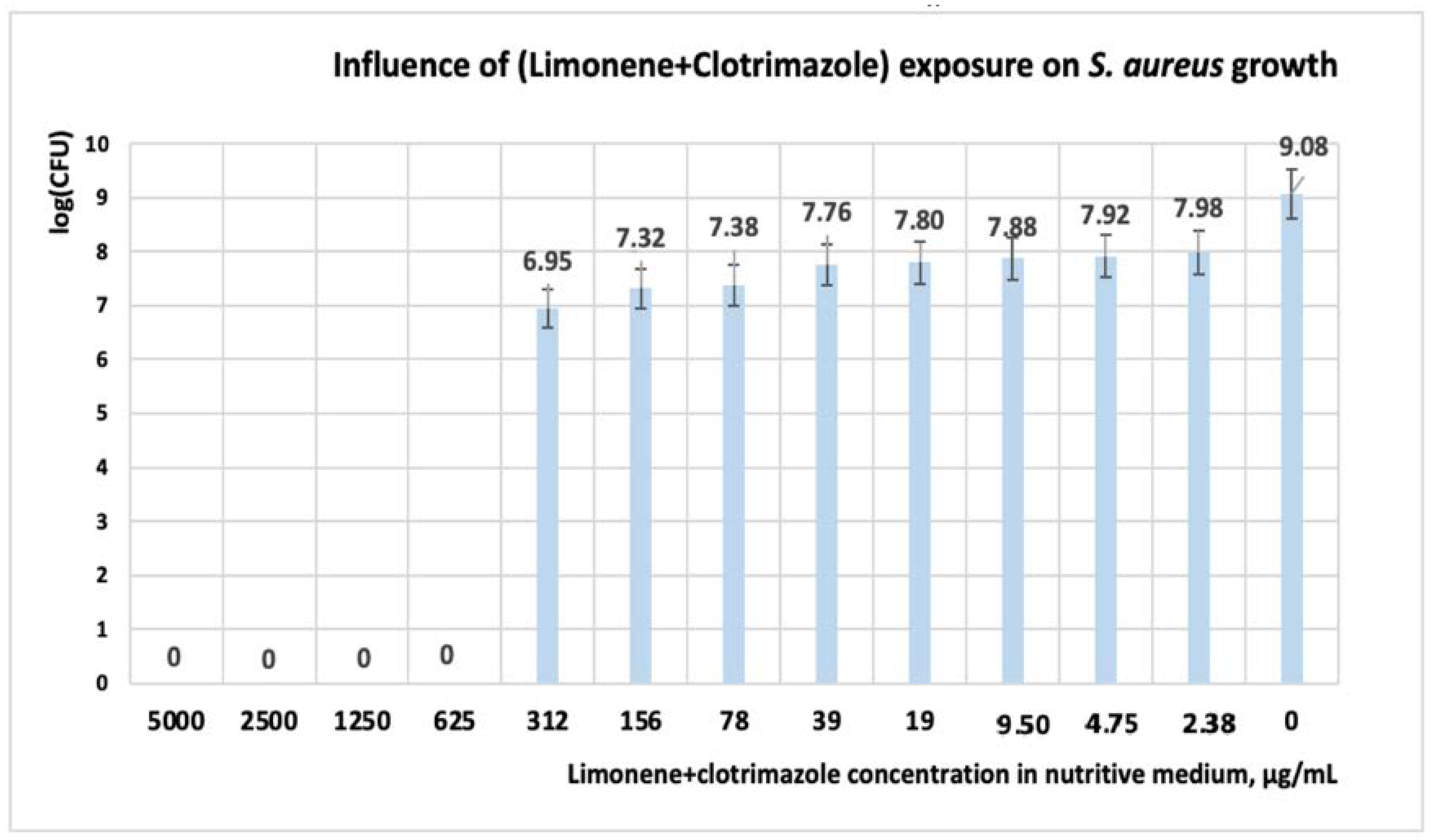
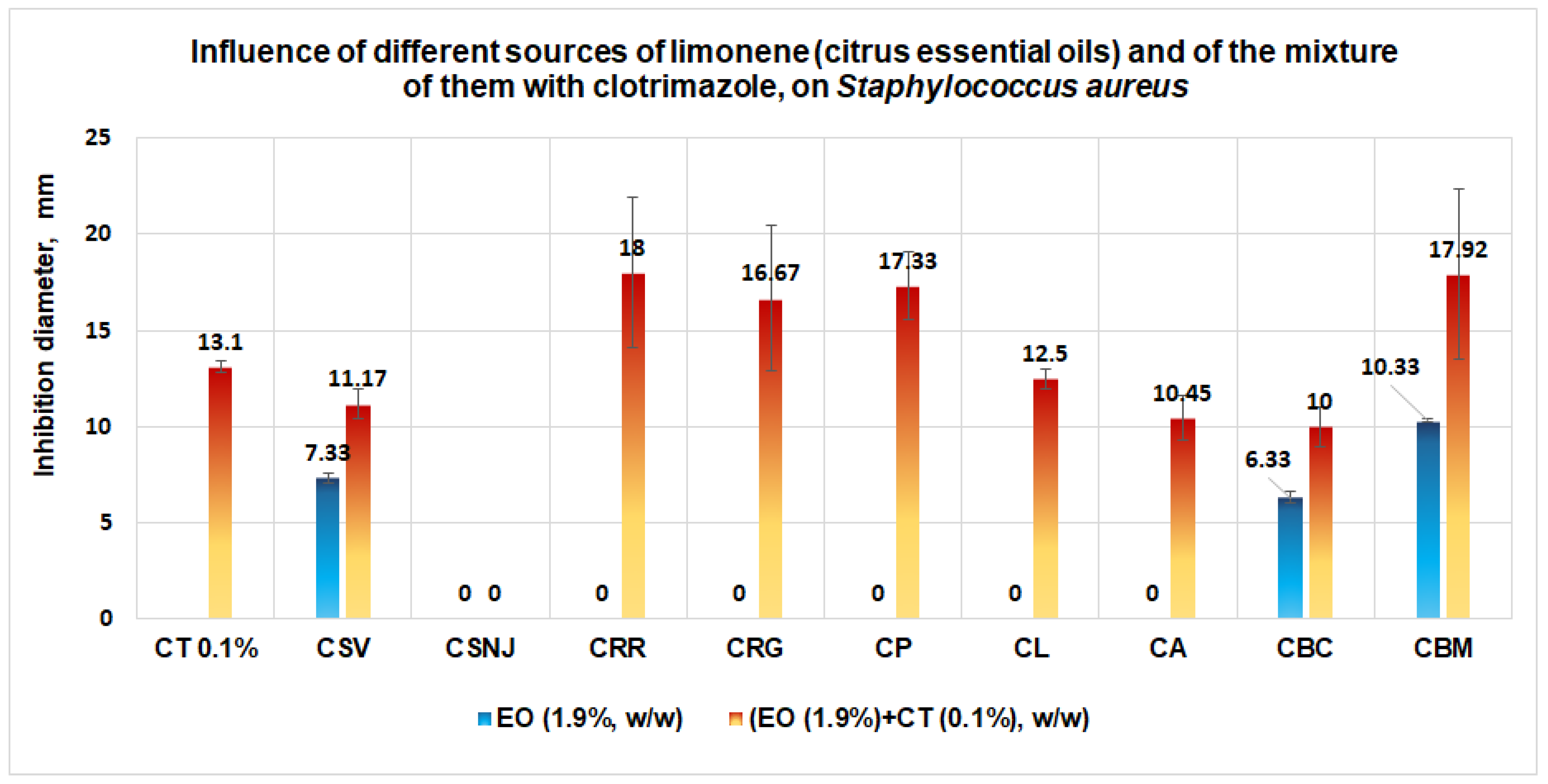
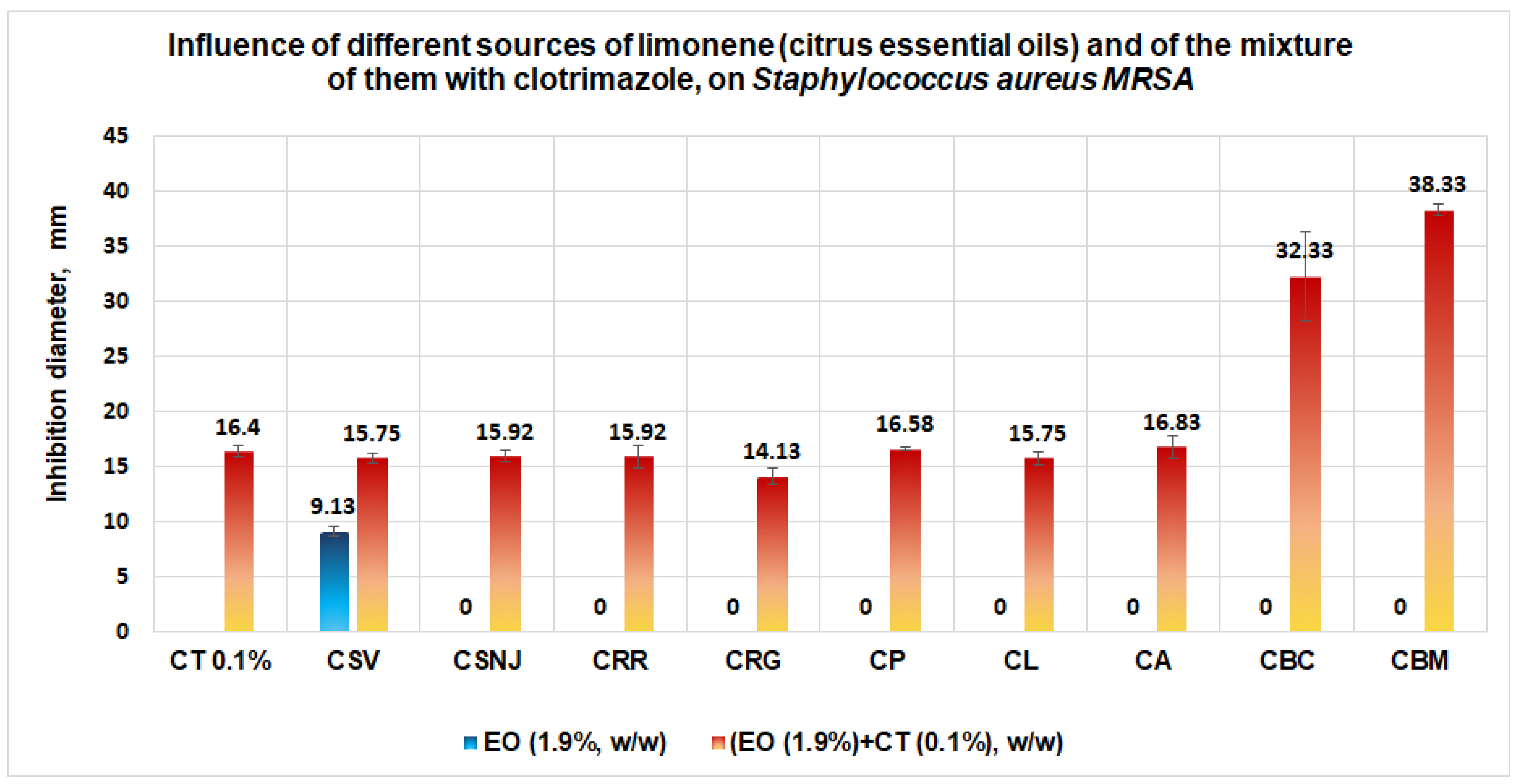
3.1.2. Studies on Microplates Performed on S. aureus
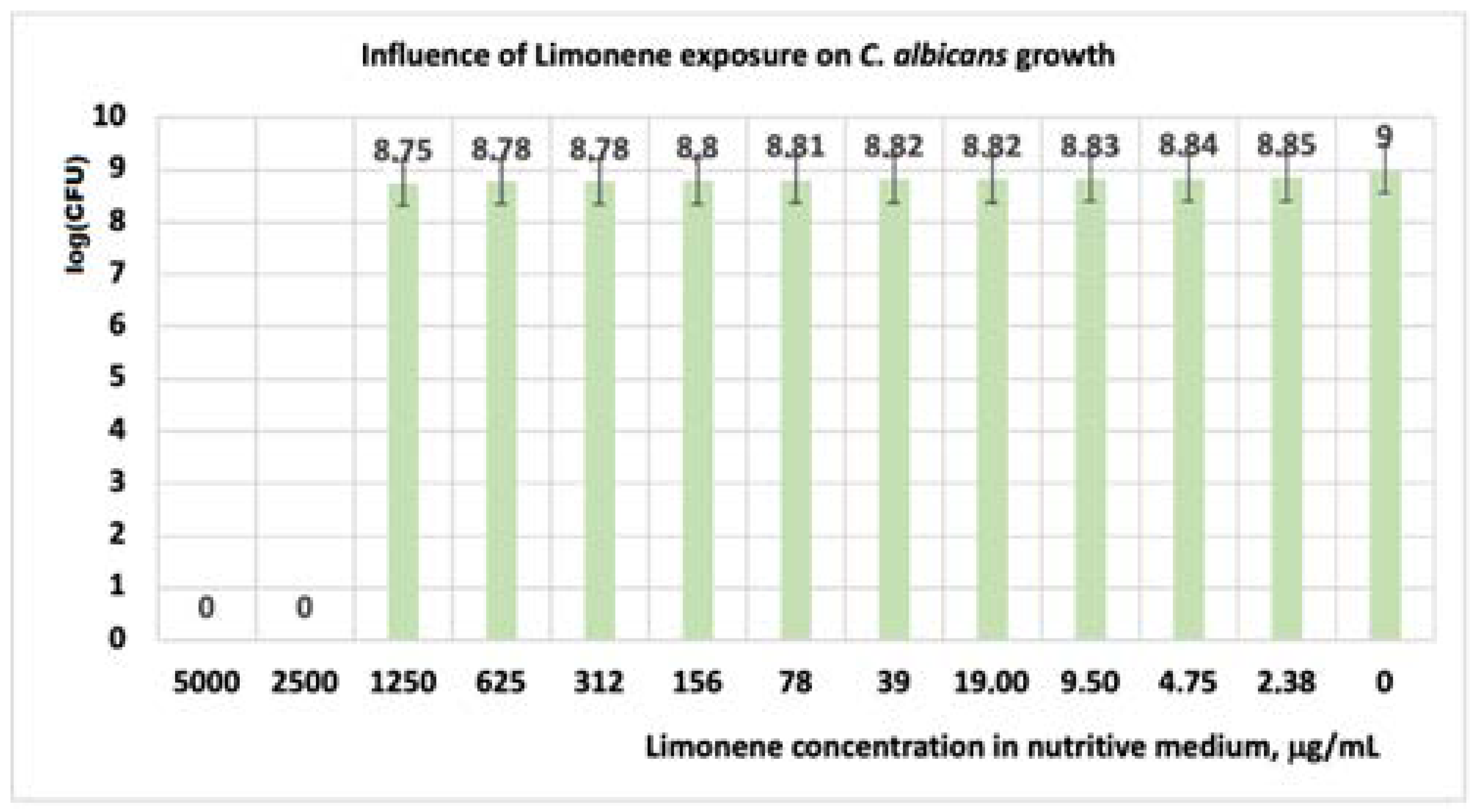
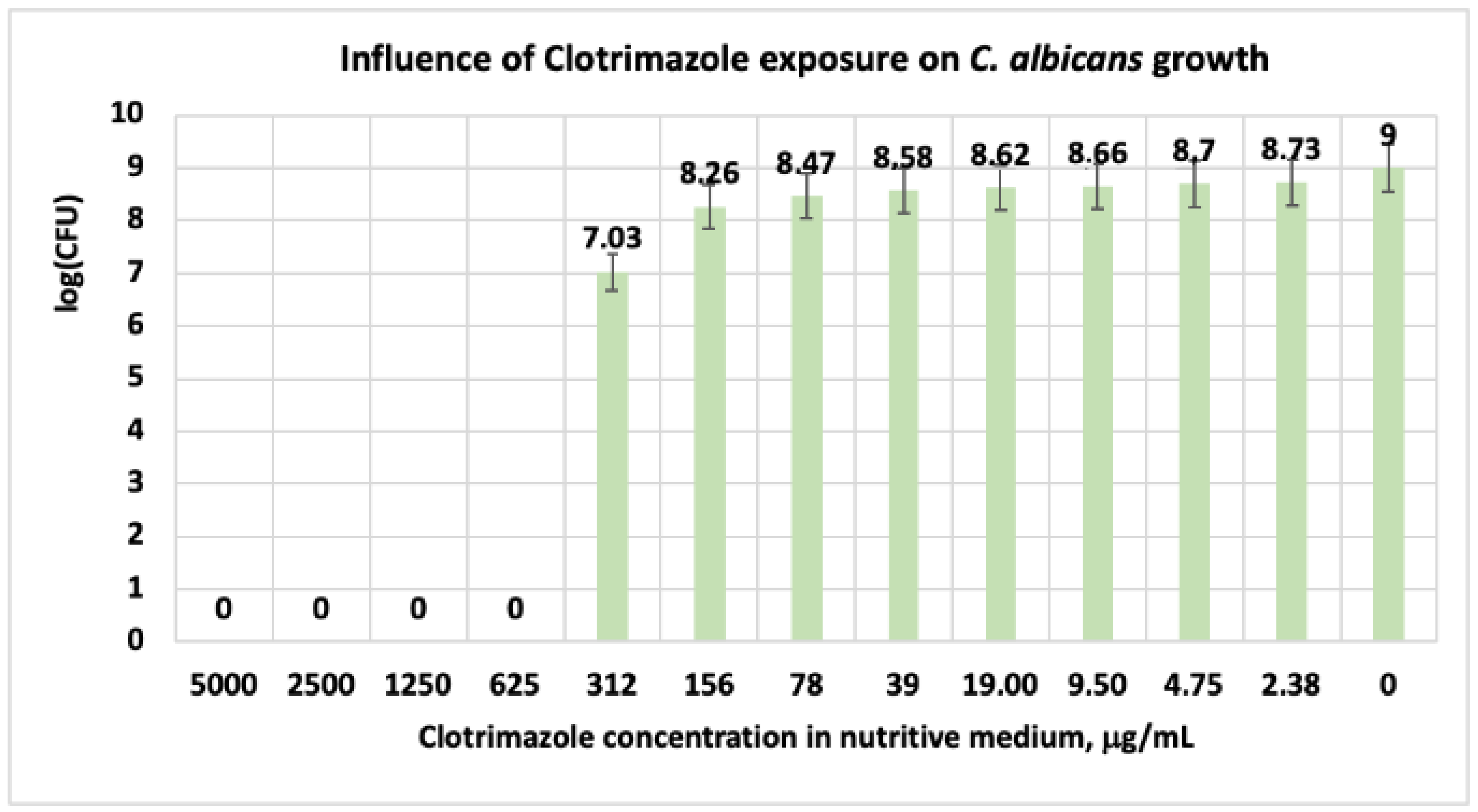
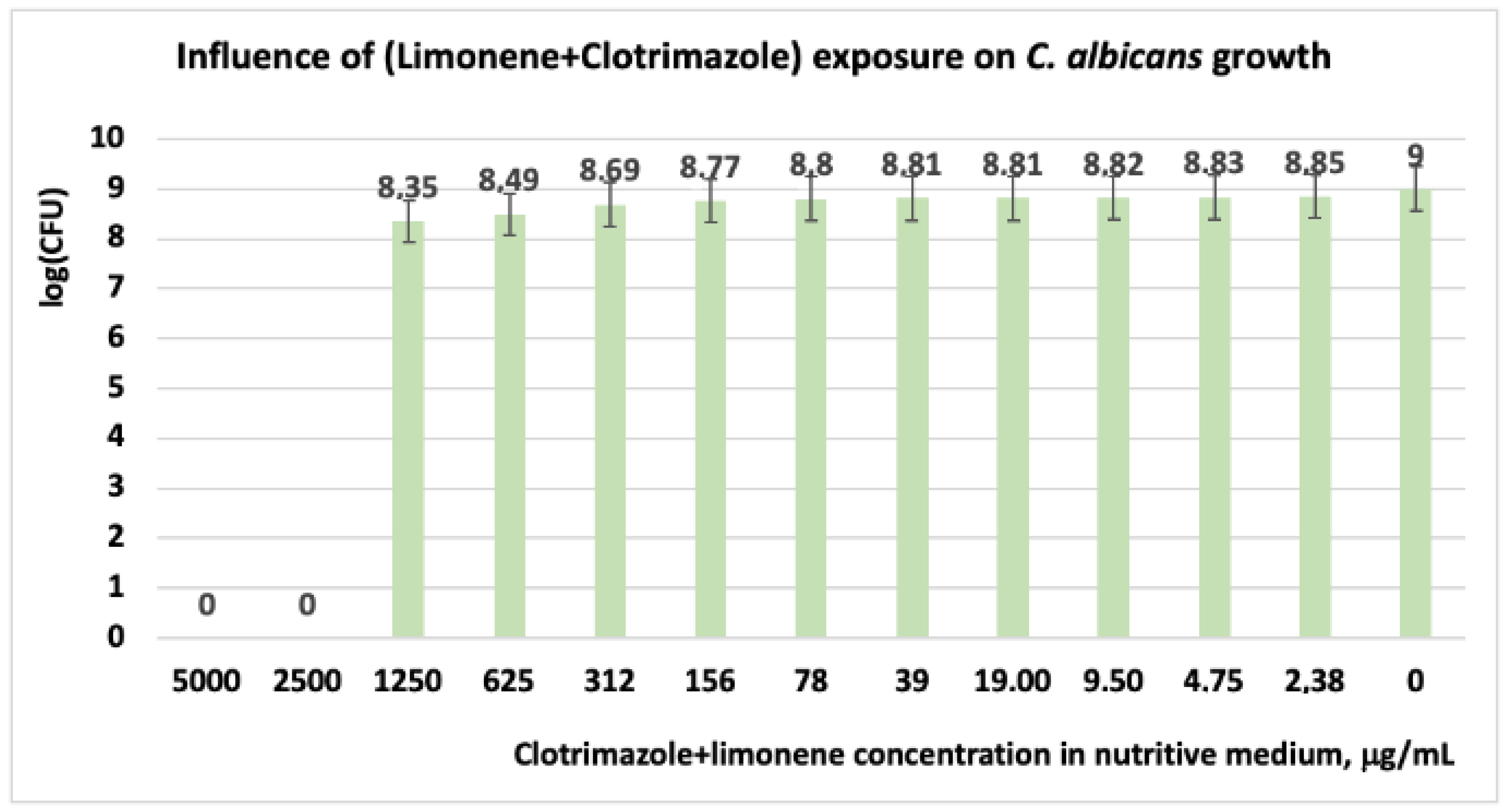
3.1.3. Studies on Microplates Performed on C. albicans
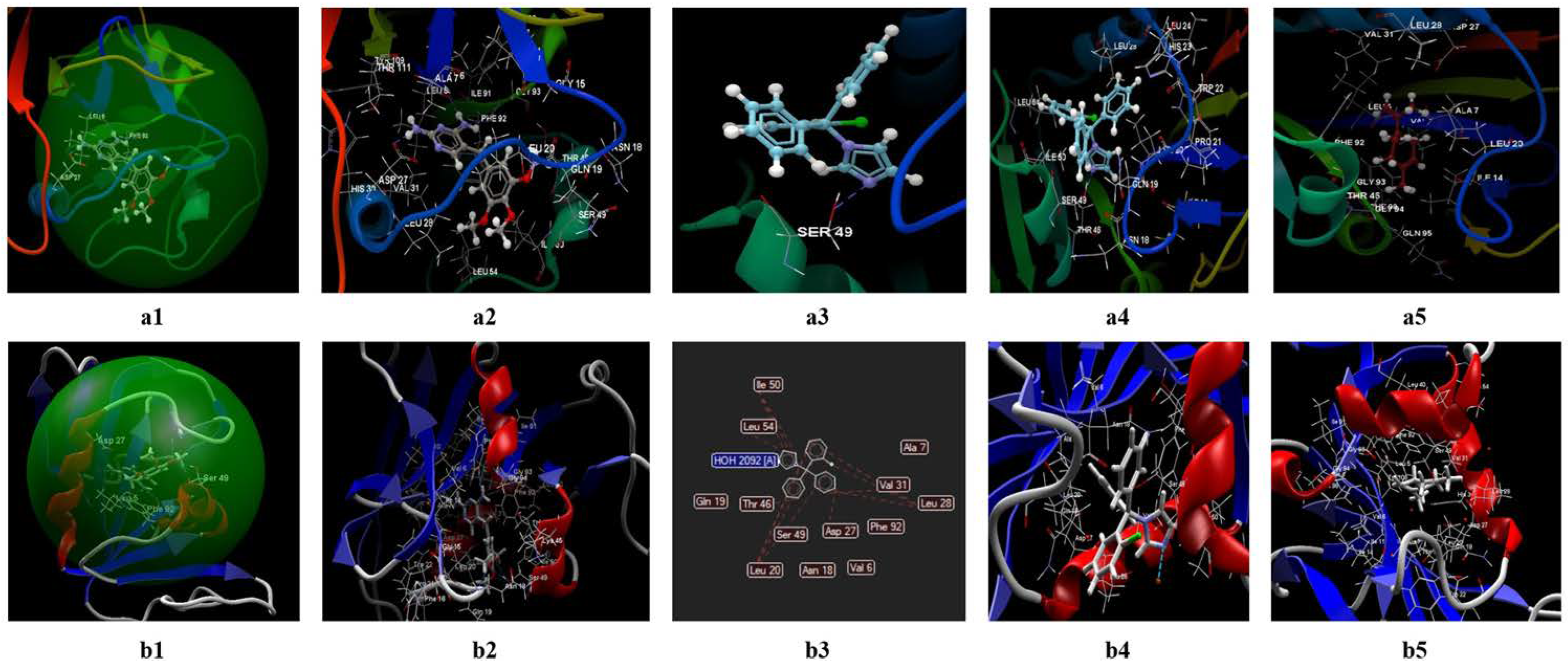
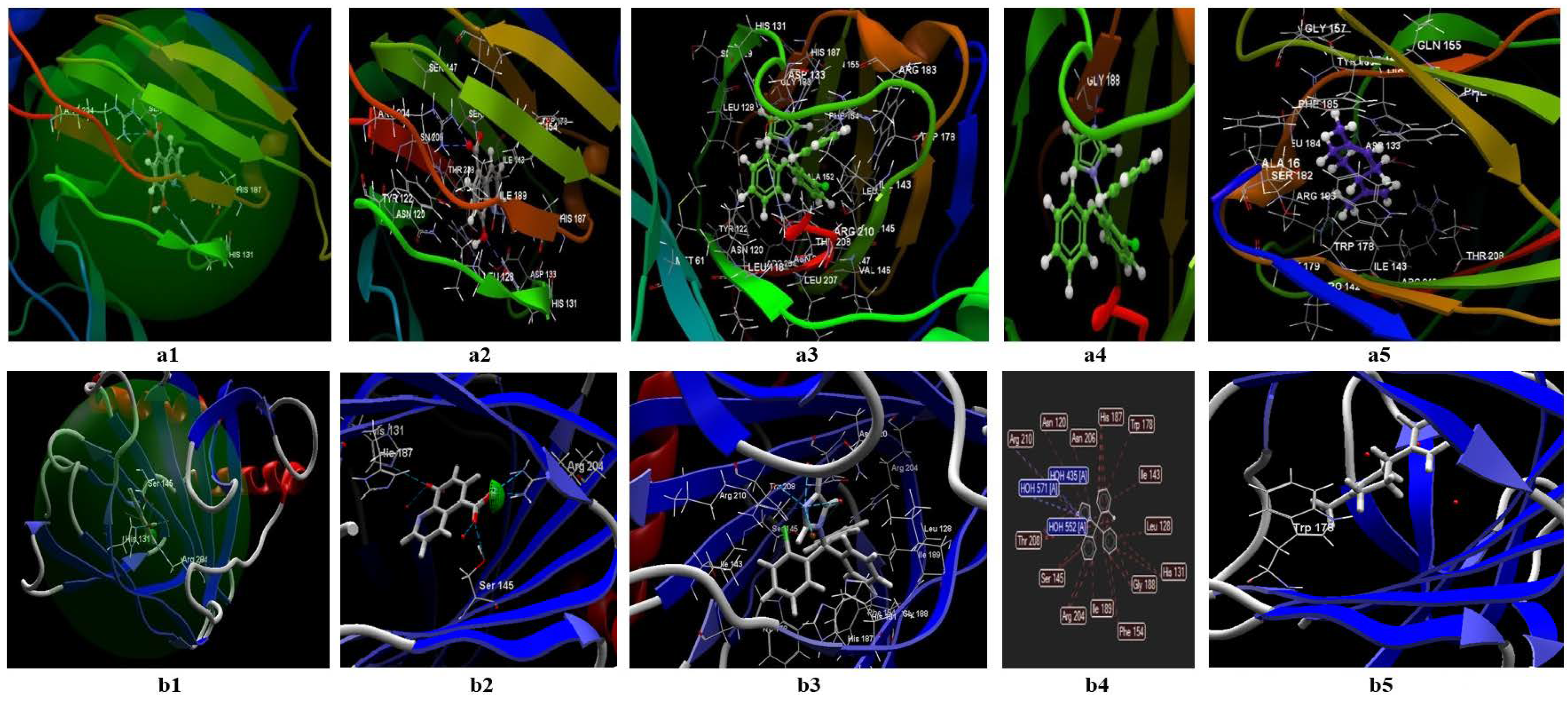
3.2. Molecular Docking Studies
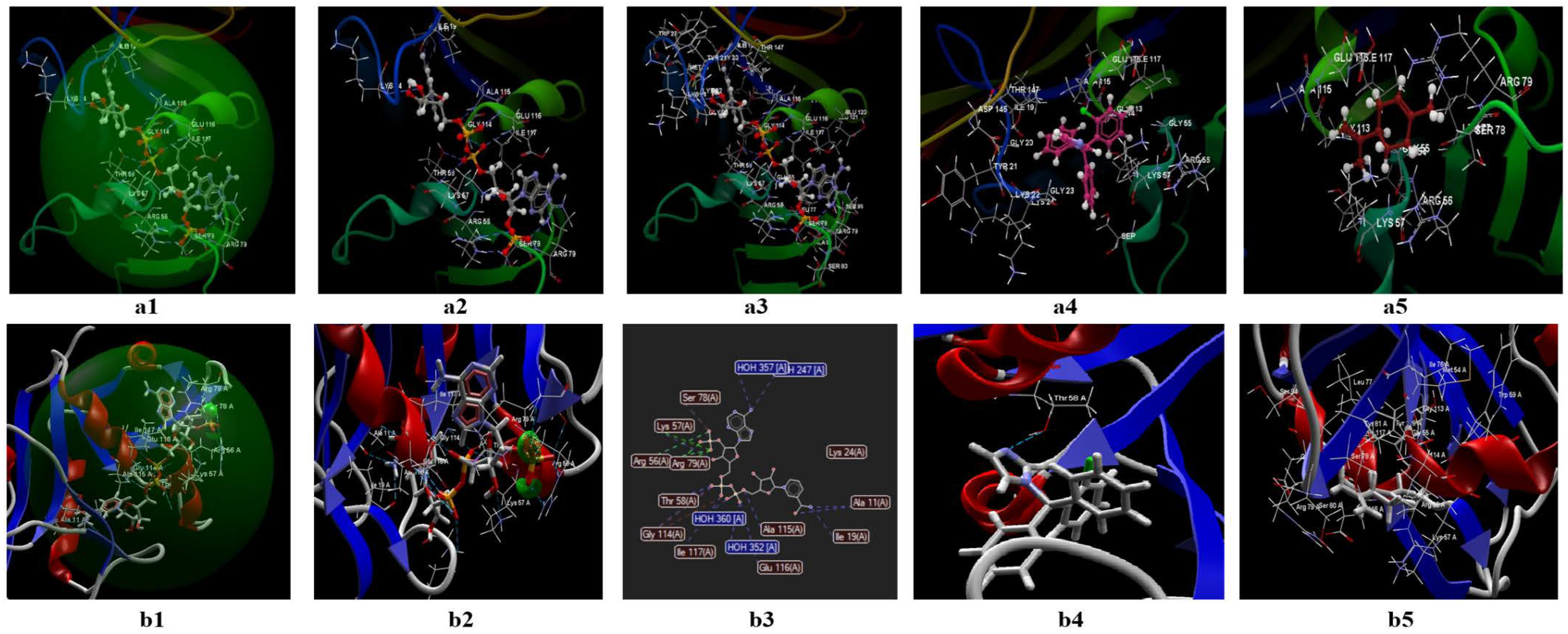
3.2.1. Docking Studies Performed on C. albicans
- The two ligands (limonene; clotrimazole) are fitting in the binding site predicted by the two models;
- Biological activities of the two ligands (clotrimazole; limonene) are similar but smaller than co-crystallized NDP;
- According to Lipinski’s rule of five [35], the values generated by the CLC model for the three ligands studied (clotrimazole, limonene, and the co-crystallized) are more favorable for delivery into the cell for limonene and clotrimazole (molecular mass less than 500) in comparison with co-crystallized (molecular mass 739.37 Da). In addition, the value obtained for the partition coefficient in octanol (logP) is more favorable for clotrimazole (logP = 3.36) and the co-crystallized (log P = −4.72) (Table 9). Analyzing the value of the partition coefficient (logP) in the two ligands (Table 9), it can be appreciated that in the case of a mixture at low concentrations between the two compounds (i.e., clotrimazole; limonene), the presence of clotrimazole is determinant in transdermal delivery, due to the absence of the hydrogen bond [48,49,50,51], and more likely will be docked first.
3.2.2. Docking Studies Performed on S. aureus
- Both ligands (limonene, clotrimazole) can be fitted in the binding site predicted by the two models;
- The partition coefficient is comparable with the co-crystallized (logP= 3.46) in the case of clotrimazole (logP = 3.36) (table from the Annex), but greater than TOP in the case of limonene (logP = 5.41);
- It is not clear if limonene and clotrimazole can act synergically.
3.2.3. Docking Studies Performed on E. coli
- The ligands can be fitted in the binding site predicted by the two models;
- The partition coefficients of the co-crystallized (logP = 2.44) are smaller than clotrimazole (logP = 3.36) (Table 9) and limonene (logP = 5.41).
- a.
- Validating the Antimicrobial Activities by Tests Performed “In Vitro”
4. Discussion
5. Conclusions
- Regarding C. albicans, data obtained suggest that, in the case of a mixture between limonene and clotrimazole, in small quantities, clotrimazole is docked first;
- Data for S. aureus show a similar docking score using both models and suggest that the limonene will be docked first in the binding site. Synergism between the two ligands (limonene; clotrimazole) in the case of S. aureus is more probable;
- In the case of E. coli, taking into account the similar docking data obtained in both models, the great numbers of interactions with amino acids from the binding site, and zero Lipinski’s violations for clotrimazole, it can be assumed that this ligand will be docked first in the binding site. The synergism between the two ligands (i.e., limonene; clotrimazole) in the case of E. coli is less probable. For E. coli, the effect of the mixture between clotrimazole and limonene is probably made by addition;
- Results obtained in validation studies indicate a clear synergism between the mixture of clotrimazole and different sources of limonene (citrus essential oils) against S. aureus, S aureus MRSA, and P. aeruginosa, with the best results for the essential oil of Citrus bergamia for S aureus MRSA and P. aeruginosa. The studies performed are important because they:
- -
- Indicate the synergism between clotrimazole and limonene against S. aureus, in the case of sources with limonene with the content of limonene and/or linalool;
- -
- Show what the best natural resources for limonene are, in order to obtain a synergistic effect;
- -
- Reveal the importance of lab studies (performed “in vitro”), which must be in agreement with the results obtained “in silico”.
- Obtaining pharmaceutical products with clotrimazole, limonene, and different sources of limonene that exhibit the best antimicrobial activity and low toxicity;
- Testing these pharmaceutical products on pathogenic microorganisms involved in skin injury.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
- Susceptible (S) (or sensitive)
- Intermediate = a bacterium is classified with intermediate susceptibility if it belongs to the group of strains that lies between the clinically susceptible and the clinically resistant. Infections caused by such strains have variable (or indeterminate) responses to chemotherapy, but they may be eliminated if the antibiotic is concentrated at the site of the infection, or the dosage is increased.
- Resistance = this term is used in two senses, microbiological and clinical.
- Co-crystallizate = native (natural) ligand.
- Score = the docking score used in the Drug Discovery Workbench is the PLANTSPLP score [63]. This score has a good balance between accuracy and evaluation time. The score mimics the potential energy change when the protein and ligand come together. This means that a very negative score corresponds to a strong binding; a less negative or even positive score corresponds to a weak or non-existent binding. (Score = S target-ligand + S ligand). The S target-ligand term is a sum of contributions from all heavy atom contacts between the ligand and the molecules included in the binding site setup. It scores the complementarity between the binding site and ligand by rewarding and punishing different types of heavy atom contacts (inter-atom distance below ~5.5 Å). Five different types of contacts are defined:
- Hydrogen bond interactions
- Lone–pair–metal ion interactions
- Non-polar interactions
- 4.
- Non-polar–polar contacts
- 5.
- Repulsive contacts:
- RMSD = root mean square deviation. The root mean square deviation (RMSD) is measured between the heavy atom positions of the ligand, in the same way as the docking between the co-crystallized and the binding site, with high accuracy (<2 Å RMSD).
- Group interaction: amino acid group within the binding site, potentially interacting with the ligand.
References
- Khan, U.M.; Sameen, A.; Aadil, R.M.; Shahid, M.; Sezen, S.; Zarrabi, A.; Ozdemir, B.; Sevindik, M.; Kaplan, D.N.; Selamoglu, Z.; et al. Citrus Genus and Its Waste Utilization: A Review on Health-Promoting Activities and Industrial Application. Evid.-Based Complement. Altern. Med. 2021, 2021, 2488804. [Google Scholar] [CrossRef] [PubMed]
- Suri, S.; Singh, A.; Nema, P.K. Current applications of citrus fruit processing waste: A scientific outlook. Appl. Food Res. 2022, 2, 100050. [Google Scholar] [CrossRef]
- Nogueira, J.O.E.; Campolina, G.A.; Batista, L.R.; Alves, E.; Caetano, A.R.S.; Brandão, R.M.; David Lee Nelson, D.L.; das Graças Cardoso, M. Mechanism of action of various terpenes and phenylpropanoids against Escherichia coli and Staphylococcus aureus. FEMS Microbiol. Lett. 2021, 368, fnab052. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, S.O.M.; Babeanu, N.; Cornea, C.P.; Radu, N. Limonene—A biomolecule with potential applications in regenerative medicine. Sci. Bulletin. Ser. F Biotechnol. 2022, 26, 139–148. [Google Scholar]
- Roman, V.; Radu, N.; Bostan, M.; Popa, O.; Tanasescu, A.H. Effect of melaleuca alternifolia bioproduct on acute monocytic leukemia cells line. Mol. Cryst. Liq. Cryst. 2019, 695, 29–36. [Google Scholar] [CrossRef]
- Bostan, M.; Radu, N.; Babeanu, N.; Zaharie, M.G.; Tanasescu, C.A.H. Biological properties of a biomaterial obtained from Syzygium aromaticum. Mol. Cryst. Liq. Cryst. 2019, 695, 45–52. [Google Scholar] [CrossRef]
- Han, Y.; Sun, Z.; Chen, W. Antimicrobial Susceptibility and Antibacterial Mechanism of Limonene against Listeria monocytogenes. Molecules 2019, 25, 33. [Google Scholar] [CrossRef]
- Gupta, A.; Jeyakumar, E.; Lawrence, R. Strategic approach of multifaceted antibacterial mechanism of limonene traced in Escherichia coli. Sci. Rep. 2021, 11, 13816. [Google Scholar] [CrossRef]
- Li, Z.H.; Cai, M.; Liu, Y.; Sun, P.L.; Luo, S.L. Antibacterial Activity and Mechanisms of Essential Oil from Citrus medica L. var. sarcodactylis. Molecules 2019, 24, 1577. [Google Scholar] [CrossRef]
- Carbone, C.; Teixeira, M.D.C.; Sousa, M.D.C.; Martins-Gomes, C.; Silva, A.M.; Souto, E.M.B.; Musumeci, T. Clotrimazole-Loaded Mediterranean Essential Oils NLC: A Synergic Treatment of Candida Skin Infections. Pharmaceutics 2019, 11, 231. [Google Scholar] [CrossRef]
- Nidhi, O.; Rolta, R.; Kumar, V.; Dev, K.; Sourirajan, A. Synergistic potential of Citrus aurantium L. essential oil with antibiotics against Candida albicans. J. Ethnopharmacol. 2020, 262, 113135. [Google Scholar] [PubMed]
- Lesgards, J.F.; Baldovini, N.; Vidal, N. Anticancer Activities of Essential Oils Constituents and Synergy with Conventional Therapies: A Review. Phytother. Res. 2014, 28, 1423–1446. [Google Scholar] [CrossRef] [PubMed]
- Radu, N.; Doncea, S.M.; Ferdes, M.; Salageanu, A.; Rau, I. Biostimulatory properties of Monascus sp. bioproducts. Mol. Cryst. Liq. Cryst. 2012, 555, 195–201. [Google Scholar] [CrossRef]
- Shojaei, S.; Kiumarsi, A.; Moghadam, A.R.; Alizadeh, J.; Marzban, H.; Ghavami, S. Perillyl alcohol (monoterpene alcohol), limonene. Enzymes 2014, 36, 7–32. [Google Scholar]
- Sobral, M.V.; Xavier, A.; Lima, T.; de Sousa, D.P. Antitumor Activity of Monoterpenes Found in Essential Oils. Sci. World J. 2014, 2014, 1–35. [Google Scholar] [CrossRef]
- Chebet, J.J.; Ehiri, J.E.; McClelland, D.J.; Taren, D.; Hakim, I.A. Effect of d-limonene and its derivatives on breast cancer in human trials: A scoping review and narrative synthesis. BMC Cancer 2021, 21, 902. [Google Scholar] [CrossRef]
- Mukhtar, Y.M.; Adu-Frimpong, M.; Xu, X.; Yu, J. Biochemical significance of limonene and its metabolites: Future prospects for designing and developing highly potent anticancer drugs. Biosci. Rep. 2018, 38, BSR20181253. [Google Scholar] [CrossRef]
- Bueno, J.; Demirci, F.; Baser, K.H.C. Antimicrobial strategies in novel drug delivery systems: Applications in the treatment of skin and soft tissue infections. In The Microbiology of Skin, Soft Tissue, Bone and Joint Infections; Academic Press: Cambridge, MA, USA, 2017; pp. 271–286. [Google Scholar]
- Popescu, G.A.; Szekely, E.; Codita, I.; Tălăpan, D.; Șerban, R.; Ruja, G. Diagnosticul, Profilaxia și Tratamentul Infecțiilor Determinate de Staphylococcus Aureus Meticilinorezistent. 2016, pp. 1–27. Available online: https://www.cnscbt.ro/index.php/ghiduri-si-protocoale/519-diagnosticul-profilaxia-si-tratamentul-infectiilor-determinate-de-staphylococcus-aureus-meticilinorezistent-mrsa/file (accessed on 10 March 2022).
- Bolla, P.K.; Meraz, C.A.; Rodriguez, V.A.; Deaguero, I.; Singh, M.; Yellepeddi, V.K.; Renukuntla, J. Clotrimazole Loaded Ufosomes for Topical Delivery: Formulation Development and In-Vitro Studies. Molecules 2019, 24, 3139. [Google Scholar] [CrossRef]
- Maheswary, T.; Nurul, A.A.; Fauzi, M.B. The Insights of Microbes’ Roles in Wound Healing: A Comprehensive Review. Pharmaceutics 2021, 13, 981. [Google Scholar] [CrossRef]
- Kalhapure, R.S.; Sonawane, S.J.; Sikwal, D.R.; Jadhav, M.; Rambharose, S.; Mocktar, C.; Govender, T. Solid lipid nanoparticles of clotrimazole silver complex: An efficient nano antibacterial against Staphylococcus aureus and MRSA. Colloids Surf. B Biointerfaces 2015, 136, 651–658. [Google Scholar] [CrossRef]
- Kumari, B.; Kesavan, K. Effect of Chitosan Coating on Microemulsion for Effective Dermal Clotrimazole Delivery. Pharm. Dev. Technol. 2017, 22, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Frosini, S.M.; Bond, R. Activity In Vitro of Clotrimazole against Canine Methicillin-Resistant and Susceptible Staphylococcus pseudintermedius. Antibiotics 2017, 6, 29. [Google Scholar] [CrossRef] [PubMed]
- Grimling, B.; Karolewicz, B.; Nawrot, U.; Włodarczyk, K.; Górniak, A. Physicochemical and Antifungal Properties of Clotrimazole in Combination with High-Molecular Weight Chitosan as a Multifunctional Excipient. Mat. Drugs 2020, 18, 591. [Google Scholar] [CrossRef] [PubMed]
- Ito, C.; Tecchio, C.; Coustan-Smith, E.; Suzuki, T.; Behm, F.M.; Raimondi, S.C.; Pui, C.H.; Campana, D. The antifungal antibiotic clotrimazole alters calcium homeostasis of leukemic lymphoblasts and induces apoptosis. Leukemia 2022, 16, 1344–1352. [Google Scholar] [CrossRef] [PubMed]
- Penso, J.; Beitner, R. Clotrimazole decreases glycolysis and the viability of lung carcinoma and colon adenocarcinoma cells. Eur. J. Pharm. 2002, 451, 227–235. [Google Scholar] [CrossRef]
- Benzaquen, L.R.; Brugnara, C.; Byers, H.R.; Gattoni-Celli, S.; Halperin, J.A. Clotrimazole inhibits cells proliferation in vitro and in vivo. Nat. Med. 1995, 1, 534–540. [Google Scholar] [CrossRef]
- Adinolfi, B.; Carpi, S.; Romanini, A.; Da Pozzo, E.; Castagna, M.; Costa, B.; Martini, C.; Olesen, S.-P.; Schmitt, N.; Breschi, M.C.; et al. Analysis of the Antitumor Activity of Clotrimazole on A375 Human Melanoma Cells. Anticancer. Res. 2015, 35, 3781–3786. [Google Scholar]
- Kadavakollu, S.; Stailey, C.; Kunapareddy, C.S.; White, S. Clotrimazole as a Cancer Drug: A Short Review. Med. Chem. 2014, 4, 722–724. [Google Scholar]
- Ochioni, A.C.; Filho, R.I.; Esteves, A.M.; Leandro, J.G.B.; Demaria, T.M.; do Nascimento Júnior, J.X.; Pereira-Dutra, F.S.; Bozza, P.T.; Sola-Penna, M.; Zancan, P. Clotrimazole presents anticancer properties against a mouse melanoma model acting as a PI3K inhibitor and inducing repolarization of tumor-associated macrophages. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2021, 1867, 1–10. [Google Scholar] [CrossRef]
- Yang, T.-L.; Hsieh, C.-M.; Meng, L.-J.; Tsai, T.; Chen, C.-T. Oleic Acid-Based Self Micro-Emulsifying Delivery System for Enhancing Antifungal Activities of Clotrimazole. Pharmaceutics 2022, 14, 478. [Google Scholar] [CrossRef]
- Groll, A.H.; Piscitelli, S.P.; Walsh, T.J. Clinical Pharmacology of Systemic Antifungal Agents: A Comprehensive Review of Agents in Clinical Use, Current Investigational Compounds, and Putative Targets for Antifungal Drug. Adv. Pharmacol. 1998, 44, 343–500. [Google Scholar] [CrossRef] [PubMed]
- Hatamleh, A.A.; Al Farraj, D.; Al-Saif, S.S.; Chidambaram, S.; Radhakrishnan, S.; Akbar, I. Synthesis, Cytotoxic Analysis, and Molecular Docking Studies of Tetrazole Derivatives via N-Mannich Base Condensation as Potential Antimicrobials. Drug Des. Dev. Ther. 2020, 14, 4477–4492. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef] [PubMed]
- wwPDB consortium. Protein Data Bank: The single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2019, 47, D520–D528. [Google Scholar] [CrossRef] [PubMed]
- Whitlow, M.; Howard, A.J.; Stewart, D.; Hardman, K.D.; Kuyper, L.F.; Baccanari, D.P.; Fling, M.E.; Tansik, R.L. X-ray crystallographic studies of Candida albicans dihydrofolate reductase. High resolution structures of the holoenzyme and an inhibited ternary complex. J. Biol. Chem. 1997, 272, 30289–30298. [Google Scholar] [CrossRef]
- Heaslet, H.; Harris, M.; Fahnoe, K.; Sarver, R.; Putz, H.; Chang, J.; Subramanian, C.; Barreiro, G.; Miller, J.R. Structural Comparison of Chromosomal and Exogenous Dihydrofolate Reductase from Staphylococcus Aureus in Complex with the Potent Inhibitor Trimethoprim. Proteins 2009, 76, 706. [Google Scholar] [CrossRef]
- Hopkinson, R.J.; Tumber, A.; Yapp, C.; Chowdhury, R.; Aik, W.; Che, K.H.; Li, X.S.; Kristensen, J.B.L.; King, O.N.F.; Chan, M.C.; et al. 5-Carboxy-8-hydroxyquinoline is a Broad Spectrum 2-Oxoglutarate Oxygenase Inhibitor which Causes Iron Translocation. Chem. Sci. 2013, 4, 3110–3117. [Google Scholar] [CrossRef]
- EUCAST The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 12.0. 2022. Available online: http://www.eucast.org (accessed on 2 March 2022).
- El-Halim, A.; Nour El-Dien, H.F.; Mohamed, G.G.; Mohmed, N.A. Metalloantibiotics: Synthesis and antimicrobial activity of clotrimazole metal chelates, spectroscopic, and thermal characterization, synthesis and reactivity. Inorg. Met.-Org. Nano-Met. Chem. 2011, 41, 544–554. [Google Scholar] [CrossRef]
- Jebakuma, S.; Kallidass, S.; Vimalan, J. Isolation, identification, and study of antimicrobial property of a bioactive compound in an Indian medicinal plant Acalypha indica. World J. Microbiol. Biotechnol. 2014, 21, 1231–1236. [Google Scholar]
- Jones, B.; Geary, I.; Lee, M. Comparison of the In Vitro Activities of Fenticonazole, Other Imidazoles, Metronidazole, and Tetracycline against Organisms Associated with Skin Infections. Antimicrob. Agents Chemother. 1989, 33, 970–972. [Google Scholar] [CrossRef]
- Schaller, K. In vitro antibacterial activity of different clotrimazole formulations. Chemotherapy 1982, 28, 32–36. [Google Scholar] [CrossRef]
- Thakre, A.; Zore, G.; Kodgire, S.; Kazi, R.; Mulange, S.; Patil, R.; Shelar, A.; Santhakumari, B.; Kulkarni, M.; Kharat, K.; et al. Limonene inhibits Candida albicans growth by inducing apoptosis. Med. Mycol. 2018, 56, 565–578. [Google Scholar]
- Ibrahim, H.; Furiga, A.; Najahi, E.; Pigasse Hénocq, C.; Nallet, J.P.; Roques, C.; Aubouy, A.; Sauvain, M.; Constant, P.; Daffé, M.; et al. Antibacterial, antifungal and antileishmanial activities of indole-N-oxide derivatives. J. Antibiot. 2012, 65, 499–504. [Google Scholar] [CrossRef]
- Alsterholm, M.; Karami, N.; Faergemann, J. Antimicrobial activity of topical skin pharmaceuticals-an in vitro study. Acta Derm. Venereol. 2010, 90, 239–245. [Google Scholar] [CrossRef]
- Rahbari, R.; Ichim, I.; Bamsey, R.; Burridge, J.; Guy, O.J.; Bolodeoku, J.; Graz, M. Characterisation of Drug Delivery Efficacy Using Microstructure-Assisted Application of a Range of APIs. Pharmaceutics 2020, 12, 1213. [Google Scholar] [CrossRef]
- Chandrashekar, N.S.; Rani, R.S. Physicochemical and pharmacokinetic parameters in drug selection and loading for transdermal drug delivery. Indian J. Pharm. Sci. 2008, 70, 94–96. [Google Scholar] [CrossRef]
- N′Da, D.D. Prodrug Strategies for Enhancing the Percutaneous Absorption of Drugs. Molecules 2014, 19, 20780–20807. [Google Scholar] [CrossRef]
- Jianu, C.; Goleț, I.; Stoin, D.; Cocan, I.; Bujancă, G.; Mișcă, C.; Mioc, M.; Mioc, A.; Șoica, C.; Lukinich-Gruia, A.T.; et al. Chemical Profile of Ruta graveolens, Evaluation of the Antioxidant and Antibacterial Potential of Its Essential Oil, and Molecular Docking Simulations. Appl. Sci. 2021, 11, 11753. [Google Scholar] [CrossRef]
- Kwiatkowski, P.; Łopusiewicz, Ł.; Kostek, M.; Drozłowska, E.; Pruss, A.; Wojciuk, B.; Sienkiewicz, M.; Zielińska-Bliźniewska, H.; Dołęgowska, B. The Antibacterial Activity of Lavender Essential Oil Alone and In Combination with Octenidine Dihydrochloride against MRSA Strains. Molecules 2020, 25, 95. [Google Scholar] [CrossRef]
- Fisher, K.; Phillips, C.A. The effect of lemon, orange and bergamot essential oils and their components on the survival of Campylobacter jejuni, Escherichia coli O157, Listeria monocytogenes, Bacillus cereus and Staphylococcus aureus in vitro and in food systems. J. Appl. Microbiol. 2006, 101, 1232–1240. [Google Scholar] [CrossRef]
- Herman, A.; Tambor, K.; Herman, A. Linalool Affects the Antimicrobial Efficacy of Essential Oils. Curr. Microbiol. 2016, 72, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Angane, M.; Swift, S.; Huang, K.; Butts, C.A.; Quek, S.Y. Essential Oils and Their Major Components: An Updated Review on Antimicrobial Activities, Mechanism of Action and Their Potential Application in the Food Industry. Foods 2022, 11, 464. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, A.C.; Meireles, L.M.; Lemos, M.F.; Guimarães, M.C.C.; Endringer, D.C.; Fronza, M.; Scherer, R. Antibacterial Activity of Terpenes and Terpenoids Present in Essential Oils. Molecules 2019, 24, 2471. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.G.; Yudice, E.D.C.; Campini, P.A.L.; Rosa, D.S. The performance evaluation of Eugenol and Linalool microencapsulated by PLA on their activities against pathogenic bacteria. Mater. Today Chem. 2021, 21, 100493. [Google Scholar] [CrossRef]
- Quirino, A.; Giorgi, V.; Palma, E.; Marascio, N.; Morelli, P.; Maletta, A.; Divenuto, F.; De Angelis, G.; Tancrè, V.; Nucera, S.; et al. Citrus bergamia: Kinetics of Antimicrobial Activity on Clinical Isolates. Antibiotics 2022, 11, 361. [Google Scholar] [CrossRef] [PubMed]
- Bona, E.; Massa, N.; Novello, G.; Pavan, M.; Rocchetti, A.; Berta, G.; Gamalero, E. Essential oil antibacterial activity against methicillin-resistant and -susceptible Staphylococcus aureus strains. Microbiol. Res. 2019, 10, 8331. [Google Scholar] [CrossRef]
- Adaszyńska-Skwirzyńska, M.; Szczerbińska, D.; Zych, S. Antibacterial activity of lavender essential oil and linalool combined with gentamicin on selected bacterial strains. Med. Weter. 2020, 76, 115–118. [Google Scholar] [CrossRef]
- Khameneh, B.; Eskin, N.A.M.; Iranshahy, M.; Fazly Bazzaz, B.S. Phytochemicals: A Promising Weapon in the Arsenal against Antibiotic-Resistant Bacteria. Antibiotics 2021, 10, 1044. [Google Scholar] [CrossRef]
- Babeanu, N.; Radu, N.; Enascuta, C.-E.; Alexandrescu, E.; Ganciarov, M.; Mohammed, M.S.O.; Suica-Bunghez, I.R.; Senin, R.; Ursu, M.; Bostan, M. Obtaining and Characterizing Composite Biomaterials of Animal Resources with Potential Applications in Regenerative Medicine. Polymers 2022, 14, 3544. [Google Scholar] [CrossRef]
- Korb, O.; Stützle, T.; Exner, T.E. Empirical scoring functions for advanced protein-ligand docking with PLANTS. J. Chem. Inf. Model. 2009, 49, 84–96. [Google Scholar] [CrossRef] [PubMed]

| No Crt | Source of Limonene | Code | Major Compounds (w/w) | Minor Compounds w/w |
|---|---|---|---|---|
| 1 | Essential oils from Citrus sinensis (Raw Valencia orange). Supplier: Life | CSV | Limonene 96.94% | - |
| 2 | Essential oils from Citrus sinensis. Supplier: nJoy Nature | CSNJ | Limonene: 96% | β-Pinene: 2.07% Linalool: 1.04 |
| 3 | Essential oil from Citrus reticulata (red mandarin). Supplier: Life | CRR | Limonene: 71.44% γ-Terpinene: 21.64% | β-Phellandrene: 1.16% α-Pinene: 1.63% o-Cymol: 1.86% |
| 4 | Essential oil from Citrus reticulata (green mandarin). Supplier: Life | CRG | Limonene: 67.56% γ-Terpinene: 20.8% | β-Pinene: 1.91% α-Pinene: 1.65% o-Cymene: 4.12% |
| 5 | Essential oils from Citrus limone. Supplier: Arom Sciences | CL | Limonene: 59.07% β-Pinene = 15.14% γ-Terpinene: 14.2% | β-Phellandrene: 3.04% α-Pinene: 2.79% |
| 6 | Essential oil from Citrus paradisi. Supplier: Adams Vision | CP | Limonene: 64.61% Bergamole: 14.12% Linalool: 9.45% | Linalool: 9.45% α-Pinene: 1.92% |
| 7 | Essential oils from Citrus aurantium. Supplier: Arom Sciences | CA | Limonene: 58.76% γ-Terpinene: 14.56% β-Pinene = 14.12% | β-Phellandrene: 2.84% α-Citral: 2.98% |
| 8 | Essential oil from Citrus bergamia. Supplier: Life | CBC | Limonene: 30.41% Bergamole: 28.92% Linalool: 23.31% | γ-Terpinene: 7.28% β-Pinene: 4.63 |
| 9 | Essential oil from Citrus bergamia. Supplier: Mayam | CBM | Limonene: 25.01% Bergamole: 39.43% β-Linalool: 20.83% | γ-Terpinene: 4.56% β-Pinene: 4.26% |
| Major compounds: c ≥ 10%; minor compounds: 1 ≤ c < 10 Beragamole = linalyl acetate | ||||
| Microorganism: E. coli | Concentration, (μg·mL−1) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5000 | 2500 | 1250 | 625 | 312 | 156 | 78 | 39 | 19 | 9.5 | 4.75 | 2.37 | 0 | |
| Limonene | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.05 | 0.01 | 0.01 | 0.43 |
| CT | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.08 | 0.06 | 0.20 | 0.43 |
| Limonene + CT | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.117 | 0.43 |
| Microorganism: E. coli | Concentration, (μg·mL−1) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5000 | 2500 | 1250 | 625 | 312 | 156 | 78 | 39 | 19 | 9.5 | 4.75 | 2.37 | |
| Limonene | S | S | S | S | S | S | S | S | I | I | I | R |
| CT | S | S | S | S | S | S | S | S | I | I | R | R |
| Limonene + CT | S | S | S | S | S | S | S | S | S | S | R | R |
| Microorganism: S. aureus | Concentration, (μg·mL−1) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5000 | 2500 | 1250 | 625 | 312 | 156 | 78 | 39 | 19 | 9.5 | 4.75 | 2.37 | 0 | |
| Limonene | 0 | 0.007 | 0.663 | 0.711 | 0.78 | 1.154 | 1.21 | 1.29 | 1.35 | 1.52 | 1.88 | 2.02 | 3 |
| CT | 0 | 0 | 0 | 0 | 0 | 0.379 | 0.40 | 0.49 | 0.53 | 0.58 | 0.62 | 0.71 | 3 |
| Limonene + CT | 0 | 0 | 0 | 0 | 0.03 | 0.07 | 0.08 | 0.19 | 0.21 | 0.25 | 0.28 | 0.32 | 3 |
| Microorganism: S. aureus | Concentration, (μg·mL−1) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5000 | 2500 | 1250 | 625 | 312 | 156 | 78 | 39 | 19 | 9.5 | 4.75 | 2.37 | |
| Limonene | S | I | R | R | R | R | R | R | R | R | R | R |
| C | S | S | S | S | S | R | R | R | R | R | R | R |
| Limonene + CT | S | S | S | S | I | I | I | R | R | R | R | R |
| Microorganism: C. albicans | Concentration, (μg·mL−1) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5000 | 2500 | 1250 | 625 | 312 | 156 | 78 | 39 | 19 | 9.5 | 4.75 | 2.37 | 0 | |
| Limonene | 0 | 0 | 8.75 | 8.78 | 8.78 | 8.80 | 8.81 | 8.82 | 8.82 | 8.83 | 8.84 | 8.85 | 9 |
| CT | 0 | 0 | 0 | 0 | 7.03 | 8.26 | 8.47 | 8.58 | 8.62 | 8.66 | 8.70 | 8.73 | 9 |
| Limonene + CT | 0 | 0 | 8.35 | 8.49 | 8.69 | 8.77 | 8.80 | 8.81 | 8.81 | 8.82 | 8.83 | 8.85 | 9 |
| Microorganism: C. albicans | Concentration, (μg·mL−1) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5000 | 2500 | 1250 | 625 | 312 | 156 | 78 | 39 | 19 | 9.5 | 4.75 | 2.37 | |
| Limonene | S | S | R | R | R | R | R | R | R | R | R | R |
| CT | S | S | S | S | I | R | R | R | R | R | R | R |
| Limonene + CT | S | S | R | R | R | R | R | R | R | R | R | R |
| Ligand | Score | RMSD, Å | Ligand | Score | RMSD, Å |
|---|---|---|---|---|---|
| The docking score and root mean square deviation between the docked ligands on the binding site 1AI9 of C. albicans. | |||||
| Model generated by CLC | Model generated by MOLEGRO | ||||
| Co-crystallized NDP | −79.35 | 2.86 | Co-crystallized NDP | −155.24 | 0.00 |
| Clotrimazole | −57.87 | 0.10 | Clotrimazole | −77.75 | 0.00 |
| Limonene | −41.94 | 0.01 | Limonene | −73.71 | 0.00 |
| The docking score and root mean square deviation between the docked ligands on the binding site 2W9H of S. aureus | |||||
| Co-crystallized TOP | −53.12 | 0.35 | Co-crystallized TOP | −100.32 | 0.00 |
| Clotrimazole | −47.56 | 0.13 | Clotrimazole | −59.82 | 0.00 |
| Limonene | −39.63 | 0.02 | Limonene | −50.58 | 0.00 |
| The docking score and root mean square deviation between the docked ligands on the binding site 4JHT of E. coli | |||||
| Co-crystallized 8XQ | −37.48 | 0.03 | Co-crystallized 8XQ | −79.94 | 0.00 |
| Clotrimazole | −10.06 | 0.01 | Clotrimazole | −64.85 | 0.00 |
| Limonene | −45.14 | 0.12 | Limonene | −65.79 | 0.00 |
| Compounds | Atoms | Weight [Daltons] | Flexible Bonds | Lipinski Violations | Hydrogen Donors | Hydrogen Acceptors | Log P |
|---|---|---|---|---|---|---|---|
| Co-crystallized NDP * | 72 | 739.37 | 13 | 3 | 6 | 24 | −4.72 |
| Co-crystallized TOP ** | 39 | 290.32 | 5 | 0 | 4 | 7 | 3.46 |
| Co-crystallized 8XQ *** | 20 | 188.16 | 1 | 0 | 1 | 4 | 2.44 |
| Clotrimazole | 42 | 344.84 | 4 | 1 | 0 | 0 | 3.36 |
| Limonene | 26 | 136.23 | 1 | 0 | 0 | 2 | 5.41 |
| Microorganism/Compound | MIC, μg/mL | Fractional Inhibitory Concentration, FIC | Fractional Inhibitory Concentration Index, FICI | Type of Interaction |
|---|---|---|---|---|
| Candida albicans | ||||
| Limonene | 1250 | 0.2496 | ||
| Clotrimazole | 312 | 1 | ||
| Limonene + Clotrimazole | 312 | 1.2496 | Indifference | |
| Escherichia coli | ||||
| Limonene | 9.5 | 0.2463 | ||
| Clotrimazole | 9.5 | 0.2463 | ||
| Limonene + Clotrimazole | 2.34 | 0.4926 | Synergy and/or Addition | |
| Staphyloccocus aureus | ||||
| Limonene | 2500 | 0.0009 | ||
| Clotrimazole | 156 | 0.015 | ||
| Limonene + Clotrimazole | 2.34 | 0.0159 | Synergy | |
| FIC limonene = MIC mixture (limonene+clotrimazole)/MIC limonene FIC clotrimazole = MIC mixture (limonene+clotrimazole)/MIC clotrimazole FICI = FIC limonene + FIC clotrimazole FICI < 0.5: Synergy 0.5 ≤ FICI ≤ 1: Addition 1.1 < FICI ≤ 400: Indiference; FICI > 4: Antagonism | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schroder, V.; Radu, N.; Cornea, P.C.; Coman, O.A.; Pirvu, L.C.; Mohammed, M.S.O.; Stefaniu, A.; Pintilie, L.; Bostan, M.; Caramihai, M.D.; et al. Studies Regarding the Antimicrobial Behavior of Clotrimazole and Limonene. Antibiotics 2022, 11, 1816. https://doi.org/10.3390/antibiotics11121816
Schroder V, Radu N, Cornea PC, Coman OA, Pirvu LC, Mohammed MSO, Stefaniu A, Pintilie L, Bostan M, Caramihai MD, et al. Studies Regarding the Antimicrobial Behavior of Clotrimazole and Limonene. Antibiotics. 2022; 11(12):1816. https://doi.org/10.3390/antibiotics11121816
Chicago/Turabian StyleSchroder, Verginica, Nicoleta Radu, Petruta Calina Cornea, Oana Andreia Coman, Lucia Camelia Pirvu, Mohammed Shaymaa Omar Mohammed, Amalia Stefaniu, Lucia Pintilie, Marinela Bostan, Mihai Dan Caramihai, and et al. 2022. "Studies Regarding the Antimicrobial Behavior of Clotrimazole and Limonene" Antibiotics 11, no. 12: 1816. https://doi.org/10.3390/antibiotics11121816
APA StyleSchroder, V., Radu, N., Cornea, P. C., Coman, O. A., Pirvu, L. C., Mohammed, M. S. O., Stefaniu, A., Pintilie, L., Bostan, M., Caramihai, M. D., & Roman, V. (2022). Studies Regarding the Antimicrobial Behavior of Clotrimazole and Limonene. Antibiotics, 11(12), 1816. https://doi.org/10.3390/antibiotics11121816









