Effect of PAS-LuxR Family Regulators on the Secondary Metabolism of Streptomyces
Abstract
1. Introduction
2. PAS-LuxR Family Regulators
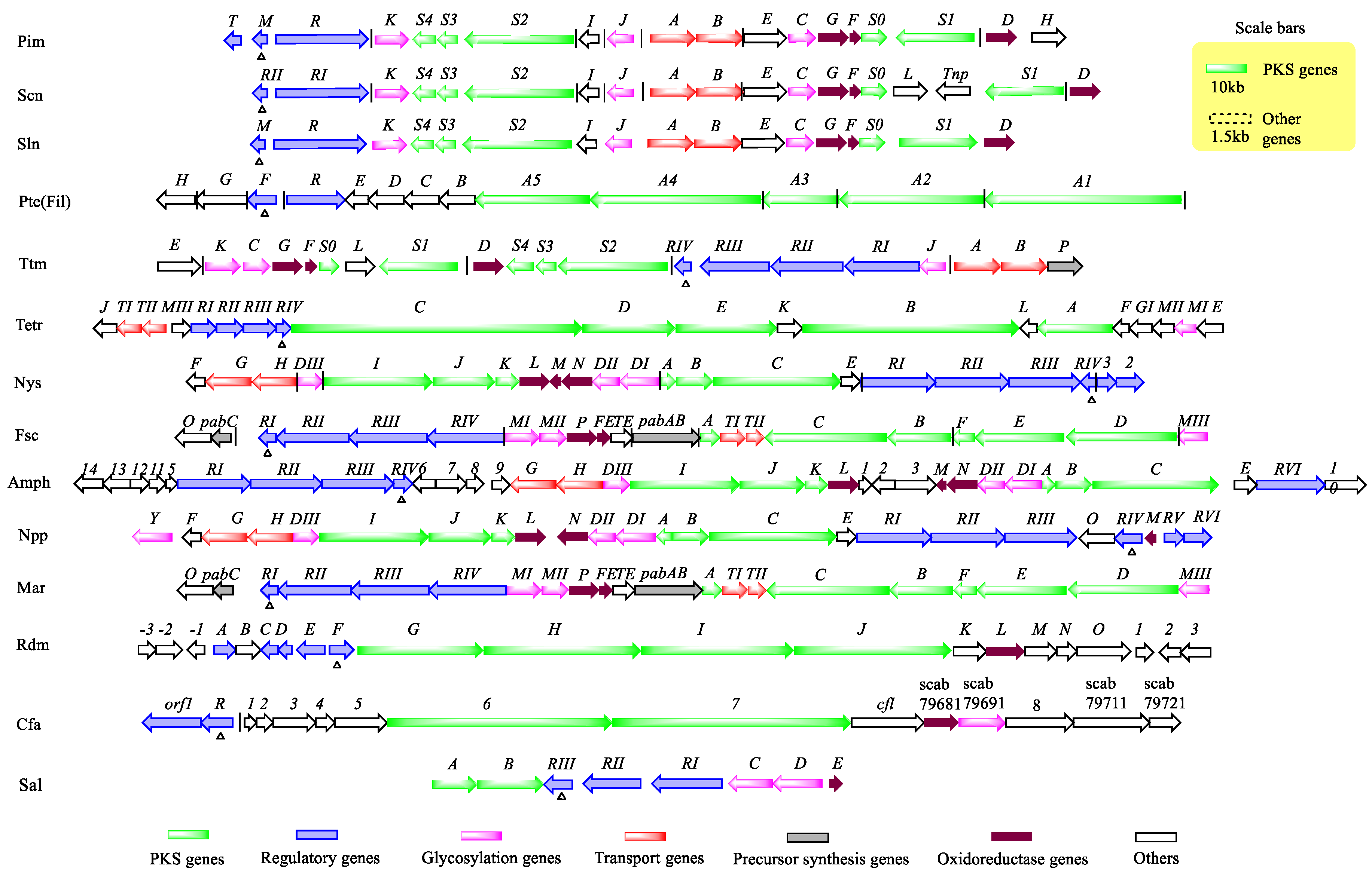
2.1. PAS-LuxR Family Regulators Are Present in the Type I Polyketide Synthase (PKS) Pathway
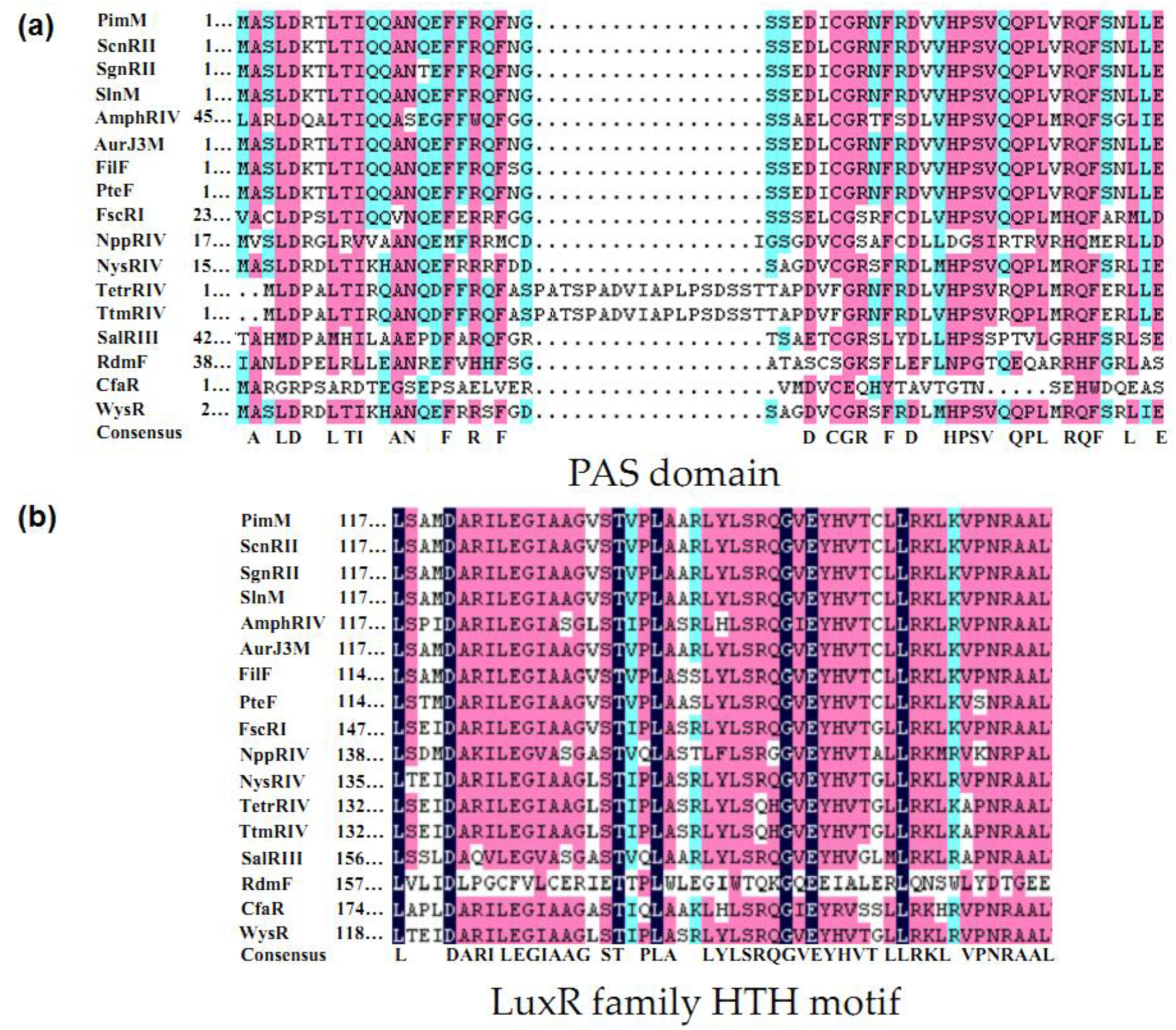
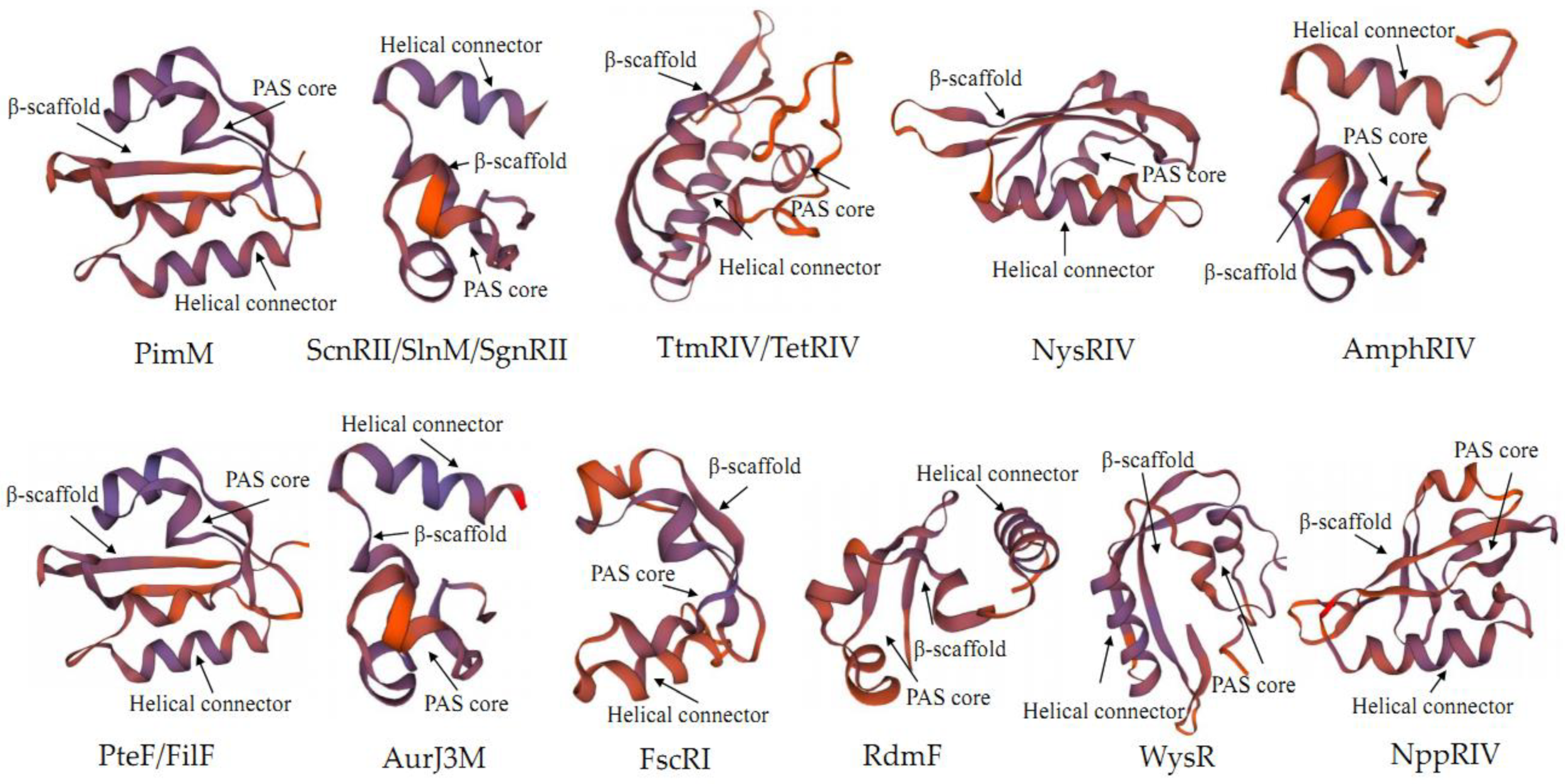
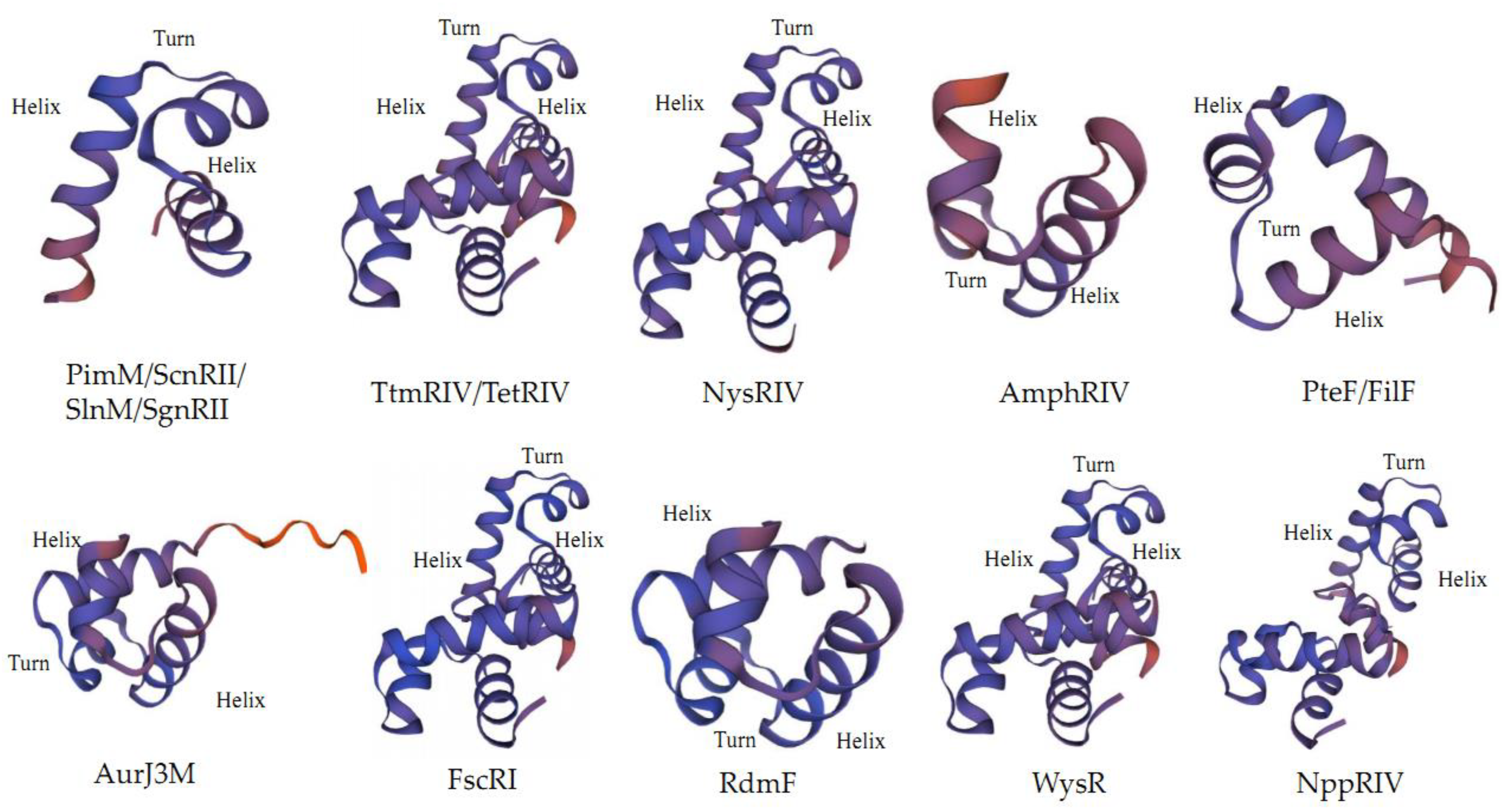
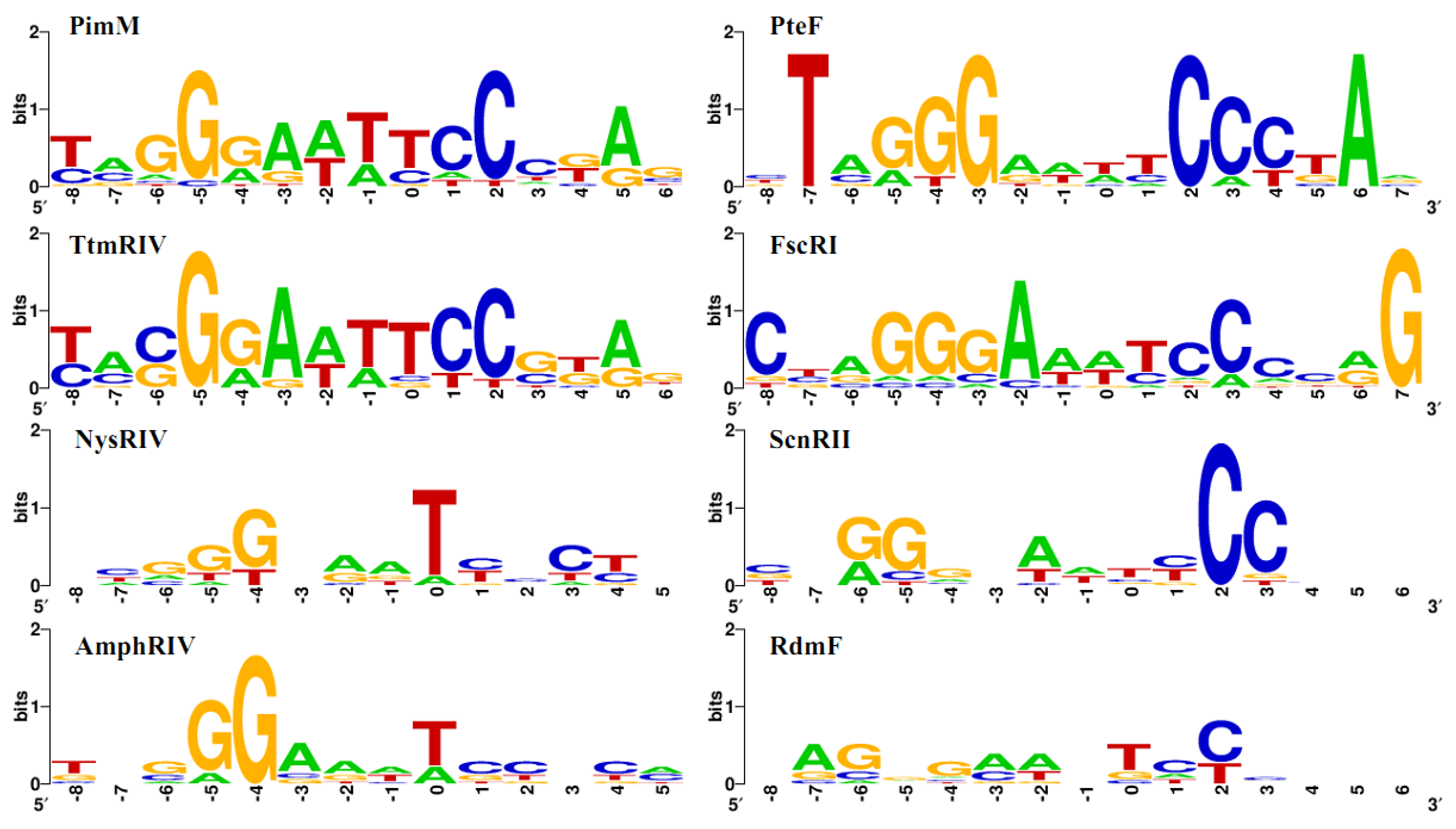
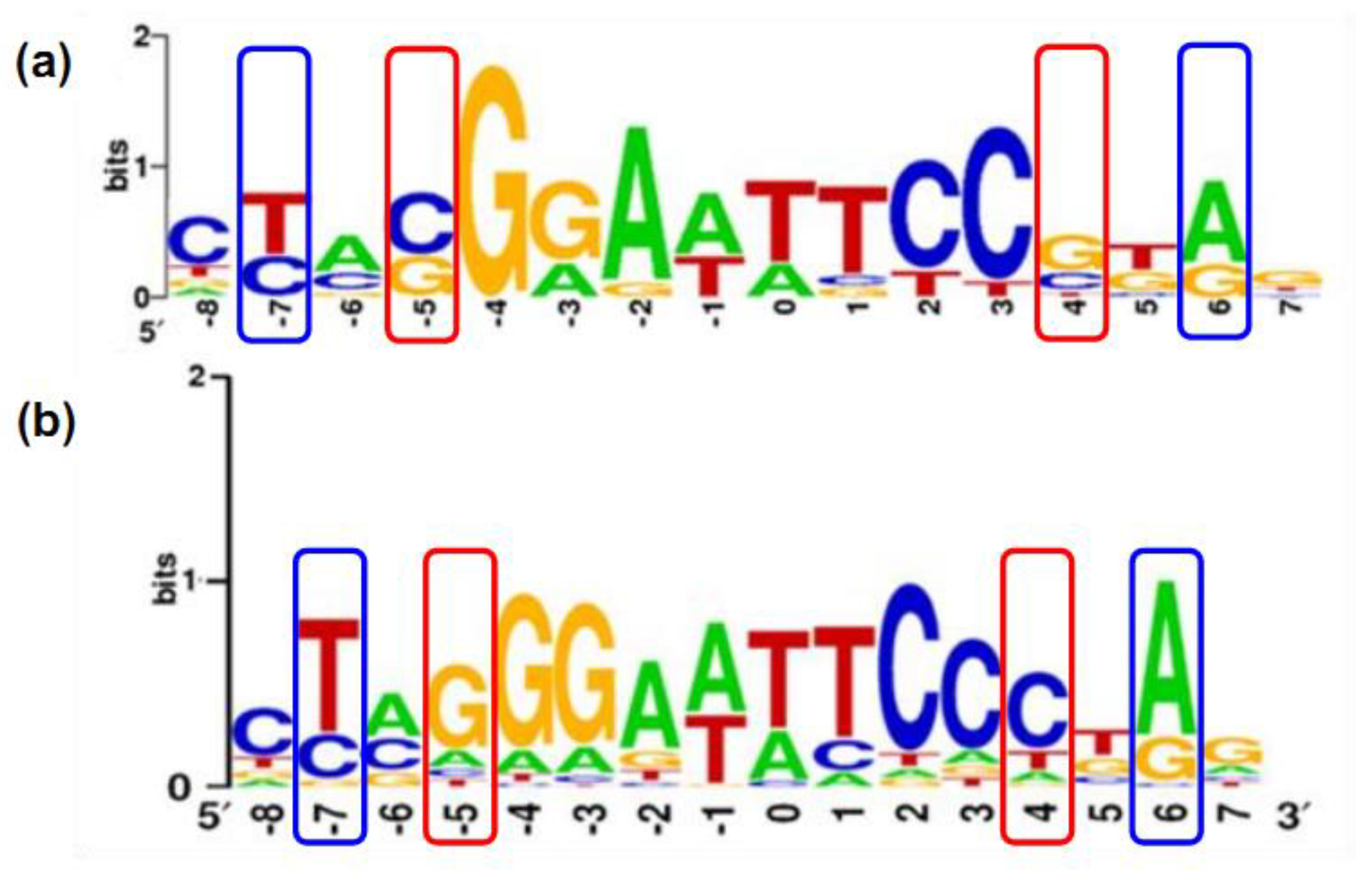
2.2. Structure of PAS-LuxR Family Regulators
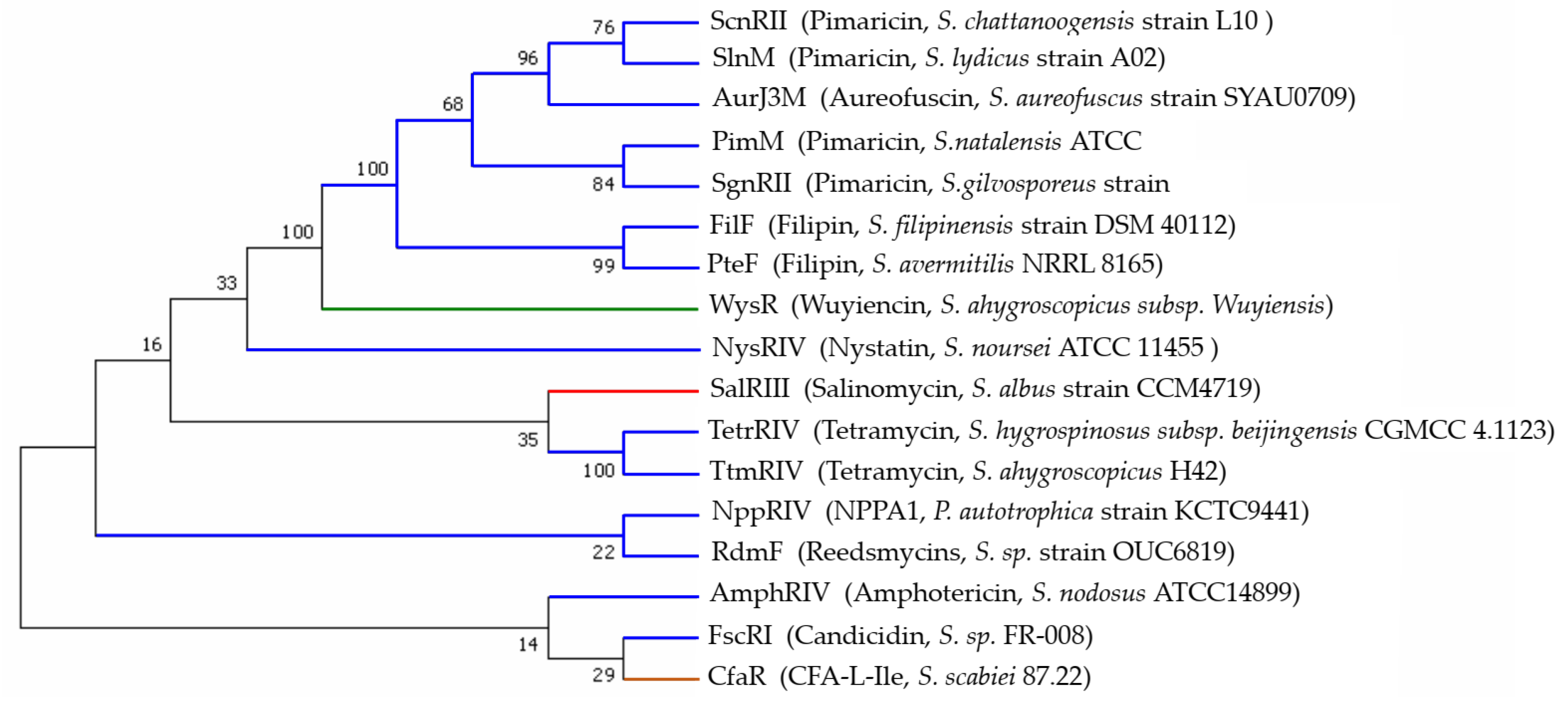
2.3. Developmental Tree Analysis
3. Regulation of Related Product Biosynthesis by PAS-LuxR Family Regulators
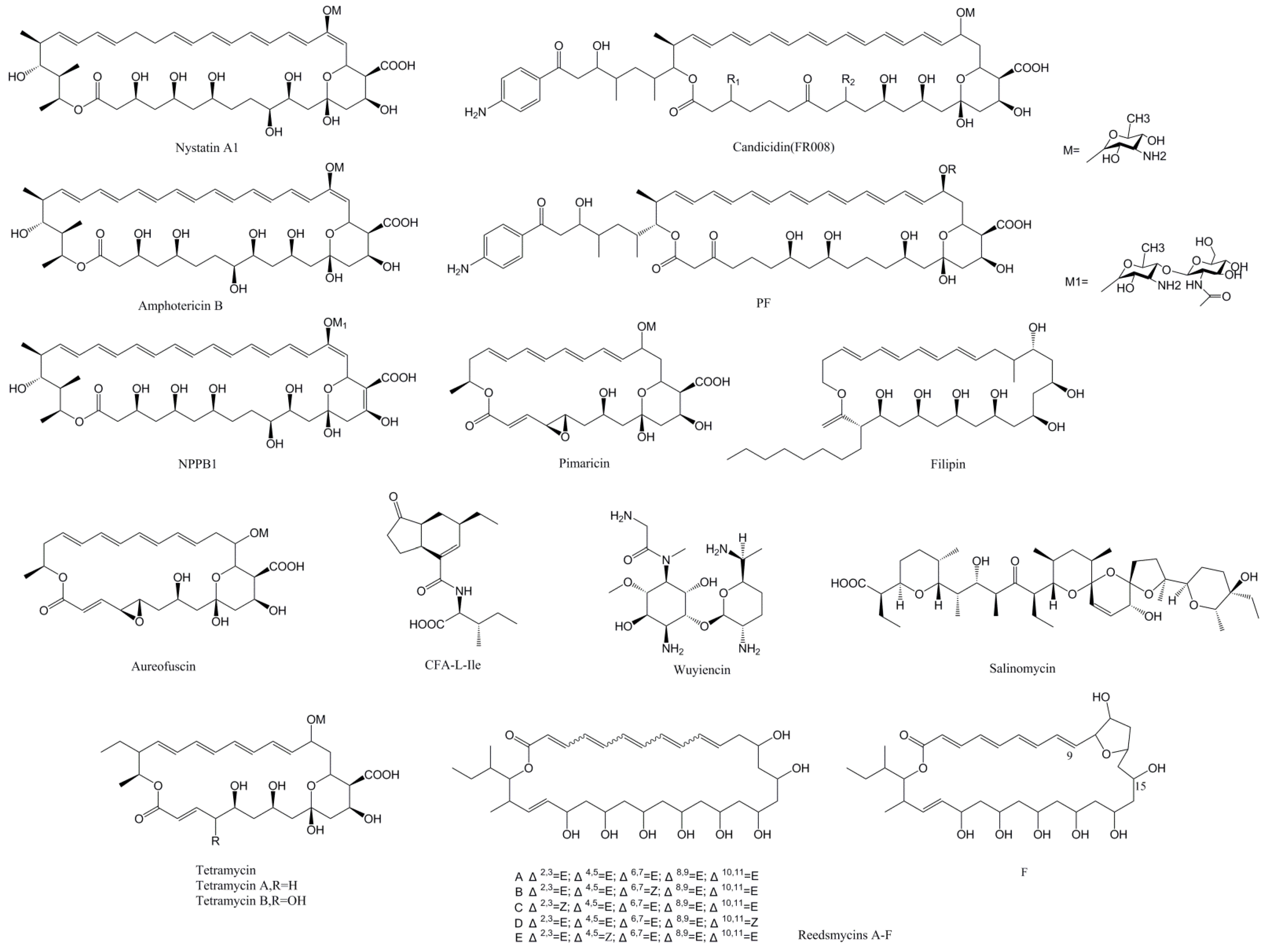
3.1. Regulation of Polyene Macrolide Biosynthesis by PAS-LuxR Family Regulators
3.1.1. Vertical Regulatory System Formed with SARP-LAL and PAS-LuxR Family Regulators
3.1.2. Network Regulatory System Formed with LAL and PAS-LuxR Family Regulators
3.1.3. Regulatory System Formed with PAS-LuxR and Other Family Regulators
| Regulator | Streptomyces | Size (aa) | Product | Type of Product | Target Genes | Yield in Overexpression Strain | Reference |
|---|---|---|---|---|---|---|---|
| PimM ScnRII SgnRII SlnM | S. natalensis (S.chattanoogensis) (S.gilvosporeus) (S. lydicus) | 192 | Pimaricin | PM | pimS0, pimS1, pimB, pimC, pimD, pimF, pimG, pimI, pimJ, pimS2, pimS3, pimS4, pimA, pimK scnA, scnE, scnD, scnI, scnJ, scnK, scnS1, scnS2 | 240–460% | [11,12,19] |
| RdmF | S. sp. CHQ-64 | 233 | Reedsmycins | NG-PM | NR | 250% | [13] |
| CfaR | S. scabiei | 152 | CFA-L-Ile | AAA | cfa1(27kb, from cfa1 to scab79721) | 1000% | [14] |
| WysR | S. wuyiensis CK-15 | 193 | Wuyiencin | AG | wysE, wysRI, wysRIII | [15] | |
| PteF FilF | S. avermitilis (S. filipinensis) (S. miharaensis) | 192 | Filipin | PM | pteA1, pteA2, pteA3, pteA4, pteA5, pteB, pteC, pteD, pteH, pteG, pteR | NR | [27,28,29] |
| TtmRIV TetrRIV | S. ahygroscopicus (S. hygrospinosus) | 201 | Tetramycin | PM | ttmK, ttmC, ttmS0, ttmS1, ttmS2, ttmS3, ttmS4, ttmE, ttmG, ttmF, ttmJ, ttmA, ttmB, ttmP | 333% (Tetramycin A) | [32,33] |
| NysRIV | S. noursei | 210 | Nystatin | PM | nysH, nysA, nysI, nysRI, nysRIV | NR | [35] |
| FscRI | S. sp. FR-008 | 222 | Candicidin | PM | fscA, fscB, fscC, fscD, fscE, fscF, fscMI, fscMII, fscMIII, fscTI, fscTII | NR | [36] |
| AmphRIV | S. nodosus | 243 | Amphotericin | PM | NR | 400% (in ΔamphNM) | [39] |
| NppRIV | P. autotrophica | 213 | NPPA1 | PM | NR | −50% | [40] |
| SalRIII | S. albus | 231 | Salinomycin | PIC | NR | NR | [42] |
| MarRI | S. marokkonensis M10 | 148 | PF | PM | NR | NR | [43] |
| AurJ3M | S.aureofuscus | 192 | Aureofuscin | PM | aurB, aurC, aurG, aurF, aurS0, aurS1, aurD, aurI, aurJ, aurA | 600% | [45] |
3.2. Regulation of the Nonpolyene Macrolides Biosynthesis by PAS-LuxR Family Regulators
3.3. Regulatory Mechanisms of PAS-LuxR Family Regulators
4. Application of PAS-LuxR Family Regulators
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Clardy, J.; Fischbach, M.A.; Walsh, C.T. New antibiotics from bacterial natural products. Nat. Biotechnol. 2006, 24, 1541–1550. [Google Scholar] [CrossRef] [PubMed]
- Novakova, R.; Rehakova, A.; Kutas, P.; Feckova, L.; Kormanec, J. The role of two SARP family transcriptional regulators in regulation of the auricin gene cluster in Streptomyces aureofaciens CCM 3239. Microbiology 2011, 157, 1629–1639. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; He, L.; Niu, G. Regulation of antibiotic biosynthesis in actinomycetes: Perspectives and challenges. Synth. Syst. Biotechnol. 2018, 3, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Bhukya, H.; Jana, A.K.; Sengupta, N.; Anand, R. Structural and dynamics studies of the TetR family protein, CprB from Streptomyces coelicolor in complex with its biological operator sequence. J. Struct. Biol. 2017, 198, 134–146. [Google Scholar] [CrossRef]
- Novichkov, P.S.; Kazakov, A.E.; Ravcheev, D.A.; Leyn, S.A.; Kovaleva, G.Y.; Sutormin, R.A.; Kazanov, M.D.; Riehl, W.; Arkin, A.P.; Dubchak, I.; et al. RegPrecise 3.0—A resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genom. 2013, 14, 745. [Google Scholar] [CrossRef] [PubMed]
- Romero-Rodriguez, A.; Robledo-Casados, I.; Sanchez, S. An overview on transcriptional regulators in Streptomyces. Biochim. Biophys. Acta 2015, 1849, 1017–1039. [Google Scholar] [CrossRef]
- Liu, G.; Chater, K.F.; Chandra, G.; Niu, G.; Tan, H. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 2013, 77, 112–143. [Google Scholar] [CrossRef]
- Taylor, B.L.; Zhulin, I.B.J.M.; Reviews, M.B. PAS domains: Internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 1999, 63, 479–506. [Google Scholar] [CrossRef]
- Santos-Aberturas, J.; Vicente, C.M.; Guerra, S.M.; Payero, T.D.; Martín, J.F.; Aparicio, J.F. Molecular control of polyene macrolide biosynthesis: Direct binding of the regulator PimM to eight promoters of pimaricin genes and identification of binding boxes. J. Boil. Chem. 2011, 286, 9150–9161. [Google Scholar] [CrossRef]
- Xiong, W.; Duan, Y.; Yan, X.; Huang, Y. Improvement of natural product production in Streptomyces by manipulating pathway-specific regulators. Sheng Wu Gong Cheng Xue Bao 2021, 37, 2127–2146. [Google Scholar] [CrossRef]
- Vicente, C.M.; Santos-Aberturas, J.; Guerra, S.M.; Payero, T.D.; Martin, J.F.; Aparicio, J.F. PimT, an amino acid exporter controls polyene production via secretion of the quorum sensing pimaricin-inducer PI-factor in Streptomyces natalensis. Microb. Cell Fact. 2009, 8, 33. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z. Proceedings of pimaricin biosynthesis. Chin. J. Antibiot. 2012, 37, 728–732. [Google Scholar] [CrossRef]
- Yao, T.; Liu, Z.; Li, T.; Zhang, H.; Liu, J.; Li, H.; Che, Q.; Zhu, T.; Li, D.; Li, W. Characterization of the biosynthetic gene cluster of the polyene macrolide antibiotic reedsmycins from a marine-derived Streptomyces strain. Microb. Cell Fact. 2018, 17, 98. [Google Scholar] [CrossRef] [PubMed]
- Bignell, D.R.D.; Seipke, R.F.; Huguet-Tapia, J.C.; Chambers, A.H.; Parry, R.J.; Loria, R. Streptomyces scabies 87-22 contains a coronafacic acid-like biosynthetic cluster that contributes to plant–microbe interactions. Mol. Plant Microbe Interact. 2010, 23, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Aung, A.; Liu, B.; Ma, J.; Shi, L.; Zhang, K.; Ge, B. Overexpression of wysR gene enhances wuyiencin production in DeltawysR3 mutant strain of Streptomyces albulus var. wuyiensis strain CK-15. J. Appl. Microbiol. 2020, 129, 565–574. [Google Scholar] [CrossRef]
- Liu, Y.; Ryu, H.; Ge, B.; Pan, G.; Sun, L.; Park, K.; Zhang, K. Improvement of Wuyiencin biosynthesis in Streptomyces wuyiensis CK-15 by identification of a key regulator, WysR. J. Microbiol. Biotechnol. 2014, 24, 1644–1653. [Google Scholar] [CrossRef]
- Mizuno, T.; Tanaka, I. Structure of the DNA-binding domain of the OmpR family of response regulators. Mol. Microbiol. 1997, 24, 665–667. [Google Scholar] [CrossRef]
- Hefti, M.H.; Francoijs, K.J.; de Vries, S.C.; Dixon, R.; Vervoort, J. The PAS fold. A redefinition of the PAS domain based upon structural prediction. Eur. J. Biochem. 2004, 271, 1198–1208. [Google Scholar] [CrossRef]
- Li, H.D.; Jin, Z.H.; Zhang, H.G.; Jin, H. Protoplast formation, regeneration and UV mutagenesis of natamycin producing Streptomyces gilvosporeus. J. Ind. Microbiol. Biot. 2008, 38, 43–46. [Google Scholar] [CrossRef]
- Santos-Aberturas, J.; Vicente, C.M.; Payero, T.D.; Martin-Sanchez, L.; Canibano, C.; Martin, J.F.; Aparicio, J.F. Hierarchical control on polyene macrolide biosynthesis: PimR modulates pimaricin production via the PAS-LuxR transcriptional activator PimM. PLoS ONE 2012, 7, e38536. [Google Scholar] [CrossRef]
- Du, Y.L.; Chen, S.F.; Cheng, L.Y.; Shen, X.L.; Tian, Y.; Li, Y.Q. Identification of a novel Streptomyces chattanoogensis L10 and enhancing its natamycin production by overexpressing positive regulator ScnRII. J. Microbiol. 2009, 47, 506–513. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.L.; Li, S.Z.; Zhou, Z.; Chen, S.F.; Fan, W.M.; Li, Y.Q. The pleitropic regulator AdpAch is required for natamycin biosynthesis and morphological differentiation in Streptomyces chattanoogensis. Microbiology 2011, 157, 1300–1311. [Google Scholar] [CrossRef]
- Du, Y.L.; Shen, X.L.; Yu, P.; Bai, L.Q.; Li, Y.Q. Gamma-butyrolactone regulatory system of Streptomyces chattanoogensis links nutrient utilization, metabolism, and development. Appl. Environ. Microbiol. 2011, 77, 8415–8426. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z. The Study of the Pathway Specific Regulation of Natamycin Biosynthesis in Streptomyces chattanoogensis. Master’s Thesis, Zhejiang University, Hangzhou, China, 2013. [Google Scholar]
- Wu, H.; Liu, W.; Dong, D.; Li, J.; Zhang, D.; Lu, C. SlnM gene overexpression with different promoters on natamycin production in Streptomyces lydicus A02. J. Ind. Microbiol. Biotechnol. 2014, 41, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tao, Z.; Zheng, H.; Zhang, F.; Long, Q.; Deng, Z.; Tao, M. Iteratively improving natamycin production in Streptomyces gilvosporeus by a large operon-reporter based strategy. Metab. Eng. 2016, 38, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, H.; Shin-ya, K.; Omura, S. Genome mining of the Streptomyces avermitilis genome and development of genome-minimized hosts for heterologous expression of biosynthetic gene clusters. J. Ind. Microbiol. Biotechnol. 2014, 41, 233–250. [Google Scholar] [CrossRef]
- Payero, T.D.; Vicente, C.M.; Rumbero, Á.; Barreales, E.G.; Santos-Aberturas, J.; de Pedro, A.; Aparicio, J.F. Functional analysis of filipin tailoring genes from Streptomyces filipinensis reveals alternative routes in filipin III biosynthesis and yields bioactive derivatives. Microb. Cell Factories 2015, 14, 114. [Google Scholar] [CrossRef]
- Kim, J.D.; Han, J.W.; Hwang, I.C.; Lee, D.; Kim, B.S. Identification and biocontrol efficacy of Streptomyces miharaensis producing filipin III against Fusarium wilt. J. Basic Microbiol. 2012, 52, 150–159. [Google Scholar] [CrossRef]
- Vicente, C.M.; Santos-Aberturas, J.; Payero, T.D.; Barreales, E.G.; de Pedro, A.; Aparicio, J.F. PAS-LuxR transcriptional control of filipin biosynthesis in S. avermitilis. Appl. Microbiol. Biotechnol. 2014, 98, 9311–9324. [Google Scholar] [CrossRef]
- Barreales, E.G.; Vicente, C.M.; de Pedro, A.; Santos-Aberturas, J.; Aparicio, J.F. Promoter Engineering Reveals the Importance of Heptameric Direct Repeats for DNA Binding by Streptomyces Antibiotic Regulatory Protein-Large ATP-Binding Regulator of the LuxR Family (SARP-LAL) Regulators in Streptomyces natalensis. Appl. Environ. Microbiol. 2018, 84, e00246-18. [Google Scholar] [CrossRef]
- Cui, H.; Ni, X.; Liu, S.; Wang, J.; Sun, Z.; Ren, J.; Su, J.; Chen, G.; Xia, H. Characterization of three positive regulators for tetramycin biosynthesis in Streptomyces ahygroscopicus. FEMS Microbiol. Lett. 2016, 363, fnw109. [Google Scholar] [CrossRef][Green Version]
- Cao, B.; Yao, F.; Zheng, X.; Cui, D.; Shao, Y.; Zhu, C.; Deng, Z.; You, D. Genome mining of the biosynthetic gene cluster of the polyene macrolide antibiotic tetramycin and characterization of a P450 monooxygenase involved in the hydroxylation of the tetramycin B polyol segment. Chembiochem 2012, 13, 2234–2242. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Ni, X.; Shao, W.; Su, J.; Su, J.; Ren, J.; Xia, H. Functional manipulations of the tetramycin positive regulatory gene ttmRIV to enhance the production of tetramycin A and nystatin A1 in Streptomyces ahygroscopicus. J. Ind. Microbiol. Biotechnol. 2015, 42, 1273–1282. [Google Scholar] [CrossRef]
- Sekurova, O.N.; Brautaset, T.; Sletta, H.; Borgos, S.E.; Jakobsen, M.O.; Ellingsen, T.E.; Strom, A.R.; Valla, S.; Zotchev, S.B. In vivo analysis of the regulatory genes in the nystatin biosynthetic gene cluster of Streptomyces noursei ATCC 11455 reveals their differential control over antibiotic biosynthesis. J. Bacteriol. 2004, 186, 1345–1354. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Huang, X.; Zhou, X.; Bai, L.; He, J.; Jeong, K.J.; Lee, S.Y.; Deng, Z. Organizational and mutational analysis of a complete FR-008/candicidin gene cluster encoding a structurally related polyene complex. Chem. Biol. 2003, 10, 1065–1076. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Xu, W.; Zhang, J.; Zhang, P.; Zhao, Z.; Sheng, D.; Ma, W.; Zhang, Y.Z.; Bai, L.; Pang, X. A Hierarchical Network of Four Regulatory Genes Controlling Production of the Polyene Antibiotic Candicidin in Streptomyces sp. Strain FR-008. Appl. Environ. Microbiol. 2020, 86, e00055-20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhao, Z.; Li, H.; Chen, X.L.; Deng, Z.; Bai, L.; Pang, X. Production of the antibiotic FR-008/candicidin in Streptomyces sp. FR-008 is co-regulated by two regulators, FscRI and FscRIV, from different transcription factor families. Microbiology 2015, 161, 539–552. [Google Scholar] [CrossRef]
- Kim, H.J.; Han, C.Y.; Park, J.S.; Oh, S.H.; Kang, S.H.; Choi, S.S.; Kim, J.M.; Kwak, J.H.; Kim, E.S. Nystatin-like Pseudonocardia polyene B1, a novel disaccharide-containing antifungal heptaene antibiotic. Sci. Rep. 2018, 8, 13584. [Google Scholar] [CrossRef]
- Jeon, H.G.; Seo, J.; Lee, M.J.; Han, K.; Kim, E.S. Analysis and functional expression of NPP pathway-specific regulatory genes in Pseudonocardia autotrophica. J. Ind. Microbiol. Biotechnol. 2011, 38, 573–579. [Google Scholar] [CrossRef]
- Han, C.Y.; Jang, J.Y.; Kim, H.J.; Choi, S.; Kim, E.S. Pseudonocardia strain improvement for stimulation of the disugar heptaene Nystatin-like Pseudonocardia polyene B1 biosynthesis. J. Ind. Microbiol. Biotechnol. 2019, 46, 649–655. [Google Scholar] [CrossRef]
- Knirschová, R.; Nováková, R.; Fecková, L.; Timko, J.; Turňa, J.; Bistáková, J.; Kormanec, J. Multiple regulatory genes in the salinomycin biosynthetic gene cluster of Streptomyces albus CCM 4719. Folia Microbiol. 2007, 52, 359–365. [Google Scholar] [CrossRef]
- Chen, L.; Lai, Y.M.; Yang, Y.L.; Zhao, X. Genome mining reveals the biosynthetic potential of the marine-derived strain Streptomyces marokkonensis M10. Synth. Syst. Biotechnol. 2016, 1, 56–65. [Google Scholar] [CrossRef]
- Tang, J.; Liu, X.; Peng, J.; Tang, Y.; Zhang, Y. Genome sequence and genome mining of a marine-derived antifungal bacterium Streptomyces sp. M10. Appl. Microbiol. Biotechnol. 2015, 99, 2763–2772. [Google Scholar] [CrossRef]
- Yang, J.; Xu, D.; Yu, W.; Hao, R.; Wei, J. Regulation of aureofuscin production by the PAS-LuxR family regulator AurJ3M. Enzym. Microb. Technol. 2020, 137, 109532. [Google Scholar] [CrossRef]
- Wei, J.; Meng, X.; Wang, Q. Enhanced production of aureofuscin by over-expression of AURJ3M, positive regulator of aureofuscin biosynthesis in Streptomyces aureofuscus. Lett. Appl. Microbiol. 2011, 52, 322–329. [Google Scholar] [CrossRef]
- Wei, J.; Yu, W.; Hao, R.; Yang, J.; Xu, D.; Gao, J.J. Regulatory effect and mechanism of aurT gene on biosynthesis of aureofuscin. Food. Sci. Technol. Mys 2021, 39, 70–77. [Google Scholar] [CrossRef]
- Han, X.; Liu, Z.; Zhang, Z.; Zhang, X.; Zhu, T.; Gu, Q.; Li, W.; Che, Q.; Li, D. Geranylpyrrol A and Piericidin F from Streptomyces sp. CHQ-64 DeltardmF. J. Nat. Prod. 2017, 80, 1684–1687. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Bown, L.; Tahlan, K.; Bignell, D.R. Regulation of coronafacoyl phytotoxin production by the PAS-LuxR family regulator CfaR in the common scab pathogen Streptomyces scabies. PLoS ONE 2015, 10, e0122450. [Google Scholar] [CrossRef]
- Santos-Aberturas, J.; Payero, T.D.; Vicente, C.M.; Guerra, S.M.; Canibano, C.; Martin, J.F.; Aparicio, J.F. Functional conservation of PAS-LuxR transcriptional regulators in polyene macrolide biosynthesis. Metab. Eng. 2011, 13, 756–767. [Google Scholar] [CrossRef] [PubMed]
- McLean Thomas, C.; Hoskisson Paul, A.; Seipke Ryan, F. Coordinate Regulation of Antimycin and Candicidin Biosynthesis. mSphere 2016, 1, e00305-16. [Google Scholar] [CrossRef] [PubMed]
- Olano, C.; García, I.; González, A.; Rodriguez, M.; Rozas, D.; Rubio, J.; Sánchez-Hidalgo, M.; Braña, A.F.; Méndez, C.; Salas, J.A. Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb. Biotechnol. 2014, 7, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Vicente, C.M.; Payero, T.D.; Rodríguez-García, A.; Barreales, E.G.; Pedro, A.d.; Santos-Beneit, F.; Aparicio, J.F.J.A. Modulation of multiple gene clusters’ expression by the PAS-LuxR transcriptional regulator PteF. Antibiotics 2022, 11, 994. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, P.; Murphy, C.D.; Caffrey, P. Exploiting the genome sequence of Streptomyces nodosus for enhanced antibiotic production. Appl. Microbiol. Biotechnol. 2016, 100, 1285–1295. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.G.; Lee, M.J.; Seo, J.; Hwang, Y.B.; Lee, M.Y.; Han, K.; Sherman, D.H.; Kim, E.S. Identification of functionally clustered nystatin-like biosynthetic genes in a rare actinomycetes, Pseudonocardia autotrophica. J. Ind. Microbiol. Biotechnol. 2009, 36, 1425–1434. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, N.; Dong, Y.; Zhou, H.; Cui, H. Effect of PAS-LuxR Family Regulators on the Secondary Metabolism of Streptomyces. Antibiotics 2022, 11, 1783. https://doi.org/10.3390/antibiotics11121783
Zhang N, Dong Y, Zhou H, Cui H. Effect of PAS-LuxR Family Regulators on the Secondary Metabolism of Streptomyces. Antibiotics. 2022; 11(12):1783. https://doi.org/10.3390/antibiotics11121783
Chicago/Turabian StyleZhang, Naifan, Yao Dong, Hongli Zhou, and Hao Cui. 2022. "Effect of PAS-LuxR Family Regulators on the Secondary Metabolism of Streptomyces" Antibiotics 11, no. 12: 1783. https://doi.org/10.3390/antibiotics11121783
APA StyleZhang, N., Dong, Y., Zhou, H., & Cui, H. (2022). Effect of PAS-LuxR Family Regulators on the Secondary Metabolism of Streptomyces. Antibiotics, 11(12), 1783. https://doi.org/10.3390/antibiotics11121783





