Abstract
The occurrence of waterborne antimicrobial-resistant (AMR) bacteria in areas of high-density oyster cultivation is an ongoing environmental and public health threat given the popularity of shellfish consumption, water-related human recreation throughout coastal Thailand, and the geographical expansion of Thailand’s shellfish industry. This study characterized the association of phenotypic and genotypic AMR, including extended-spectrum β-lactamase (ESBL) production, and virulence genes isolated from waterborne Escherichia coli (E. coli) (n = 84), Salmonella enterica (S. enterica) subsp. enterica (n = 12), Vibrio parahaemolyticus (V. parahaemolyticus) (n = 249), and Vibrio cholerae (V. cholerae) (n = 39) from Thailand’s coastal aquaculture regions. All Salmonella (100.0%) and half of V. cholerae (51.3%) isolates harbored their unique virulence gene, invA and ompW, respectively. The majority of isolates of V. parahaemolyticus and E. coli, ~25% of S. enterica subsp. enterica, and ~12% of V. cholerae, exhibited phenotypic AMR to multiple antimicrobials, with 8.9% of all coastal water isolates exhibiting multidrug resistance (MDR). Taken together, we recommend that coastal water quality surveillance programs include monitoring for bacterial AMR for food safety and recreational water exposure to water for Thailand’s coastal water resources.
1. Introduction
AMR is an important One Health concept involving interconnectedness between humans, animals, and their shared environment, that can pose serious health threats to humans and animals. Every year, more than 700,000 deaths are attributed to AMR infection, and it is estimated that the number of deaths from MDR bacteria will increase to 10 million people by 2050 [1]. The extensive use of antimicrobials in human medicine, veterinary medicine, agriculture, and aquaculture has been implicated in the emergence and dissemination of AMR in the environment. The global consumption of antimicrobials in humans increased more than 60% between 2000 and 2015, especially in low- and middle-income countries, due to simple accessibility and irrational use [2]. The excessive use of antimicrobials directly affects the bacterial community and contributes to the overall selective pressure for AMR in aquatic environments [3]. Moreover, contamination from biocides and heavy metals in aquatic environments can also contribute to AMR selection due to their cross-resistance [4].
Studies of AMR in humans and animals have been widely investigated in contrast to environmental monitoring and surveillance of AMR. Currently, AMR bacteria derived from environmental pollutants are of particular concern since they have become a significant emerging health threat, according to the United Nations Environment Program (UNEP) [5]. AMR and virulence bacteria from anthropogenic activities have been circulated in groundwater and surface water supplied to human communities [6,7]. Water treatment plants are implicated as giant reservoirs of AMR bacteria due to the diversity of bacteria, which includes virulent bacterial strains, AMR, heavy metals, and bioactive ingredients that can promote the likelihood of AMR development [8,9]. The terminal discharge for human wastewater and sewage can often be in coastal areas [10,11]. As a consequence, marine aquatic environments have been identified as critical regional hotspots for investigating how AMR can develop, persist, and disseminate throughout these locations.
Coastal aquaculture is often decentralized and of small scale, but growing consumer demand for cultivated seafood can increase water pollution and enhance the spread of emerging diseases and bacterial contaminants [12]. For example, E. coli and Salmonella spp. are often found in estuarine water from potential sources such as vertebrate animals, terrestrial runoff from rainfall, or human activities such as sewage or aquaculture, while Vibrio spp. are natural inhabitants of marine environments [13,14]. Shiga toxin-producing E. coli (STEC) are considered major pathogenic strains that can cause adverse health effects in humans and normally carry stx1 and/or stx2, which encode for Shiga toxins [15]. STEC infection in humans can cause stomach cramps, diarrhea, vomiting, and may be associated with severe consequences [16]. Pathogenic Salmonella spp. carry invA, which allow bacteria to invade epithelium cells of the human gastrointestinal tract [17]. Therefore, the invA gene has been widely used as a genetic marker to confirm the presence of Salmonella isolates [18].
Vibrio spp. that can cause human gastrointestinal infection are V. cholerae and V. parahaemolyticus. Both species produce virulent toxins that cause disease in humans: V. cholerae uses ctxA to encode cholerae toxin while V. parahaemolyticus uses tdh and trh to encode thermostable direct hemolysin and TDH-related hemolysin [19,20]. V. cholerae also harbors specific outer membrane proteins and virulence factors encoded from ompW, which can be used as a specific molecular indicator of the species [19]. These virulent bacteria can also harbor AMR genes and become AMR reservoirs that can potentially transfer resistant genes to neighboring bacteria through various horizontal gene transfer mechanisms [21]. Moreover, bacteria that harbor integrons or integrative and conjugative elements can function as effective reservoirs and donors of AMR genes that collectively increase the widespread dissemination of AMR [22,23].
It is estimated that up to 90% of antimicrobial use can be excreted as an active compound or metabolite into the environment, which in turn can foster emergence of AMR and dissemination of resistance determinants in both freshwater and coastal systems [24]. Additionally, the high traditional usage of antimicrobials in Thailand’s aquaculture industry can further select for AMR and disperse associated genetic determinants in estuaries and coastal environments used for shellfish cultivation. Therefore, the objectives of this study were to examine the distribution of bacterial phenotypic and genotypic AMR, their resistance determination, ESBL production, and virulence genes in E. coli, Salmonella spp., V. cholerae, and V. parahaemolyticus isolated from coastal water, and to characterize the association between resistance, virulence factors, and ESBL production in these bacterial species from the coastal aquaculture regions.
2. Results
2.1. Bacterial Confirmation and Identification of Virulence Factors
A total of 384 bacterial isolates including E. coli (n = 84), S. enterica subsp. enterica (subspecies I) (n = 12), V. parahaemolyticus (n = 249), and V. cholerae (n = 39) were isolated from seawater samples from Thailand’s coastal aquaculture regions and subjected to antimicrobial susceptibility testing (AST). All S. enterica subsp. enterica isolates were positive for invA, and 98.4% (n = 245/249) of V. parahaemolyticus isolates were positive for tlh, which is a species-specific gene for V. parahaemolyticus. The two virulence factors tdh and trh were absent from this group of V. parahaemolyticus isolates. More than half of the V. cholerae isolates (51.3%, n = 20) were positive for ompW, which is a species-specific gene and virulence factor; in contrast, all 20 isolates were negative for the virulence gene ctx, which encodes the cholera toxin. Neither stx1 nor stx2 were detected in the 84 E. coli isolates.
2.2. Serotyping of S. enterica subsp. enterica and V. cholerae
The distribution of serovars among the twelve seawater isolates of S. enterica subsp. enterica were Bolton (n = 1), Braenderup (n = 1), Bruebach (n = 1), Chester (n = 1), Lamberhurst (n = 2), Othmarschen (n = 2), Paratyphi B (n = 1), Wentworth (n = 1), Litchfield (n = 1), and Orion (n = 1). None of the 20 isolates of V. cholerae had O1, O139, or O141.
2.3. Phenotypic Resistance
The relative frequency of phenotypic AMR for the 384 bacterial isolates (n = 384) was ampicillin (AMP) (52.1%), followed by trimethoprim (TRI) (18.2%), sulfamethoxazole (SUL) (15.9%), streptomycin (STR) (3.4%), and ceftazidime (CAZ) (2.3%) (Table 1). Very few isolates (0.5%) exhibited resistance to ciprofloxacin (CIP), cefotaxime (CTX), and cefpodoxime (CPD), with only 0.3% exhibiting resistance to gentamicin (GEN).

Table 1.
Phenotypic characterization of AMR in E. coli, Salmonella, V. parahaemolyticus, and V. cholerae isolated from seawater from coastal Thailand.
Twenty-five AMR patterns were observed. Among these patterns, AMP (27.6%), AMP-TRI (8.1%), AMP-SUL (5.7%), and TRI (3.4%) were predominant (Table 2). Resistance to a variety of antimicrobial classes was found in E. coli and V. parahaemolyticus (Figure 1); in contrast, V. cholerae (82.1%) and S. enterica subsp. enterica (75.0%) isolates were mostly susceptible to antimicrobials. MDR was observed in 8.9% of all the bacterial isolates, with E. coli (25.0%) and S. enterica subsp. enterica (16.7%) as the main species conferring the most MDR.

Table 2.
Phenotypic resistance patterns of E. coli, S. enterica subsp. enterica, V. parahaemolyticus, and V. cholerae isolated from seawater from coastal Thailand.
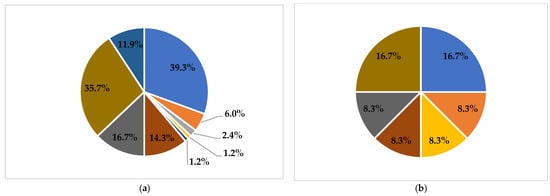
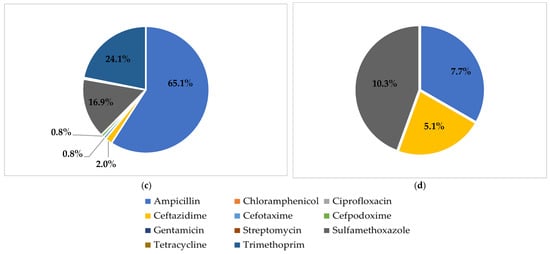
Figure 1.
Proportion of phenotypic AMR for different species of bacteria. (a): E. coli; (b): S. enterica subsp. enterica; (c): V. parahaemolyticus; (d): V. cholerae.
2.4. ESBL Production
No ESBL production was detected in any bacterial isolate from the coastal water samples. However, the prevalence of resistance to ceftazidime (2.3%) was higher than for cefotaxime and cefpodoxime (0.5%).
2.5. Genotypic Resistance
The most common AMR genes detected in this study were blaTEM (5.5%), tetA (3.7%), qnrS (1.8%), strA (1.6%), and floR (1.3%) (Table 3), which were predominately found in E. coli. The prevalence of mobile genetic elements such as integrase (int1) was low, at 1.8% (n = 7 of E. coli isolates), while no int2 and int3 were detected (Table 3). Interestingly, four out of seven of the positive int1 isolates were MDR. ICEs (intSXT) were detected in one isolate of V. cholerae. None of the bacterial isolates had mcr-1, mcr-2, and mcr-3.

Table 3.
Genotypic characterization of AMR and their determinants.
2.6. Association between Phenotypic and Genotypic Characterization of Resistance
The association between resistance phenotype, genotype, and resistance determinants was examined for all isolates using Cohen’s kappa coefficient (Table 4). The highest strength of association was observed in four pairs of MDR vs. TET (61%), STR vs. strA (0.62), CHL vs. floR (72%), and MDR vs. floR (72%) (p < 0.0001).

Table 4.
Kappa analysis among phenotype, genotype, and resistance determinants (n = 384).
Based on multivariate logistic regression, ampicillin-resistant isolates were positively associated with the presence of blaTEM (OR = 20.3; p < 0.0001). In another analysis, trimethoprim-resistant isolates were associated with the presence of MDR (OR = 5.7; p < 0.0001) and int1 (OR = 4.7; p = 0.015) (Table 5).

Table 5.
Multivariate logistic regression analysis of factors associated with ampicillin resistance and MDR in coastal seawater samples stratified by bacterial species (n = 384).
3. Discussion
Investigation of AMR in the environment is challenging because the bacteria have acquired multiple mechanisms to confer AMR and MDR. This study focused on the distribution of AMR phenotypes and genotypes, and the virulence of E. coli, S. enterica subsp. enterica, V. parahaemolyticus, and V. cholerae isolated from coastal seawater in dense cultivation areas. Tracking resistant bacteria from coastal seawater is important under a One Health perspective, especially for aquatic products that are usually consumed raw or partially cooked, such as oysters. Therefore, contaminated water with resistant bacteria that is used for coastal aquaculture can pose a serious health risk to human health. Among the 384 bacterial isolates from coastal waters from Thailand’s oyster-producing regions, the most common resistance phenotype was to ampicillin (52.1%), which is consistent with previous studies that also found a high prevalence of ampicillin resistance in coastal water [25,26,27].
The overall prevalence of MDR in this study was less than 10%, with the majority of the MDR bacteria being E. coli (25.0%) and S. enterica subsp. enterica (16.7%). In this study, E. coli also had the highest diversity of resistance patterns compared to other bacterial species, indicating that different bacterial species from the same coastal environment can acquire and/or maintain different resistance traits. This finding indicates that E. coli is potentially a reservoir and/or mode of introduction of drug resistance for coastal environments used for shellfish production in Thailand. A recent study indicated that the sources of bacterial contamination that causes marine pollution were anthropogenic activities, aquaculture, agriculture, industry, etc. [28]. In low- and middle-income countries, untreated water from inadequate and ineffective facilities has been shown to be a significant source of bacterial contamination [29]. Thermotolerant coliforms are the main bacteria in fecal coliforms that are usually present in the intestinal tracts of human and warm-blooded animals, which can indicate fecal contamination in humans, animals, water, food, and the environment [30,31]. In Thailand, the Department of Pollution Control sets the microbiological standards of coastal seawater (total coliforms, thermotolerant coliforms, and Enterococci), but does not regulate for the presence of resistant bacteria. Therefore, guidelines may be needed to reduce AMR bacterial contamination in coastal environments, especially for regions used for shellfish production and human recreation.
Among the 384 isolates, all S. enterica subsp. enterica isolates (n = 12) harbored invA. A diversity of S. enterica subsp. enterica serovars was observed in this study, indicating that multiple sources of contamination may exist for this bacterial pathogen. For example, S. enterica subsp. enterica serovars Lamberhurst and Othmarschen were reported in poultry and humans [32,33]. S. enterica subsp. enterica serovars Braenderup, Bruebach, Chester, Paratyphi B, Wentworth, Litchfield, and Orion have been isolated from shell eggs, papaya, wastewater, companion animals, livestock animals, and humans [34,35,36,37,38]. The stx1 and stx2 genes are bacterial toxins found in various serogroups of E. coli, but neither stx1 nor stx2 was detected in this study. Given our sample size of 84 E. coli, perhaps it is not surprising that stx genes are not found in this bank of E. coli, given that it has been estimated that one in a 1000 fecal coliform isolates harbors the stx gene [39,40].
The tdh (thermostable direct hemolysin) and trh (TDH-related hemolysin) are major virulence factors of V. parahaemolyticus. In this study, both genes were not present in our coastal water isolates. However, other virulence indicators, including type 3 secretion systems T3SS1 and T3SS2β found in pathogenic V. parahaemolyticus, have been reported in seafood samples [41]. For V. cholerae, more than half (51.3%) of the isolates were positive for ompW, which codes for an outer membrane protein, but none harbored the ctx gene which codes for the cholera toxin. This result agrees with previous studies showing that isolates of environmental V. cholerae are often not positive for the ompW gene [42,43]. The V. cholerae serogroups O1, O139, and O141 were also not found in this study. The distribution of serogroup O1 has been reported in environmental samples in northern Thailand, which contrasts with the results of this study [44]. However, non O1/O139 isolated from coastal area of southern Thailand was reported in humans with gastroenteritis [45].
ESBL-producing bacteria have been implicated in impacts on public health due to limited therapeutic options following human infections. The rapid spread of ESBL-producing bacteria in coastal environments increases concerns because of the widespread occurrence of gram-negative bacteria. In this study, ESBL producers were not detected in any of our coastal bacterial isolates. However, the cefotaxime-hydrolysing β-lactamase isolated in Munich (CTX-M) families, including CTX-M-15, CTX-M-14, and CTX-M-27, have been commonly found worldwide [46]. Previous studies reported ESBL producers in lagoon, recreational water, and wastewater [47,48]. Moreover, the spread of carbapenem-resistant Enterobacterales has been an emerging threat in coastal and estuarine water [49]. The blaTEM (5.5%) was the main resistance gene detected in this study, which agrees with a previous study [50]. Inversely, a previous study of E. coli isolated from coastal water contained a variety of blaCTX-M genes [51]. Other genes, including tetA (3.7%), qnrS (1.8%), strA (1.6%), and floR (1.3%), were detected at low prevalence this study. On the other hand, sul1 and sul2 were the most abundant AMR genes in the coastal mariculture system in China [52]. The absence of mcr-1, mcr-2, and mcr-3 was observed in this study, which is in contrast to a recent study in Brazil which found mcr-1 in E. coli isolated from coastal waters [53]. These differences in the geographical distribution of AMR contamination for coastal environments suggest that the processes of AMR contamination and persistence can vary widely from region to region.
Novel resistance mechanisms are associated with mobile genetic elements, which can facilitate the widespread dissemination of resistance determinants in the environment [54]. In this study, integrase (int1) was observed at a low level (1.8%), limited mostly to isolates of MDR E. coli. This observation raises concern of transferable MDR genes between intra- and inter-bacterial species, because integrons are located in transferable plasmids and conserved DNA sequences carrying gene cassettes with resistance genes, which can facilitate the spread of multiple resistant genes simultaneously [55]. In this study, one isolate of V. cholerae had intSXT. As a consequence, it may be prudent for coastal water quality monitoring programs to also include AMR surveillance for these mobile genetic elements.
Cohen’s kappa analyses are generally used to test the agreement between two test methods. This study found strong associations (kappa agreement: 0.61–0.80) between MDR vs. TET, STR vs. strA, CHL vs. floR, and MDR vs. floR, each with statistical significance. For example, the MDR isolates were associated with the presence of resistance to tetracycline and floR, while streptomycin- and chloramphenicol-resistant bacteria were associated with their corresponding genes (strA and floR). Numerous other pairs of association were also observed, indicating relatively common association between resistant phenotypes, genotypes, and their determinants. Regarding the inferences from the logistic regression analyses, ampicillin-resistant isolates exhibited 20-times higher odds of carrying blaTEM (OR = 20.3) compared to ampicillin-susceptible isolates (p < 0.0001). This finding indicates that ampicillin-resistant isolates collected from Thailand’s coastal aquaculture regions may preferentially harbor blaTEM. A previous study recommended that blaTEM might be a good indicator for AMR resistance genes in wastewater [56]. Additional regression analyses found that the trimethoprim-resistant isolates were positively associated with MDR (OR = 5.7; p < 0.0001) and int1 (OR = 4.7; p = 0.015). This observation suggests that mobile genetic elements may play a significant role in MDR bacterial development. Further studies using whole genome sequencing are recommended in order to more fully characterize the genomic basis of bacterial AMR resistance in Thailand’s coastal regions used for shellfish cultivation.
4. Materials and Methods
4.1. Seawater Sample Collection and Bacterial Isolation
Coastal water samples of 500 mL were collected in sterile bottles from different oyster cultivation areas in Thailand, including Surat Thani (9°12′737″ N, 99°27′276″ E), Chanthaburi (8°22′587″ N, 98°35′846″ E), Trat (12°04′610″ N, 102°35′843″ E), Phetchaburi (13°15′843″ N, 99°59′299″ E), Chonburi (13°20′482″ N, 100°55′054″ E), and Phang Nga (8°22′587″ N, 98°35′846″ E) provinces, during February 2021 to January 2022.
Bacterial confirmation followed standard methods. E. coli was determined using Levine’s Eosin Methylene Blue (L-EMB) agar (Difco, Becton, Dickinson and Company, Sparks, MD, USA). The colonies showing flat, dark center, with or without metallic sheen, were collected and confirmed by biochemical tests, including triple sugar iron (TSI) agar and indole test [57]. Xylose Lysine Deoxycholate (XLD) (Difco) and MacConkey Agar (Difco) were used for Salmonella determination. Presumptive colonies showing pink color with or without black center were confirmed biochemically as Salmonella using TSI and citrate test [58]. CHROMagar™ Vibrio Agar (HiMedia Laboratories Ltd., Mumbai, India) was used for Vibrio spp. determination with colonies showing mauve and green-blue color identified as V. parahaemolyticus and V. cholerae, respectively. Oxidase test and arginine glucose slants were used to confirm isolates [59]. The positive control strains were E. coli ATCC™ 25,922, S. enterica serovar Typhimurium ATCC™ 14,028, V. parahaemolyticus ATCC™ 17,802, and V. cholerae non-O1/non-O139 ATCC™ 14,733, respectively.
One confirmed bacterial isolate of E. coli, V. parahaemolyticus, and V. cholerae per one positive sample, and up to five Salmonella isolates per one positive sample were sub-cultured and stored in 20% glycerol at −80 °C in the Department of Veterinary Public health, Faculty of Veterinary Science from Chulalongkorn University.
4.2. Serotyping of S. enterica subsp. enterica and V. cholerae
All Salmonella isolates were serotyped by detection of somatic (O) and flagella (H) antigens using slide agglutination, according to the Kauffmann–White scheme [60] with available commercial antiserum (S&A Reagents Lab, Bangkok, Thailand).
All V. cholerae isolates were serotyped using a slide agglutination test with polyvalent V. cholerae O1, monoclonal V. cholerae O139, and monoclonal V. cholerae O141 antiserum (S&A Reagents Lab, Bangkok, Thailand).
4.3. Antimicrobial Susceptibility Testing
AST was performed on all isolates using a standard agar dilution method [61]. Eight antimicrobial drugs, including ampicillin, chloramphenicol, ciprofloxacin, gentamicin, streptomycin, sulfamethoxazole, tetracycline, and trimethoprim, were selected based on their importance in human and veterinary medicine. Clinical breakpoints and epidemiological cut-off values of E. coli, S. enterica subsp. enterica, V. parahaemolyticus, and V. cholerae were based on available standard protocols. The antimicrobials with their clinical breakpoints for E. coli and S. enterica subsp. enterica (in parentheses) are ampicillin (32 µg/mL), chloramphenicol (32 µg/mL), ciprofloxacin (>1 µg/mL), gentamicin (>8 µg/mL), streptomycin (32 µg/mL), sulfamethoxazole (512 µg/mL), tetracycline (16 µg/mL), and trimethoprim (16 µg/mL) [61]. For V. parahaemolyticus and V. cholerae, the MIC breakpoints are ampicillin (32 µg/mL), chloramphenicol (32 µg/mL), ciprofloxacin (4 µg/mL), gentamicin (>8 µg/mL), streptomycin (64 µg/mL), sulfamethoxazole (76 µg/mL), tetracycline (16 µg/mL), and trimethoprim (4 µg/mL) [62]. Staphylococcus aureus ATCC 29213, E. coli ATCC 25922, and Pseudomonas aeruginosa ATCC 27853 were used as positive control strains. Resistance to at least three groups of antimicrobials was considered MDR.
4.4. Determination of ESBL Production
The disc diffusion method was performed on all bacterial isolates according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [61]. The susceptibility to ceftazidime (30 µg), cefotaxime (30 µg), and cefpodoxime (10 µg) (Oxoid, Basingstoke, UK) was used for the screening test. Resistance to at least one of these cephalosporins was then confirmed using a combination disk diffusion method of ceftazidime (30 µg), cefotaxime (30 µg), and these two disks combined with clavulanic acid. A difference in the inhibition zone between single cephalosporin and cephalosporins containing clavulanic acid greater than 5 mm was classified as a positive ESBL-producing isolate.
4.5. Genotypic Characterization of AMR and Virulence Genes by Polymerase Chain Reaction (PCR)
The DNA template was prepared using the whole cell boiling method [63]. Briefly, the bacterial isolate was streaked onto nutrient agar (Difco) and incubated overnight at 37 °C. An individual colony was transferred to an Eppendorf tube containing 150 µL of rNase-free water. The suspension was mixed, heated, and immediately placed on ice, after which the suspension was centrifuged at 11,000 rpm for 5 min. DNA-containing supernatant was collected for PCR template.
All primers for resistance genes, their determinants, and virulence-encoded genes are listed in Table 6 (21 antimicrobials, 4 resistance determinants, 7 virulence genes). Resistance genes were selected to correspond with AMR phenotypes, β-lactam and ESBL production (blaTEM, blaSHV, blaCTX-M, and blaPSE); carbapenem (blaNDM and blaOXA); phenicols (floR and cmlA); erythromycin (ermB); quinolones (qnrS); gentamicin (aadA1); tetracyclines (tetA and tetB); streptomycin (strA); sulfonamide (sul1 and sul2); trimethoprim (dfrA1 an
d dfrA12); and colistin (mcr-1, mcr-2, and mcr-3). AMR determinants such as integrons (int1, int2, and int3) and the SXT element (intSXT) were also screened for. The presence of stx1 and stx2 genes in E. coli, invA in S. enterica subsp. enterica, tdh and trh in V. parahaemolyticus, and the outer membrane protein gene (ompW) and cholera toxin (ctx) V. cholerae were also determined. Bacterial isolates, which had been confirmed by PCR and sequencing of relevant genes and elements in previous studies, were used as positive control strains [14,64,65,66,67].

Table 6.
Primers used in this study.
Table 6.
Primers used in this study.
| Gene | Primer | Oligonucleotide Sequences (5′-3′) | Product Size (bp) | Reference |
|---|---|---|---|---|
| Genotype | ||||
| blaTEM | blaTEM-F | GCGGAACCCCTATTT | 964 | [68] |
| blaTEM-R | TCTAAAGTATATATGAGTAAACTTGGTCTGAC | |||
| blaSHV | blaSHV-F | TTCGCCTGTGTATTATCTCCCTG | 854 | [69] |
| blaSHV-R | TTAGCGTTGCCAGTGYTG | |||
| blaCTX-M | blaCTX-M-F | CGATGTGCAGTACCAGTAA | 585 | [70] |
| blaCTX-M-R | AGTGACCAGAATCAGCGG | |||
| blaPSE | BlaPSE-F | GCTCGTATAGGTGTTTCCGTTT | 575 | [71] |
| blaPSE-R | CGATCCGCAATGTTCCATCC | |||
| blaNDM | blaNDM-F | GGTTTGGCGATCTGGTTTTC | 621 | [72] |
| blaNDM-R | CGGAATGGCTCATCACGATC | |||
| blaOXA | blaOXA-F | ACACAATACATATCAACTTCGC | 813 | [73] |
| blaOXA-R | AGTGTGTGTTTAGAATGGTGATC | |||
| floR | floR-F | ATGGTGATGCTCGGCGTGGGCCA | 800 | [74] |
| floR-R | GCGCCGTTGGCGGTAACAGACACCGTGA | |||
| cmlA | cmlA-F | TGGACCGCTATCGGACCG | 641 | [64] |
| cmlA-R | CGCAAGACACTTGGGCTGC | |||
| ermB | ermB-F | AGACACCTCGTCTAACCTTCGCTC | 640 | [75] |
| ermB-R | TCCATGTACTACCATGCCACAGG | |||
| qnrS | qnrS-F | GCAAGTTCATTGAACAGGGT | 428 | [76] |
| qnrS-R | TCTAAACCGTCGAGTTCGGCG | |||
| addA1 | addA1-F | CTCCGCAGTGGATGGCGG | 631 | [64] |
| addA1-R | GATCTGCGCGCGAGGCCA | |||
| tetA | tetA-F | GCTGTCGGATCGTTTCGG | 658 | [64] |
| tetA-R | CATTCCGAGCATGAGTGCC | |||
| tetB | tetB-F | CTGTCGCGGCATCGGTCAT | 615 | [64] |
| tetB-R | CAGGTAAAGCGATCCCACC | |||
| strA | strA-F | TGGCAGGAGGAACAGGAGG | 405 | [64] |
| strA-R | AGGTCGATCAGACCCGTGC | |||
| sul1 | sul1-F | CGGCGTGGGCTACCTGAACG | 433 | [77] |
| sul1-R | GCCGATCGCGTGAAGTTCCG | |||
| sul2 | sul2-F | CGGCATCGTCAACATAACCT | 721 | [77] |
| sul2-R | TGTGCGGATGAAGTCAGCTC | |||
| dfrA1 | dfrA1-F | GGAGTGCCAAAGGTGAACAGC | 367 | [78] |
| dfrA1-R | GAGGCGAAGTCTTGGGTAAAAAC | |||
| dfrA12 | dfrA12-F | TTCGCAGACTCACTGAGGG | 330 | [79] |
| dfrA12-R | CGGTTGAGACAAGCTCGAAT | |||
| mcr-1 | mcr-1-F | AGTCCGTTTGTTCTTGTGGC | 320 | [79] |
| mcr-1-R | AGATCCTTGGTCTCGGCTTG | |||
| mcr-2 | mcr-2-F | CAAGTGTGTTGGTCGCAGTT | 715 | [79] |
| mcr-2-R | TCTAGCCCGACAAGCATACC | |||
| mcr-3 | mcr-3-F | AAATAAAAATTGTTCCGCTTATG | 929 | [79] |
| mcr-3-R | AATGGAGATCCCCGTTTTT | |||
| Integrons | ||||
| int1 | int1-F | CCTGCACGGTTCGAATG | 497 | [80] |
| Int1-R | TCGTTTGTTCGCCCAGC | |||
| int2 | int2-F | GGCAGACAGTTGCAAGACAA | 247 | [80] |
| int2-R | AAGCGATTTTCTGCGTGTTT | |||
| int3 | int3-F | CCGGTTCAGTCTTTCCTCAA | 155 | [80] |
| int3-R | GAGGCGTGTACTTGCCTCAT | |||
| Integrative and conjugative elements | ||||
| intsxt | intSXT-F | GCTGGATAGGTTAAGGGCGG | 592 | [80] |
| intSXT-R | CTCTATGGGCACTGTCCACATTG | |||
| Virulence genes of E. coli | ||||
| stx1 | stx-1-F | CAACACTGGATGATCTCAG | 349 | [81] |
| stx-1-R | CCCCCTCAACTGCTAATA | |||
| stx2 | stx-2-F | ATCAGTCGTCACTCACTGGT | 110 | [81] |
| stx-2-R | CTGCTGTCACAGTGACAAA | |||
| Species-specific and virulence genes of S. enterica subsp. enterica | ||||
| invA | invA-F | GTGAAATTATCGCCACGTTCGGGCAA | 284 | [82] |
| invA-R | TCATCGCACCGTCAAAGGAACC | |||
| Species-specific * and virulence genes of V. parahaemolyticus | ||||
| tlh * | tlh-F | AAAGCGGATTATGCAGAAGCACTG | 450 | [83] |
| tlh-R | GCTACTTTCTAGCATTTTCTCTGC | |||
| tdh | tdh-F | GTAAAGGTCTCTGACTTTTGGAC | 269 | [83] |
| tdh-R | TGGAATAGAACCTTCATCTTCACC | |||
| trh | trh-F | TTGGCTTCGATATTTTCAGTATCT | 500 | [83] |
| trh-R | CATAACAAACATATGCCCATTTCCG | |||
| Species-specific * and virulence genes of V. cholerae | ||||
| ompW * | ompW-F | CACCAAGAAGGTGACTTTATTGTG | 588 | [42] |
| ompW-R | GAACTTATAACCACCCGCG | |||
| ctx | ctx-F | CAGTCAGGTGGTCTTATGCCAAGAGG | 167 | [84] |
| ctx-R | CCCACTAAGTGGGCACTTCTCAAACT | |||
* Indicted specie-specific genes for V. parahaemolyticus and V. cholerae.
PCR was conducted following the manufacturer’s instructions. A 5 µL of DNA template, 25 µL of TopTaq Master Mix (Qiagen®, Stockach, Germany), 5 µL of coralLoad, 2 µL of each forward and reverse primer, and 11 µL of sterile rNase free water were utilized. The PCR amplification was performed based on Tpersonal combi model (Biometra®, Göttingen, Germany). PCR products were then separated on 1.5% (w/v) agarose gel, stained with RedsafeTM nucleic acid staining solution (Intron Biotechnology, Seongnam, Republic of Korea), and photographed using Omega Fluor™ gel documentation system (Aplegen, CA, USA).
4.6. Statistical Analyses
Descriptive statistics were used to characterize the prevalence of resistance, AMR distribution, ESBL production, virulence genes, integrons, and SXT element in E. coli, Salmonella, V. parahaemolyticus, and V. cholerae. Cohen’s kappa coefficient was used to determine the agreement between pairs of phenotypes and genotypes of AMR and resistance determinants for all isolates. The interpretation of the kappa coefficient, expressed as a strength of agreement, was: <0.00: poor; 0.00–0.20: slight; 0.21–0.40: fair; 0.41–0.60: moderate; 0.61–0.80: substantial; 0.81–1.00: almost perfect [85].
Multivariate logistic regression analysis was performed to characterize the association between the most common resistant phenotypes, AMR determinants, virulence genes, and ESBL production. Odds ratios (OR) were used to identify the magnitude of the observed association. The interpretation of ORs was indicated as OR > 1: positive association; OR < 1: negative association; OR = 1: no association. Two-sided hypothesis testing together with likelihood ratio test were used with a p ≤ 0.05 to decide statistical significance. All statistical analyses were performed with Stata version 14.0 (StataCorp, College Station, TX, USA).
5. Conclusions
The occurrence of waterborne AMR bacteria in areas of high-density oyster cultivation is an ongoing environmental and public health threat consistent with the One Health concept, especially given the popularity of shellfish consumption, water-related human recreation throughout coastal Thailand, and geographical expansion of the shellfish industry. Waterborne isolates of E. coli, S. enterica subsp. enterica, V. parahaemolyticus, and, to a lesser extent, V. cholerae from coastal Thailand exhibited phenotypic AMR for a wide variety of antimicrobials and for E. coli possessing concurrent genomic AMR. Although coastal seawater is regulated for excessive total coliforms, fecal coliforms, and Enterococci, current water quality monitoring does not include bacteria surveillance for phenotypic and/or genotypic AMR. Therefore, in addition to tracking and preventing key sources of bacterial contamination and promoting proper treatment of wastewater before release to Thailand’s coastal water resources, we recommend that water quality surveillance programs also include monitoring excessive levels of bacterial AMR to better protect shellfish food safety, water-related body contact recreation, and to reduce AMR contamination for Thailand’s coastal water resources.
Author Contributions
Conceptualization, S.J. and E.R.A.; methodology, S.A., V.T., N.R., and S.J.; validation, S.J.; formal analysis, S.J. and S.A.; investigation, S.J. and E.R.A.; resources, S.J.; funding acquisition S.J.; writing—original draft preparation, S.A., V.T., and N.R.; writing—review and editing, S.J. and W.H. with final drafts by S.J. and E.R.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Thailand Science Research and Innovation (TSRI), grant number CU_FRB640001_01_31_9.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors thank Jarukorn Sripradite for laboratory assistance. This project has been reviewed and certified by Chulalongkorn University, Faculty of Veterinary Science Biosafety Committee (IBC 2131029).
Conflicts of Interest
The authors declare no conflict of interest.
References
- O’Neill, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations, Review on Antimicrobial Resistance, Chaired by Jim O’Neill, December 2014; Wellcome Trust: London, UK, 2016. [Google Scholar]
- Sriram, A.; Kalanxhi, E.; Kapoor, G.; Craig, J.; Balasubramanian, R.; Brar, S.; Criscuolo, N.; Hamilton, A.; Klein, E.; Tseng, K. The State of the World’s Antibiotics 2021: A Global Analysis of Antimicrobial Resistance and Its Drivers; The Center for Disease Dynamics, Economics & Policy: Washington, DC, USA, 2021. [Google Scholar]
- Kusi, J.; Ojewole, C.O.; Ojewole, A.E.; Nwi-Mozu, I. Antimicrobial resistance development pathways in surface waters and public health implications. Antibiotics 2022, 11, 821. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Malak, H.A.; Abulreesh, H.H. Environmental antimicrobial resistance and its drivers: A potential threat to public health. J. Glob. Antimicrob. Resist. 2021, 27, 101–111. [Google Scholar]
- The United Nations Environment Programme (UNEP). Antimicrobial Resistance from Environmental Pollution among Biggest Emerging Health Threats, Says UN Environment. Available online: https://www.unep.org/news-and-stories/press-release/antimicrobial-resistance-environmental-pollution-among-biggest (accessed on 6 October 2022).
- Szekeres, E.; Chiriac, C.M.; Baricz, A.; Szőke-Nagy, T.; Lung, I.; Soran, M.L.; Rudi, K.; Dragos, N.; Coman, C. Investigating antibiotics, antibiotic resistance genes, and microbial contaminants in groundwater in relation to the proximity of urban areas. Environ. Pollut. 2018, 236, 734–744. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Jackson, C.R.; Frye, J.G. The prevalence and antimicrobial resistance phenotypes of Salmonella, Escherichia coli and Enterococcus sp. in surface water. Lett. Appl. Microbiol. 2020, 71, 3–25. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Cao, M.; Yuan, D.; Zhang, Y.; He, Q. Hydrogeological characterization and environmental effects of the deteriorating urban karst groundwater in a karst trough valley: Nanshan, SW China. Hydrogeol. J. 2018, 26, 1487–1497. [Google Scholar] [CrossRef]
- Nadimpalli, M.L.; Marks, S.J.; Montealegre, M.C.; Gilman, R.H.; Pajuelo, M.J.; Saito, M.; Tsukayama, P.; Njenga, S.M.; Kiiru, J.; Swarthout, J.; et al. Urban informal settlements as hotspots of antimicrobial resistance and the need to curb environmental transmission. Nat. Microbiol. 2020, 5, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Yin, G.; Liu, M.; Chen, C.; Jiang, Y.; Hou, L.; Zheng, Y. A systematic review of antibiotics and antibiotic resistance genes in estuarine and coastal environments. Sci. Total Environ. 2021, 777, 146009. [Google Scholar] [CrossRef]
- Miłobedzka, A.; Ferreira, C.; Vaz-Moreira, I.; Calderón-Franco, D.; Gorecki, A.; Purkrtova, S.; Bartacek, J.; Dziewit, L.; Singleton, C.M.; Nielsen, P.H.; et al. Monitoring antibiotic resistance genes in wastewater environments: The challenges of filling a gap in the one-health cycle. J. Hazard. Mater. 2022, 424, 127407. [Google Scholar] [CrossRef]
- Brooks, B.W.; Conkle, J.L. Commentary: Perspectives on aquaculture, urbanization and water quality. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2019, 217, 1–4. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, H.J.; Myung, G.E.; Choi, E.J.; Kim, I.A.; Jeong, Y.I.; Park, G.J.; Soh, S.M. Distribution of pathogenic Vibrio species in the coastal seawater of South Korea (2017–2018). Osong Public Health Res. Perspect. 2019, 10, 337–342. [Google Scholar] [CrossRef]
- Jeamsripong, S.; Khant, W.; Chuanchuen, R. Distribution of phenotypic and genotypic antimicrobial resistance and virulence genes in Vibrio parahaemolyticus isolated from cultivated oysters and estuarine water. FEMS Microbiol. Ecol. 2020, 96, fi-aa081. [Google Scholar] [CrossRef]
- Eklund, M.; Leino, K.; Siitonen, A. Clinical Escherichia coli strains carrying stx genes: Stx variants and stx-positive virulence profiles. J. Clin. Microb. 2002, 40, 4585–4593. [Google Scholar] [CrossRef]
- European Food Safety Authority. Pathogenicity assessment of Shiga toxin-producing Escherichia coli (STEC) and the public health risk posed by contamination of food with STEC. EFSA J. 2020, 18, 5967. [Google Scholar] [CrossRef]
- Rahn, K.; De Grandis, S.A.; Clarke, R.C.; McEwen, S.A.; Galán, J.E.; Ginocchio, C.; Curtiss, R.; Gyles, C.L. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes. 1992, 6, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Abd-Elghany, S.M.; Fathy, T.M.; Zakaria, A.I.; Imre, K.; Morar, A.; Herman, V.; Pașcalău, R.; Șmuleac, L.; Morar, D.; Imre, M.; et al. Prevalence of multidrug-resistant Salmonella enterica serovars in buffalo meat in Egypt. Foods 2022, 11, 2924. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Chaturvedi, A.N. Prevalence of virulence genes (ctxA, stn, OmpW and tcpA) among non-O1 Vibrio cholerae isolated from freshwater environment. Int. J. Hyg. Environ. Health 2006, 209, 521–526. [Google Scholar] [CrossRef]
- Gutierrez West, C.K.; Klein, S.L.; Lovell, C.R. High frequency of virulence factor genes tdh, trh, and tlh in Vibrio parahaemolyticus strains isolated from a pristine estuary. Appl. Environ. Microbiol. 2013, 79, 2247–2252. [Google Scholar] [CrossRef]
- Sun, D. Pull in and push out: Mechanisms of horizontal gene transfer in bacteria. Front. Microbiol. 2018, 9, 2154. [Google Scholar] [CrossRef]
- Mazel, D. Integrons: Agents of bacterial evolution. Nat. Rev. Microbiol. 2006, 4, 608–620. [Google Scholar] [CrossRef]
- Wozniak, R.; Waldor, M. Integrative and conjugative elements: Mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 2010, 8, 552–563. [Google Scholar] [CrossRef]
- World Health Organization (WHO); Food and Agriculture Organization (FAO); World Organization for Animal Health (OIE). Technical Brief on Water, Sanitation, Hygiene and Wastewater Management to Prevent Infections and Reduce the Spread of Antimicrobial Resistance; World Health Organization: Geneva, Switzerland; Food Agriculture Organization: Rome, Italy; World Organization for Animal Health: Paris, France, 2020; ISBN 9789240006416. [Google Scholar]
- Zhao, J.Y.; Dang, H. Coastal seawater bacteria harbor a large reservoir of plasmid-mediated quinolone resistance determinants in Jiaozhou Bay, China. Microb. Ecol. 2012, 64, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Ghenem, L.; Elhadi, N. Isolation, molecular characterization, and antibiotic resistance patterns of Vibrio parahaemolyticus isolated from coastal water in the Eastern province of Saudi Arabia. J. Water Health 2018, 16, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Alibi, S.; Beltifa, A.; Hassen, W.; Jaziri, A.; Soussia, L.; Zbidi, F.; Mansour, H.B. Coastal surveillance and water quality monitoring in the Rejiche Sea-Tunisia. Water Environ. Res. 2021, 93, 2025–2033. [Google Scholar] [CrossRef] [PubMed]
- Nunes, M.; Leston, S. Coastal Pollution: An Overview. Life Below Water; Springer: Cham, Switzerland, 2022; pp. 155–165. ISBN 978-3-319-98536-7. [Google Scholar]
- Prüss-Ustün, A.; Bartram, J.; Clasen, T.; Colford, J.M.; Cumming, O.; Curtis, V.; Bonjour, S.; Dangour, A.D.; De France, J.; Fewtrell, L.; et al. Burden of disease from inadequate water, sanitation and hygiene in low- and middle-income settings: A retrospective analysis of data from 145 countries. Trop. Med. Int. Health 2014, 19, 894–905. [Google Scholar] [CrossRef] [PubMed]
- Hammad, A.M.; Eltahan, A.; Hassan, H.A.; Abbas, N.H.; Hussien, H.; Shimamoto, T. Loads of coliforms and fecal coliforms and characterization of thermotolerant Escherichia coli in fresh raw milk cheese. Foods 2022, 11, 332. [Google Scholar] [CrossRef] [PubMed]
- Hartinger, S.M.; Medina-Pizzali, M.L.; Salmon-Mulanovich, G.; Larson, A.J.; Pinedo-Bardales, M.; Verastegui, H.; Riberos, M.; Mäusezahl, D. Antimicrobial resistance in humans, animals, water and household environs in rural Andean Peru: Exploring dissemination pathways through the One Health Lens. Int. J. Environ. Res. Public Health 2021, 18, 4604. [Google Scholar] [CrossRef]
- Sorour, H.K.; Amer, F. Detection of biofilm formation and antibiotic resistance of Salmonella in broiler chicken. Assiut. Vet. Med. J. 2018, 64, 146–153. [Google Scholar]
- Jha, B.; Kim, C.M.; Kim, D.M.; Chung, J.H.; Yoon, N.R.; Jha, P.; Kim, S.W.; Jang, S.J.; Kim, S.G.; Chung, J.K. First report of iliacus abscess caused by Salmonella enterica serovar Othmarschen. J. Infect. Chemother. 2016, 22, 117–119. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Multistate Outbreak of Salmonella Braenderup Infections Linked to Rose Acre Farms Shell Eggs (Final Update). Available online: https://www.cdc.gov/salmonella/braenderup-04-18/index.html (accessed on 6 October 2022).
- Wawa, A.I. Challenges facing wastewater management in fast growing cities in Tanzania: A case of Dodoma city council. Huria J. 2020, 27, 168–185. [Google Scholar]
- Aoki, Y.; Watanabe, Y.; Kitazawa, K.; Ando, N.; Hirai, S.; Yokoyama, E. Emergence of Salmonella enterica subsp. enterica serovar Chester in a rural area of Japan. J. Vet. Med. Sci. 2020, 82, 580–584. [Google Scholar] [CrossRef]
- Khanam, F.; Rajib, N.H.; Tonks, S.; Khalequzzaman, M.; Pollard, A.J.; Clemens, J.D.; Qadri, F.; The STRATAA Study Team. Case report: Salmonella enterica serovar Paratyphi B infection in a febrile Ill child during enhanced passive surveillance in an urban slum in Mirpur, Dhaka. Am. J. Trop. Med. Hyg. 2020, 103, 231–233. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; McMillan, E.A.; Jackson, C.R.; Desai, P.T.; Porwollik, S.; McClelland, M.; Hiott, L.M.; Humayoun, S.B.; Frye, J.G. Draft genome sequence of Salmonella enterica subsp. enterica serovar Orion strain CRJJGF_00093 (Phylum Gammaproteobacteria). Genome Announc. 2016, 4, e01063–e01116. [Google Scholar] [CrossRef] [PubMed]
- García-Aljaro, C.; Muniesa, M.; Jofre, J.; Blanch, A.R. Prevalence of the stx2 gene in coliform populations from aquatic environments. Appl. Environ. Microbiol. 2004, 70, 3535–3540. [Google Scholar] [CrossRef]
- Mauro, S.A.; Koudelka, G.B. Shiga toxin: Expression, distribution, and its role in the environment. Toxins 2011, 3, 608–625. [Google Scholar] [CrossRef] [PubMed]
- Prabhakaran, D.M.; Ramamurthy, T. Genetic and virulence characterisation of Vibrio parahaemolyticus isolated from Indian coast. BMC Microbiol. 2020, 20, 1–14. [Google Scholar] [CrossRef]
- Sathiyamurthy, K.; Baskaran, A.; Subbaraj, D.K. Prevalence of Vibrio cholerae and other vibrios from environmental and seafood sources, Tamil Nadu, India. Br. Microbiol. Res. J. 2013, 3, 538–549. [Google Scholar] [CrossRef]
- Praja, R.A.K.; Sukrama, I.D.M.; Fatmawati, N.N.D. Detection of genes encoding ompW and ctxA of Vibrio cholerae isolated from shrimp and shellfish at Kedonganan fish market, Bali-Indonesia. Oceana Biomed. J. 2019, 2, 1–14. [Google Scholar] [CrossRef][Green Version]
- Mala, W.; Faksri, K.; Samerpitak, K.; Yordpratum, U.; Kaewkes, W.; Tattawasart, U.; Chomvarin, C. Antimicrobial resistance and genetic diversity of the SXT element in Vibrio cholerae from clinical and environmental water samples in northeastern Thailand. Infect. Genet. Evol. 2017, 52, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Tulatorn, S.; Preeprem, S.; Vuddhakul, V.; Mittraparp-Arthorn, P. Comparison of virulence gene profiles and genomic fingerprints of Vibrio cholerae O1 and non-O1/non-O139 isolates from diarrheal patients in southern Thailand. Trop. Med. Health 2018, 46, 31. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-spectrum β-lactamases: An update on their characteristics, epidemiology and detection. JAC Antimicrob. Resist. 2021, 3, dlab092. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, S.B.; Søraas, A.V.; Arnesen, L.S.; Leegaard, T.M.; Sundsfjord, A.; Jenum, P.A. A comparison of extended spectrum β-lactamase producing Escherichia coli from clinical, recreational water and wastewater samples associated in time and location. PLoS ONE 2017, 12, e0186576. [Google Scholar] [CrossRef] [PubMed]
- Hassen, B.; Jouini, A.; Elbour, M.; Hamrouni, S.; Maaroufi, A. Detection of extended-spectrum β-lactamases (ESBL) producing enterobacteriaceae from fish trapped in the lagoon area of Bizerte, Tunisia. Biomed. Res. Int. 2020, 2020, 7132812. [Google Scholar] [CrossRef]
- Cohen, R.; Paikin, S.; Rokney, A.; Rubin-Blum, M.; Astrahan, P. Multidrug-resistant Enterobacteriaceae in coastal water: An emerging threat. Antimicrob. Resist. Infect. Control 2020, 9, 169. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Hu, Y.; Wen, H.; Wu, J.; Liu, Y.; Zhang, Y.; Wu, H. Occurrence and driving mechanism of antibiotic resistance genes in marine recreational water around Qinhuangdao, China. Front. Mar. Sci. 2022, 9, 976438. [Google Scholar] [CrossRef]
- Leonard, A.F.C.; Zhang, L.; Balfour, A.J.; Garside, R.; Hawkey, P.M.; Murray, A.K.; Ukoumunne, O.C.; Gaze, W.H. Exposure to and colonisation by antibiotic-resistant E. coli in UK coastal water users: Environmental surveillance, exposure assessment, and epidemiological study (Beach Bum Survey). Environ. Int. 2018, 114, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Lu, J.; Wu, J.; Zhang, Y.; Zhang, C. Proliferation of antibiotic resistance genes in coastal recirculating mariculture system. Environ. Pollut. 2019, 248, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro-Moura, J.R.; Kraychete, G.B.; Longo, L.; Corrêa, L.L.; da Silva, N.M.V.; Campana, E.H.; Oliveira, C.J.B.; Picão, R.C. Description and comparative genomic analysis of a mcr-1-carrying Escherichia coli ST683/CC155 recovered from touristic coastal water in Northeastern Brazil. Infect. Genet. Evol. 2022, 97, 105196. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.H.; Moore, L.S.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karley, A.; Guerin, P.; Piddock, L.J. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Akrami, F.; Rajabnia, M.; Pournajaf, A. Resistance integrons—A mini review. Caspian J. Intern. Med. 2019, 10, 370–376. [Google Scholar] [PubMed]
- Narciso-da-Rocha, C.; Varela, A.R.; Schwartz, T.; Nunes, O.C.; Manaia, C.M. blaTEM and vanA as indicator genes of antibiotic resistance contamination in a hospital–urban wastewater treatment plant system. J. Glob. Antimicrob. Resist. 2014, 2, 309–315. [Google Scholar] [CrossRef]
- Food and Drug Administration. BAM Chapter 4: Enumeration of Escherichia coli and the Coliform Bacteria. Available online: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-4-enumeration-escherichia-coli-and-coliform-bacteria#conventional (accessed on 28 October 2022).
- Food and Drug Administration. BAM Chapter 9: Vibrio. Available online: https://www.fda.gov/food/foodscienceresearch/laboratorymethods/ucm070830.htm (accessed on 28 October 2022).
- Food and Drug Administration. BAM Chapter 5: Salmonella. Available online: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-5-salmonella (accessed on 28 October 2022).
- Grimont, P.A.D.; Weill, F.X. Antigenic Formulae of the Salmonella Serovars, 9th ed.; WHO Collaborating Center for Reference and Research on Salmonella; Institut Pasteur: Paris, France, 2007. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Test for Bacteria Isolated from Animals, 31st ed.; CLSI Guideline M100; Clinical and Laboratory Standard Institute: Wayne, PA, USA, 2015. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria, 3rd ed.; CLSI Guideline M45; Clinical and Laboratory Standard Institute: Wayne, PA, USA, 2016. [Google Scholar]
- Lévesque, C.; Piché, L.; Larose, C.; Roy, P.H. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 1995, 39, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Chuanchuen, R.; Padungtod, P. Antimicrobial resistance genes among Salmonella enterica isolates from poultry and swine in Thailand. Int. J. Infect. Dis. 2009, 12, e117. [Google Scholar] [CrossRef][Green Version]
- Thaotumpitak, V.; Sripradite, J.; Atwill, E.R.; Tepaamorndech, S.; Jeamsripong, S. Bacterial pathogens and factors associated with Salmonella contamination in hybrid red tilapia (Oreochromis spp.) cultivated in a cage culture system. Food Qual. Saf. 2022, 6, fyac036. [Google Scholar] [CrossRef]
- Chuanchuen, R.; Pathanasophon, P.; Khemtong, S.; Wannaprasat, W.; Padungtod, P. Susceptibilities to antimicrobials and disinfectants in Salmonella isolates obtained from poultry and swine in Thailand. J. Vet. Med. Sci. 2008, 70, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Pungpian, C.; Lee, S.; Trongjit, S.; Sinwat, N.; Angkititrakul, S.; Prathan, R.; Srisanga, S.; Chuanchuen, R. Colistin resistance and plasmid-mediated mcr genes in Escherichia coli and Salmonella isolated from pigs, pig carcass and pork in Thailand, Lao PDR and Cambodia border provinces. J. Vet. Sci. 2021, 22, 5. [Google Scholar] [CrossRef]
- Olesen, I.; Hasman, H.; Aarestrup, F.M. Prevalence of ß-lactamases among ampicillin-resistant Escherichia coli and Salmonella isolated from food animals in Denmark. Microb. Drug Resist. 2004, 10, 334–340. [Google Scholar] [CrossRef]
- Hasman, H.; Mevius, D.; Veldman, K.; Olesen, I.; Aarestrup, F.M. ß-Lactamases among extended-spectrum ß-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J. Antimicrob. Chemother. 2005, 56, 115–121. [Google Scholar] [CrossRef]
- Batchelor, M.; Hopkins, K.; Threlfall, E.J.; Clifton-Hadley, F.A.; Stallwood, A.D.; Davies, R.H.; Liebana, E. blaCTX-M genes in clinical Salmonella isolates recovered from humans in England and Wales from 1992 to 2003. Antimicrob. Agents Chemother. 2005, 49, 1319–1322. [Google Scholar] [CrossRef]
- Li, R.; Lai, J.; Wang, Y.; Liu, S.; Li, Y.; Liu, K.; Shen, J.; Wu, C. Prevalence and characterization of Salmonella species isolated from pigs, ducks and chickens in Sichuan Province, China. Int. J. Food Microbiol. 2013, 163, 14–18. [Google Scholar] [CrossRef]
- Poirel, L.; Walsh, T.R.; Cuvillier, V.; Nordmann, P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 2011, 70, 119–123. [Google Scholar] [CrossRef]
- Costa, D.; Poeta, P.; Sáenz, Y.; Vinué, L.; Rojo-Bezares, B.; Jouini, A.; Zarazaga, M.; Rodrigues, J.; Torres, C. Detection of Escherichia coli harbouring extended-spectrum ß-lactamases of the CTX-M, TEM and SHV classes in faecal samples of wild animals in Portugal. J. Antimicrob. Chemother. 2006, 58, 1311–1312. [Google Scholar] [CrossRef] [PubMed]
- Ying, Y.; Wu, F.; Wu, C.; Jiang, Y.; Yin, M.; Zhou, W.; Zhu, X.; Cheng, C.; Zhu, L.; Li, K.; et al. Florfenicol resistance in Enterobacteriaceae and whole-genome sequence analysis of florfenicol-resistant Leclercia adecarboxylata strain R25. Int. J. Genom. 2019, 2019, 9828504. [Google Scholar] [CrossRef]
- Raissy, M.; Moumeni, M.; Ansari, M.; Rahimi, E. Antibiotic resistance pattern of some Vibrio strains isolated from seafood. Iran J. Fish Sci. 2012, 11, 618–626. [Google Scholar]
- Cattoir, V.; Poirel, L.; Rotimi, V.; Soussy, C.J.; Nordmann, P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 2007, 60, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.B.; Khan, M.A.; Ahmad, I.; Rehman, T.; Ullah, S.; Dad, R.; Sultan, A.; Memon, A.M. Phentotypic, gentotypic antimicrobial resistance and pathogenicity of Salmonella enterica serovars Typimurium and Enteriditis in poultry and poultry products. Microb. Pathog. 2019, 129, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Shahrani, M.; Dehkordi, F.S.; Momtaz, H. Characterization of Escherichia coli virulence genes, pathotypes and antibiotic resistance properties in diarrheic calves in Iran. Biol. Res. 2014, 47, 28. [Google Scholar] [CrossRef] [PubMed]
- Rebelo, A.R.; Bortolaia, V.; Kjeldgaard, J.S.; Pedersen, S.K.; Leekitcharoenphon, P.; Hansen, I.M.; Guerra, B.; Malorny, B.; Borowiak, M.; Hammerl, J.A. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. 2018, 23, 17–00672. [Google Scholar] [CrossRef] [PubMed]
- Kitiyodom, S.; Khemtong, S.; Wongtavatchai, J.; Chuanchuen, R. Characterization of antibiotic resistance in Vibrio spp. isolated from farmed marine shrimps (Penaeus monodon). FEMS Microbiol. Ecol. 2010, 72, 219–227. [Google Scholar] [CrossRef]
- Khan, A.; Yamasaki, S.; Sato, T.; Ramamurthy, T.; Pal, A.; Datta, S.; Chowdhury, N.R.; Das, S.C.; Sikdar, A.; Tsukamoto, T.; et al. Prevalence and genetic profiling of virulence determinants of non-O157 shiga toxin-producing Escherichia coli isolated from cattle, beef, and humans, Calcutta, India. Emerg. Infect. Dis. 2002, 8, 54–62. [Google Scholar] [CrossRef]
- Kumar, R.; Datta, T.K.; Lalitha, K.V. Salmonella grows vigorously on seafood and expresses its virulence and stress genes at different temperature exposure. BMC Microbiol. 2015, 15, 254. [Google Scholar] [CrossRef] [PubMed]
- Bej, A.K.; Patterson, D.P.; Brasher, C.W.; Vickery, M.C.L.; Jones, D.D.; Kaysner, C.A. Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh, and trh. J. Microbiol. Methods 1999, 36, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.C.; You, W.Y.; Chen, S.Y. Detection of toxigenic Vibrio cholerae, V. parahaemolyticus and V. vulnificus in oyster by multiplex-PCR with internal amplification control. J. Food Drug Anal. 2012, 20, 48–58. [Google Scholar] [CrossRef]
- Landis, J.R.; Koch, G.G. The measurement of observer agreement for categorical data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).