Irrigation Ponds as Sources of Antimicrobial-Resistant Bacteria in Agricultural Areas with Intensive Use of Poultry Litter
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area and Sampling Design
2.2. DNA Extraction from Water Samples
2.3. Detection of β-Lactam, Sulfonamide and Fluoroquinolone Resistance Genes
2.4. Amplicon Sequencing of 16S rRNA Genes from Water Samples
2.5. Bioinformatics Analysis
2.6. Statistical Analyses
2.7. Isolation of ARB from Water Samples
3. Results
3.1. PCR Amplification of Antimicrobial Resistance Genes in Water Samples
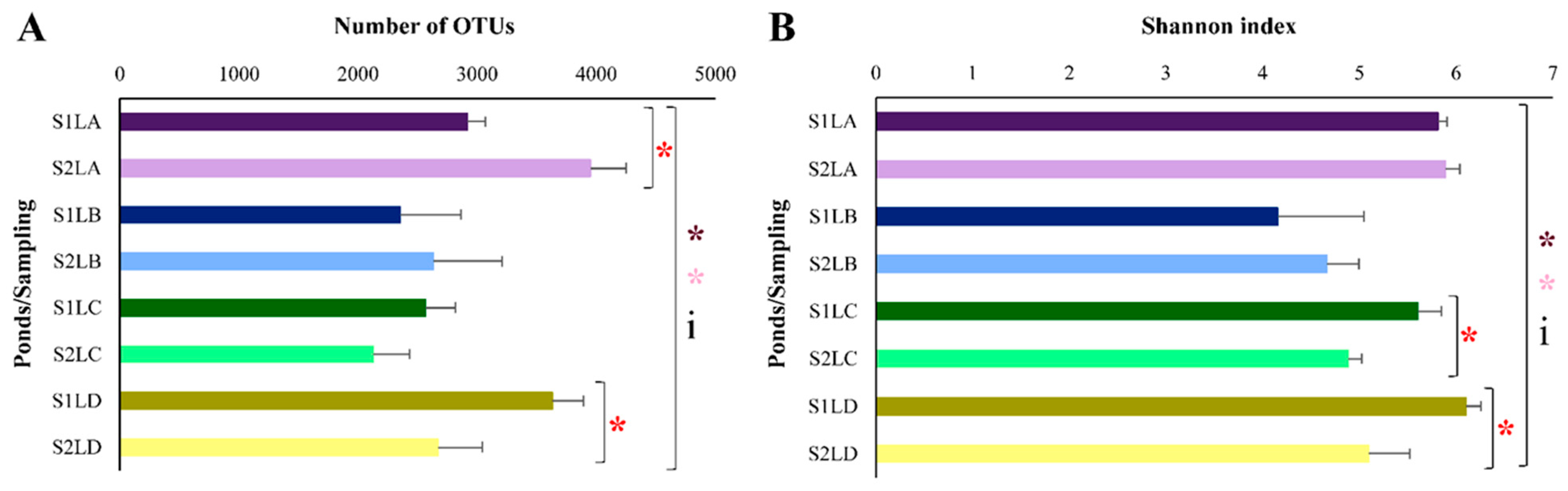
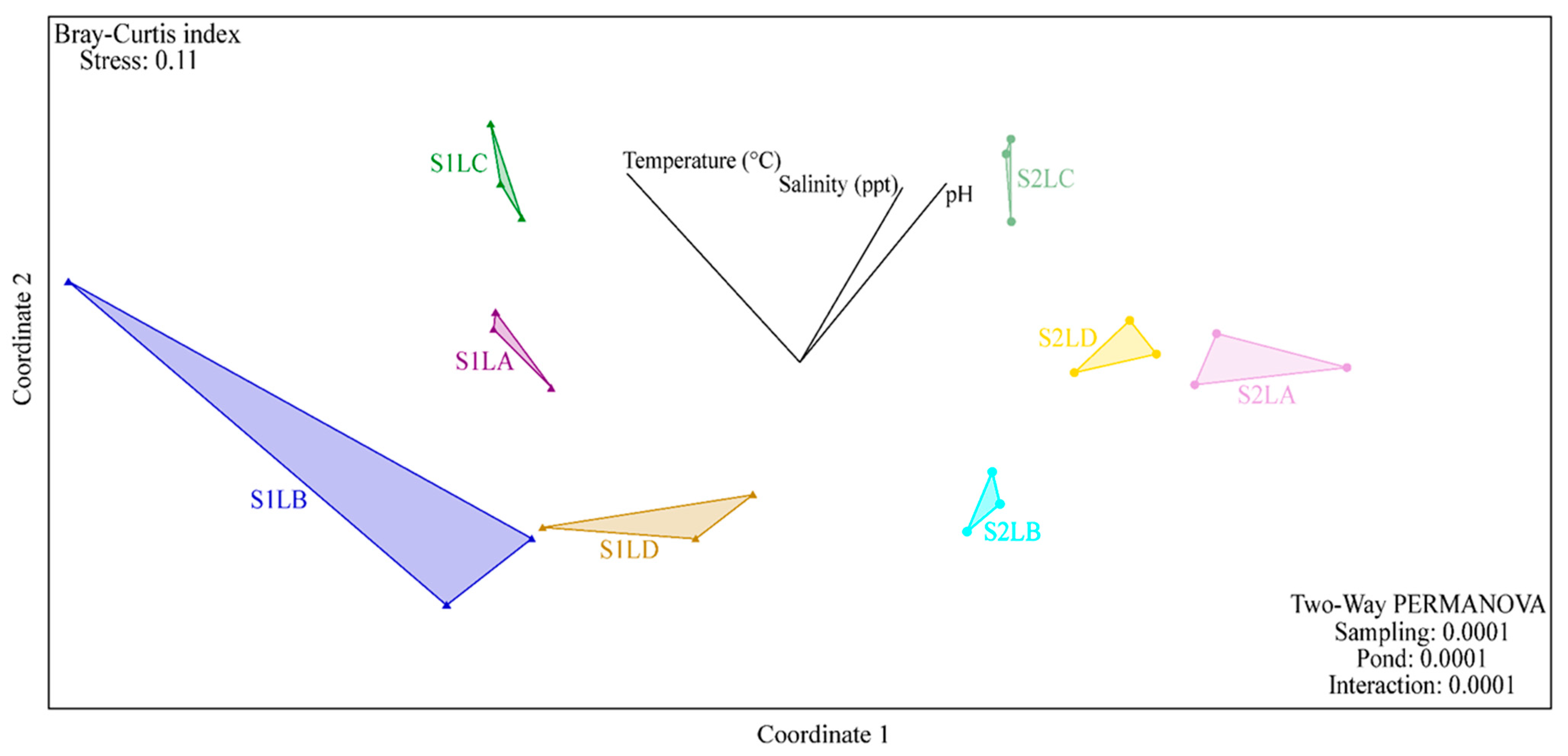
3.2. Structure and Composition of the Bacterial Communities of Water Samples
3.3. Strains of ARB Identified by MALDI-TOF
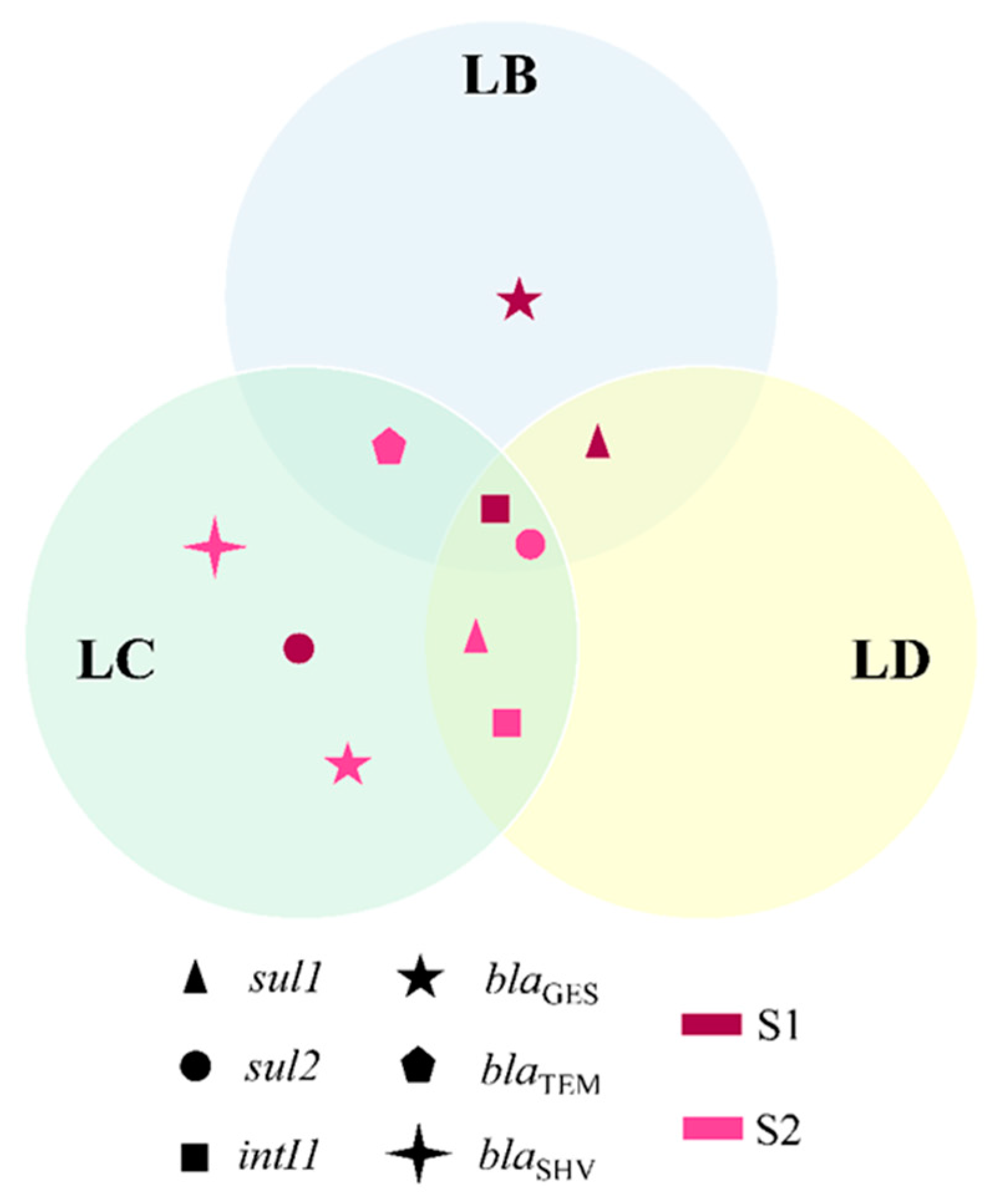
3.4. Comparison of Bacterial Community Composition with the Prevalence of ARB
3.5. ARGs in Isolated Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Antimicrobial Resistance-Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014; Volume 61, pp. 383–394. [Google Scholar]
- Chow, L.K.M.; Ghaly, T.M.; Gillings, M.R. A Survey of Sub-Inhibitory Concentrations of Antibiotics in the Environment. J. Environ. Sci. 2021, 99, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Maillard, J.Y.; Bloomfield, S.F.; Courvalin, P.; Essack, S.Y.; Gandra, S.; Gerba, C.P.; Rubino, J.R.; Scott, E.A. Reducing Antibiotic Prescribing and Addressing the Global Problem of Antibiotic Resistance by Targeted Hygiene in the Home and Everyday Life Settings: A Position Paper. Am. J. Infect. Control 2020, 48, 1090–1099. [Google Scholar] [CrossRef] [PubMed]
- Rabello, R.F.; Bonelli, R.R.; Penna, B.A.; Albuquerque, J.P.; Souza, R.M.; Cerqueira, A.M.F. Antimicrobial Resistance in Farm Animals in Brazil: An Update Overview. Animals 2020, 10, 552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global Trends in Antimicrobial Use in Food Animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gouvêa, R.; Dos Santos, F.F.; De Aquino, M.H.C. Fluoroquinolones in Industrial Poultry Production, Bacterial Resistance and Food Residues: A Review. Braz. J. Poult. Sci. 2015, 17, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Meletis, G. Carbapenem Resistance: Overview of the Problem and Future Perspectives. Ther. Adv. Infect. Dis. 2016, 3, 15–21. [Google Scholar] [CrossRef] [Green Version]
- Morales-Gutiérrez, F.J.; Barbosa, J.; Barrón, D. Metabolic Study of Enrofloxacin and Metabolic Profile Modifications in Broiler Chicken Tissues after Drug Administration. Food Chem. 2015, 172, 30–39. [Google Scholar] [CrossRef]
- van Duijkeren, E.; Schink, A.-K.; Roberts, M.C.; Wang, Y.; Schwarz, S. Mechanisms of Bacterial Resistance to Antimicrobial Agents. Microbiol. Spectr. 2018, 6, 52–82. [Google Scholar] [CrossRef]
- Cardoso, M. Antimicrobial Use, Resistance and Economic Benefits and Costs to Livestock Producers in Brazil. OECD Food Agric. Fish. Pap. 2019, 135, 1–44. [Google Scholar]
- Palma, E.; Tilocca, B.; Roncada, P. Antimicrobial Resistance in Veterinary Medicine: An Overview. Int. J. Mol. Sci. 2020, 21, 1914. [Google Scholar] [CrossRef] [Green Version]
- Sarmah, A.K.; Meyer, M.T.; Boxall, A.B.A. A Global Perspective on the Use, Sales, Exposure Pathways, Occurrence, Fate and Effects of Veterinary Antibiotics (VAs) in the Environment. Chemosphere 2006, 65, 725–759. [Google Scholar] [CrossRef] [PubMed]
- Kemper, N. Veterinary Antibiotics in the Aquatic and Terrestrial Environment. Ecol. Indic. 2008, 8, 1–13. [Google Scholar] [CrossRef]
- Vollú, R.E.; Cotta, S.R.; Jurelevicius, D.; Leite, D.C.D.A.; Parente, C.E.T.; Malm, O.; Martins, D.C.; Resende, Á.V.; Marriel, I.E.; Seldin, L. Response of the Bacterial Communities Associated with Maize Rhizosphere to Poultry Litter as an Organomineral Fertilizer. Front. Environ. Sci. 2018, 6, 118. [Google Scholar] [CrossRef] [Green Version]
- Ljubojevic, D.; Puvača, N.; Pelic, M.; Todorovic, D.; Pajic, M.; Milanov, D.; Velhner, M. Epidemiological Significance of Poultry Litter for Spreading the Antibiotic-Resistant Strains of Escherichia coli. Worlds Poult. Sci. J. 2016, 72, 485–494. [Google Scholar] [CrossRef]
- Lamshöft, M.; Sukul, P.; Zühlke, S.; Spiteller, M. Behaviour of 14C-Sulfadiazine and 14C-Difloxacin during Manure Storage. Sci. Total Environ. 2010, 408, 1563–1568. [Google Scholar] [CrossRef]
- Jechalke, S.; Heuer, H.; Siemens, J.; Amelung, W.; Smalla, K. Fate and Effects of Veterinary Antibiotics in Soil. Trends Microbiol. 2014, 22, 536–545. [Google Scholar] [CrossRef]
- Xie, W.Y.; Shen, Q.; Zhao, F.J. Antibiotics and Antibiotic Resistance from Animal Manures to Soil: A Review. Eur. J. Soil Sci. 2018, 69, 181–195. [Google Scholar] [CrossRef] [Green Version]
- Muhammad, J.; Khan, S.; Su, J.Q.; Hesham, A.E.L.; Ditta, A.; Nawab, J.; Ali, A. Antibiotics in Poultry Manure and Their Associated Health Issues: A Systematic Review. J. Soils Sediments 2020, 20, 486–497. [Google Scholar] [CrossRef]
- Wright, G.D. Antibiotic Resistance in the Environment: A Link to the Clinic? Curr. Opin. Microbiol. 2010, 13, 589–594. [Google Scholar] [CrossRef]
- Jones, L.A.; Worobo, R.W.; Smart, C.D. Plant-Pathogenic Oomycetes, Escherichia coli Strains, and Salmonella spp. Frequently Found in Surface Water Used for Irrigation of Fruit and Vegetable Crops in New York State. Appl. Environ. Microbiol. 2014, 80, 4814–4820. [Google Scholar] [CrossRef] [Green Version]
- Dourado, F.; Arraes, T.C.; Silva, M.F. The “Megadesastre” in the Mountain Region of Rio de Janeiro State-Causes, Mechanisms of Mass Movements and Spatial Allocation of Investments for Reconstruction Post Disaster. Anuário Inst. Geociências-UFRJ 2012, 35, 43–54. [Google Scholar] [CrossRef]
- Rosi, A.; Canavesi, V.; Segoni, S.; Dias Nery, T.; Catani, F.; Casagli, N. Landslides in the Mountain Region of Rio de Janeiro: A Proposal for the Semi-Automated Definition of Multiple Rainfall Thresholds. Geosciences 2019, 9, 203. [Google Scholar] [CrossRef] [Green Version]
- Parente, C.E.T.; Azeredo, A.; Vollú, R.E.; Zonta, E.; Azevedo-Silva, C.E.; Brito, E.M.S.; Seldin, L.; Torres, J.P.M.; Meire, R.O.; Malm, O. Fluoroquinolones in Agricultural Soils: Multi-Temporal Variation and Risks in Rio de Janeiro Upland Region. Chemosphere 2019, 219, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Parente, C.E.T.; Brusdzenski, G.S.; Zonta, E.; Lino, A.S.; Azevedo-Silva, C.E.; Dorneles, P.R.; Azeredo, A.; Torres, J.P.M.; Meire, R.O.; Malm, O. Fluoroquinolones and Trace Elements in Poultry Litter: Estimation of Environmental Load Based on Nitrogen Requirement for Crops. J. Environ. Sci. Health B 2020, 55, 1087–1098. [Google Scholar] [CrossRef] [PubMed]
- Instituto Nacional de Meteorologia Histórico de Dados Meteorológicos. Available online: https://portal.inmet.gov.br/ (accessed on 5 March 2021).
- Xu, X.; Kong, F.; Cheng, X.; Yan, B.; Du, X.; Gai, J.; Ai, H.; Shi, L.; Iredell, J. Integron Gene Cassettes in Acinetobacter spp. Strains from South China. Int. J. Antimicrob. Agents 2008, 32, 441–445. [Google Scholar] [CrossRef] [PubMed]
- Campana, E.H.; Xavier, D.E.; Petrolini, F.V.B.; Cordeiro-Moura, J.R.; de Araujo, M.R.E.; Gales, A.C. Carbapenem-Resistant and Cephalosporin-Susceptible: A Worrisome Phenotype among Pseudomonas aeruginosa Clinical Isolates in Brazil. Braz. J. Infect. Dis. 2017, 21, 57–62. [Google Scholar] [CrossRef] [Green Version]
- Picão, R.C.; Poirel, L.; Gales, A.C.; Nordmann, P. Diversity of β-Lactamases Produced by Ceftazidime-Resistant Pseudomonas aeruginosa Isolates Causing Bloodstream Infections in Brazil. Antimicrob. Agents Chemother. 2009, 53, 3908–3913. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Pérez, F.J.; Hanson, N.D. Detection of Plasmid-Mediated AmpC β-Lactamase Genes in Clinical Isolates by Using Multiplex PCR. J. Clin. Microbiol. 2002, 40, 2153–2162. [Google Scholar] [CrossRef] [Green Version]
- Toleman, M.A.; Bennett, P.M.; Bennett, D.M.C.; Jones, R.N.; Walsh, T.R. Global Emergence of Trimethoprim/Sulfamethoxazole Resistance in Stenotrophomonas maltophilia Mediated by Acquisition of sul Genes. Emerg. Infect. Dis. 2007, 13, 559–565. [Google Scholar] [CrossRef]
- Cattoir, V.; Poirel, L.; Rotimi, V.; Soussy, C.J.; Nordmann, P. Multiplex PCR for Detection of Plasmid-Mediated Quinolone Resistance Qnr Genes in ESBL-Producing Enterobacterial Isolates. J. Antimicrob. Chemother. 2007, 60, 394–397. [Google Scholar] [CrossRef] [Green Version]
- Kraychete, G.B.; Botelho, L.A.B.; Campana, E.H.; Picão, R.C.; Bonelli, R.R. Updated Multiplex PCR for Detection of All Six Plasmid-Mediated Qnr Gene Families. Antimicrob. Agents Chemother. 2016, 60, 7524–7526. [Google Scholar] [CrossRef] [PubMed]
- Yamane, K.; Wachino, J.I.; Suzuki, S.; Kimura, K.; Shibata, N.; Kato, H.; Shibayama, K.; Konda, T.; Arakawa, Y. New Plasmid-Mediated Fluoroquinolone Efflux Pump, QepA, Found in an Escherichia coli Clinical Isolate. Antimicrob. Agents Chemother. 2007, 51, 3354–3360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Apprill, A.; Mcnally, S.; Parsons, R.; Weber, L. Minor Revision to V4 Region SSU rRNA 806R Gene Primer Greatly Increases Detection of SAR11 Bacterioplankton. Aquatic. Microbial. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global Patterns of 16S rRNA Diversity at a Depth of Millions of Sequences per Sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing Mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Cardenas, E.; Fish, J.; Chai, B.; Farris, R.J.; Kulam-Syed-Mohideen, A.S.; McGarrell, D.M.; Marsh, T.; Garrity, G.M.; et al. The Ribosomal Database Project: Improved Alignments and New Tools for rRNA Analysis. Nucleic Acids Res. 2009, 37, 141–145. [Google Scholar] [CrossRef] [Green Version]
- Hammer, R.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Box, G.E.P.; Cox, D.R. An Analysis of Transformations. J. R. Stat. Soc. Ser. B 1964, 26, 211–243. [Google Scholar] [CrossRef]
- Suzuki, S.; Ogo, M.; Miller, T.W.; Shimizu, A.; Takada, H.; Siringan, M.A.T. Who Possesses Drug Resistance Genes in the Aquatic Environment?: Sulfamethoxazole (SMX) Resistance Genes among the Bacterial Community in Water Environment of Metro-Manila, Philippines. Front. Microbiol. 2013, 4, 102. [Google Scholar] [CrossRef] [Green Version]
- Röderova, M.; Halova, D.; Papousek, I.; Dolejska, M.; Masarikova, M.; Hanulik, V.; Pudova, V.; Broz, P.; Htoutou-Sedlakova, M.; Sauer, P.; et al. Characteristics of Quinolone Resistance in Escherichia coli Isolates from Humans, Animals, and the Environment in the Czech Republic. Front. Microbiol. 2017, 7, 2147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoll, C.; Sidhu, J.P.S.; Tiehm, A.; Toze, S. Prevalence of Clinically Relevant Antibiotic Resistance Genes in Surface Water Samples Collected from Germany and Australia. Environ. Sci. Technol. 2012, 46, 9716–9726. [Google Scholar] [CrossRef] [PubMed]
- Christou, A.; Agüera, A.; Bayona, J.M.; Cytryn, E.; Fotopoulos, V.; Lambropoulou, D.; Manaia, C.M.; Michael, C.; Revitt, M.; Schröder, P.; et al. The Potential Implications of Reclaimed Wastewater Reuse for Irrigation on the Agricultural Environment: The Knowns and Unknowns of the Fate of Antibiotics and Antibiotic Resistant Bacteria and Resistance Genes—A Review. Water Res. 2017, 123, 448–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, Z.G.; Zhang, K.; Zhang, Y. Occurrence and Distribution of Antibiotic Resistance Genes in the Coastal Area of the Bohai Bay, China. Mar. Pollut. Bull. 2016, 107, 245–250. [Google Scholar] [CrossRef]
- Proia, L.; von Schiller, D.; Sànchez-Melsió, A.; Sabater, S.; Borrego, C.M.; Rodríguez-Mozaz, S.; Balcázar, J.L. Occurrence and Persistence of Antibiotic Resistance Genes in River Biofilms after Wastewater Inputs in Small Rivers. Environ. Pollut. 2016, 210, 121–128. [Google Scholar] [CrossRef]
- Amato, M.; Dasí, D.; González, A.; Ferrús, M.A.; Castillo, M.Á. Occurrence of Antibiotic Resistant Bacteria and Resistance Genes in Agricultural Irrigation Waters from Valencia City (Spain). Agric. Water Manag. 2021, 256, 107097. [Google Scholar] [CrossRef]
- Lai, F.Y.; Muziasari, W.; Virta, M.; Wiberg, K.; Ahrens, L. Profiles of Environmental Antibiotic Resistomes in the Urban Aquatic Recipients of Sweden Using High-Throughput Quantitative PCR Analysis. Environ. Pollut. 2021, 287, 117651. [Google Scholar] [CrossRef]
- Jurelevicius, D.; Cotta, S.R.; Montezzi, L.F.; Dias, A.C.F.; Mason, O.U.; Picão, R.C.; Jansson, J.K.; Seldin, L. Enrichment of Potential Pathogens in Marine Microbiomes with Different Degrees of Anthropogenic Activity. Environ. Pollut. 2021, 268, 115757. [Google Scholar] [CrossRef]
- Amarasiri, M.; Sano, D.; Suzuki, S. Understanding Human Health Risks Caused by Antibiotic Resistant Bacteria (ARB) and Antibiotic Resistance Genes (ARG) in Water Environments: Current Knowledge and Questions to Be Answered. Crit. Rev. Environ. Sci. Technol. 2020, 50, 2016–2059. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Z.; Zhang, L.; Fan, W.; Yang, C.; Li, E.; Du, Y.; Wang, X. Inconsistent Seasonal Variation of Antibiotics between Surface Water and Groundwater in the Jianghan Plain: Risks and Linkage to Land Uses. J. Environ. Sci. 2021, 109, 102–113. [Google Scholar] [CrossRef]
- Furlan, J.P.R.; Stehling, E.G. Detection of β-Lactamase Encoding Genes in Feces, Soil and Water from a Brazilian Pig Farm. Environ. Monit. Assess. 2018, 190, 76. [Google Scholar] [CrossRef] [PubMed]
- Queenan, A.M.; Bush, K. Carbapenemases: The Versatile β-Lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef]
- Yang, Y.; Song, W.; Lin, H.; Wang, W.; Du, L.; Xing, W. Antibiotics and Antibiotic Resistance Genes in Global Lakes: A Review and Meta-Analysis. Environ. Int. 2018, 116, 60–73. [Google Scholar] [CrossRef] [Green Version]
- Poirel, L.; Madec, J.-Y.; Lupo, A.; Schink, A.-K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial Resistance in Escherichia coli. Microbiol. Spectr. 2018, 6, 289–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riaz, L.; Mahmood, T.; Khalid, A.; Rashid, A.; Ahmed Siddique, M.B.; Kamal, A.; Coyne, M.S. Fluoroquinolones (FQs) in the Environment: A Review on Their Abundance, Sorption and Toxicity in Soil. Chemosphere 2018, 191, 704–720. [Google Scholar] [CrossRef] [PubMed]
- van Doorslaer, X.; Dewulf, J.; van Langenhove, H.; Demeestere, K. Fluoroquinolone Antibiotics: An Emerging Class of Environmental Micropollutants. Sci. Total Environ. 2014, 500–501, 250–269. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Wang, S.; Fan, J.; Long, S.; Wang, L.; Tang, C.; Tam, N.F.; Yang, Y. Predicting Distribution Coefficients for Antibiotics in a River Water–Sediment Using Quantitative Models Based on Their Spatiotemporal Variations. Sci. Total Environ. 2019, 655, 1301–1310. [Google Scholar] [CrossRef]
- Leal, R.M.P.; Alleoni, L.R.F.; Tornisielo, V.L.; Regitano, J.B. Sorption of Fluoroquinolones and Sulfonamides in 13 Brazilian Soils. Chemosphere 2013, 92, 979–985. [Google Scholar] [CrossRef] [Green Version]
- Brooks, J.P.; Adeli, A.; Read, J.J.; McLaughlin, M.R. Rainfall Simulation in Greenhouse Microcosms to Assess Bacterial-Associated Runoff from Land-Applied Poultry Litter. J. Environ. Qual. 2009, 38, 218–229. [Google Scholar] [CrossRef]
- Singh, P.; Karimi, A.; Devendra, K.; Waldroup, P.W.; Cho, K.K.; Kwon, Y.M. Influence of Penicillin on Microbial Diversity of the Cecal Microbiota in Broiler Chickens. Poult. Sci. 2013, 92, 272–276. [Google Scholar] [CrossRef]
- Shin, H.W.; Lim, J.; Kim, S.; Kim, J.; Kwon, G.C.; Koo, S.H. Characterization of Trimethoprim-Sulfamethoxazole Resistance Genes and Their Relatedness to Class 1 Integron and Insertion Sequence Common Region in Gram-Negative Bacilli. J. Microbiol. Biotechnol. 2015, 25, 137–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eliopoulos, G.M.; Huovinen, P. Resistance to Trimethoprim-Sulfamethoxazole. Antimicrob. Resist. 2001, 32, 1608–1614. [Google Scholar] [CrossRef] [PubMed]
- Ugbo, E.N.; Anyamene, C.O.; Moses, I.B.; Iroha, I.R.; Babalola, O.O.; Ukpai, E.G.; Chukwunwejim, C.R.; Egbule, C.U.; Emioye, A.A.; Okata-Nwali, O.D.; et al. Prevalence of blaTEM, blaSHV, and blaCTX-M Genes among Extended Spectrum Beta-Lactamase-Producing Escherichia coli and Klebsiella pneumoniae of Clinical Origin. Gene Rep. 2020, 21, 100909. [Google Scholar] [CrossRef]
- Fadil Saedii, A.A.; Abdelraheim, A.R.; Abdel Aziz, A.A.; Swelam, S.H. ESBL-Producing E. coli and Klebsiella among Patients Treated at Minia University Hospitals. J. Infect. Dis. Prev. Med. 2017, 5, 156. [Google Scholar] [CrossRef]
- Zongo, K.J.; Dabire, A.M.; Compaore, L.G.; Sanou, I.; Sangare, L.; Simpore, J.; Zeba, B. First Detection of bla TEM, SHV and CTX-M among Gram Negative Bacilli Exhibiting Extended Spectrum -Lactamase Phenotype Isolated at University Hospital Center, Yalgado Ouedraogo, Ouagadougou, Burkina Faso. Afr. J. Biotechnol. 2015, 14, 1174–1180. [Google Scholar] [CrossRef] [Green Version]
- Elshafiee, E.A.; Kadry, M.; Nader, S.M.; Ahmed, Z.S. Extended-Spectrum-Beta-Lactamases and Carbapenemase-Producing Klebsiella pneumoniae Isolated from Fresh Produce Farms in Different Governorates of Egypt. Vet. World 2022, 15, 1191–1196. [Google Scholar] [CrossRef]
- Schuetz, A.N. Emerging Agents of Gastroenteritis: Aeromonas, Plesiomonas, and the Diarrheagenic Pathotypes of Escherichia coli. Semin. Diagn Pathol. 2019, 36, 187–192. [Google Scholar] [CrossRef]
- Meurant, A.; Guérin, F.; le Hello, S.; Saint-Lorant, G.; de La Blanchardière, A. Cefepime Use: A Need for Antimicrobial Stewardship. Infect. Dis. Now 2021, 51, 445–450. [Google Scholar] [CrossRef]
- Mançano, S.M.C.N.; Campana, E.H.; Felix, T.P.; Barrueto, L.R.L.; Pereira, P.S.; Picão, R.C. Frequency and Diversity of Stenotrophomonas Spp. Carrying blaKPC in Recreational Coastal Waters. Water Res. 2020, 185, 116210. [Google Scholar] [CrossRef]
- Falgenhauer, L.; Schwengers, O.; Schmiedel, J.; Baars, C.; Lambrecht, O.; Heß, S.; Berendonk, T.U.; Falgenhauer, J.; Chakraborty, T.; Imirzalioglu, C. Multidrug-Resistant and Clinically Relevant Gram-Negative Bacteria Are Present in German Surface Waters. Front. Microbiol. 2019, 10, 2779. [Google Scholar] [CrossRef] [Green Version]
- WHO. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis; WHO/EMP/IAU/2017.12; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Lapidot, A.; Yaron, S. Transfer of Salmonella enterica Serovar Typhimurium from Contaminated Irrigation Water to Parsley Is Dependent on Curli and Cellulose, the Biofilm Matrix Components. J. Food Prot. 2009, 72, 618–623. [Google Scholar] [CrossRef] [PubMed]
- Marques, R.Z.; Wistuba, N.; Brito, J.C.M.; Bernardoni, V.; Rocha, D.C.; Gomes, M.P. Crop Irrigation (Soybean, Bean, and Corn) with Enrofloxacin-Contaminated Water Leads to Yield Reductions and Antibiotic Accumulation. Ecotoxicol. Environ. Saf. 2021, 216, 112193. [Google Scholar] [CrossRef] [PubMed]
- Uyttendaele, M.; Jaykus, L.A.; Amoah, P.; Chiodini, A.; Cunliffe, D.; Jacxsens, L.; Holvoet, K.; Korsten, L.; Lau, M.; McClure, P.; et al. Microbial Hazards in Irrigation Water: Standards, Norms, and Testing to Manage Use of Water in Fresh Produce Primary Production. Compr. Rev. Food Sci. Food Saf. 2015, 14, 336–356. [Google Scholar] [CrossRef]

| Ceftriaxone Resistant Strains | Antimicrobial Resistance Genes | |||
|---|---|---|---|---|
| blaTEM | blaSHV | |||
| Escherichia sp. (CCA 1.2) * | 1 | 0 | ||
| Escherichia sp. (CCA 1.3) | 1 | 0 | ||
| Klebsiella sp. (CCA 2.1) | 1 | 1 | ||
| Klebsiella sp. (CCA 2.2) | 1 | 1 | ||
| Klebsiella sp. (CCA 2.3) | 1 | 1 | ||
| Sulfamethoxazole resistant strains | sul1 | sul2 | intI1 | intI2 |
| Escherichia sp. (CSA 1.1) * | 0 | 1 | 1 | 0 |
| Proteus sp. (CSA 2.1A) | 0 | 1 | 0 | 0 |
| Aeromonas sp. (CSA 2.2) | 0 | 1 | 0 | 0 |
| Enterobacter sp. (CSA 2.3) | 1 | 0 | 1 | 0 |
| Enterobacter sp. (CSA 5.1) | 0 | 0 | 0 | 1 |
| Enterobacter sp. (CSA 5.2) | 0 | 0 | 0 | 1 |
| Pantoea sp. (CSB 1.1) | 0 | 0 | 0 | 1 |
| Escherichia sp. (CSD 1.2) | 0 | 1 | 0 | 0 |
| Escherichia sp. (CSD 1.3) | 0 | 1 | 1 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopes, E.S.; Parente, C.E.T.; Picão, R.C.; Seldin, L. Irrigation Ponds as Sources of Antimicrobial-Resistant Bacteria in Agricultural Areas with Intensive Use of Poultry Litter. Antibiotics 2022, 11, 1650. https://doi.org/10.3390/antibiotics11111650
Lopes ES, Parente CET, Picão RC, Seldin L. Irrigation Ponds as Sources of Antimicrobial-Resistant Bacteria in Agricultural Areas with Intensive Use of Poultry Litter. Antibiotics. 2022; 11(11):1650. https://doi.org/10.3390/antibiotics11111650
Chicago/Turabian StyleLopes, Eliene S., Cláudio E. T. Parente, Renata C. Picão, and Lucy Seldin. 2022. "Irrigation Ponds as Sources of Antimicrobial-Resistant Bacteria in Agricultural Areas with Intensive Use of Poultry Litter" Antibiotics 11, no. 11: 1650. https://doi.org/10.3390/antibiotics11111650
APA StyleLopes, E. S., Parente, C. E. T., Picão, R. C., & Seldin, L. (2022). Irrigation Ponds as Sources of Antimicrobial-Resistant Bacteria in Agricultural Areas with Intensive Use of Poultry Litter. Antibiotics, 11(11), 1650. https://doi.org/10.3390/antibiotics11111650









