Abstract
Bacteria with carbapenem or tigecycline resistance have been spreading widely among humans, animals and the environment globally, being great threats to public health. However, bacteria co-carrying drug resistance genes of carbapenem and tigecycline in Shewanella and Acinetobacter species remain to be investigated. Here, we detected nine blaNDM-1-carrying Shewanella spp. isolates as well as three A. portensis isolates co-harboring tet(X3) and blaNDM-1 from seventy-two samples collected from a dairy farm in China. To explore their genomic characteristic and transmission mechanism, we utilized various methods, including PCR, antimicrobial susceptibility testing, conjugation experiment, whole-genome sequencing, circular intermediate identification and bioinformatics analysis. Clonal dissemination was found among three A. portensis, of which tet(X3) and blaNDM-1 were located on a novel non-conjugative plasmid pJNE5-X3_NDM-1 (333,311 bp), and the circular intermediate ΔISCR2-tet(X3)-blaNDM-1 was identified. Moreover, there was another copy of tet(X3) on the chromosome of A. portensis. It was verified that blaNDM-1 could be transferred to Escherichia coli C600 from Shewanella spp. by conjugation, and self-transmissible IncA/C2 plasmids mediated the transmission of blaNDM-1 in Shewanella spp. strains. Stringent surveillance was warranted to curb the transmission of such vital resistance genes.
1. Introduction
Carbapenems are essential treatment options for clinically significant, multidrug-resistant (MDR) Gram-negative bacteria infections because they have a broad antibacterial spectrum and high antibacterial activity [1]. On the other hand, the populations of carbapenem-resistant bacteria have been quickly growing worldwide in recent years, posing a severe threat to public health [2]. Diverse carbapenemases, which are typically encoded on transmissible plasmids, are the fundamental mechanism of carbapenem resistance. The New Delhi metallo-β-lactamase (NDM), Klebsiella pneumoniae carbapenemase (KPC) and OXA-48-type oxacillinase are the three most common carbapenemases [3]. The NDM-1 was initially discovered in India and has now spread throughout the world. It has the ability to hydrolyze practically all β-lactam antibiotics, resulting in the development of MDR bacteria [4]. Because of their excellent therapeutic action on extended-spectrum β-lactamases (ESBLs) and the AmpC enzyme-producing bacteria, meropenem and imipenem are frequently used to treat severe Gram-negative bacteria infections. However, due to the wide prevalence of carbapenem-resistant Gram-negative bacteria in recent years, effective antibiotics against drug-resistant bacteria remain scarce, with tigecycline serving as the last-resort option [5].
Tigecycline is used to treat a variety of clinical infections caused by Gram-positive and Gram-negative bacteria with multidrug resistance. However, the discoveries of plasmid-mediated tet(X3) and tet(X4) have limited its utility, owing to the capacity of tet(X) to catalyze the degradation of tigecycline [6,7]. According to previous retrospective screening, plasmid-borne tet(X) genes were present in bacteria of different settings, including food animals, migratory birds, clinical specimens and environmental samples in China [7,8]. Importantly, tet(X3) also had a high carriage rate in the gastrointestinal tract of cows [9]. Considering that there are many cow-related food products, antibiotic resistance genes could have been transmitted to humans through vocational contact and transfer among humans.
Known as environmental bacteria, Shewanella spp. was widely distributed in marine ecosystems and could also be recovered from food-producing animals and human active areas such as hospitals [10,11,12]. Although most of the human infections reported linked with Shewanella spp. were opportunistic and sporadic, disease syndromes and multidrug resistance have still increased in recent years [13]. Previous reports about Shewanella related to multidrug resistance were usually associated with blaOXA rather than blaNDM. A blaOXA-416-carrying extensively resistant isolate of Shewanella xiamenensis was isolated in Algeria from hospital effluents [14], blaOXA-55-carrying Shewanella algae was isolated from a patient in the hospital in Marseille, France [15] and several chromosome-based blaOXA-48-like variants were found in Shewanella spp. from ornamental fish in the Netherlands [10]. In 2017, Shewanella putrefaciens with chromosomal blaOXA-436 and plasmid-borne blaNDM-1 was isolated from a hospital in Pakistan [11].
Acinetobacter portensis was a novel Acinetobacter species originally identified from raw meat [16]. It is now further described as one MDR pathogen in this study for the first time. The emergence of Acinetobacter spp. co-harboring tet(X3) and blaNDM-1 from animal samples in China has already been reported, but A. portensis was not investigated [5,17].
In this study, we looked at the prevalence and molecular characteristics of MDR bacteria with resistance to carbapenem and tigecycline from a dairy farm in China in 2021, and we totally identified twelve NDM-1-producing isolates, including nine Shewanella spp. strains and three A. portensis strains carrying two copies of tet(X) simultaneously, all of which were collected from milking environment samples. The genomic epidemiology of drug-resistant strains and the characteristics of resistance plasmids were also analyzed, with the goal of learning about the molecular genetic characteristics of carbapenem-resistant and tigecycline-resistant bacteria of dairy farm origin to infer the underlying potential public health concerns.
2. Results and Discussion
2.1. Collection of Antimicrobial-Resistant Strains and Resistance Phenotypes
Thirteen meropenem-resistant strains were isolated from seventy-two non-duplicated samples collected from a dairy farm in China, including three A. portensis (23.08%), nine Shewanella spp. (69.23%) and one Stenotrophomonas maltophilia (7.69%). Except for the strain Stenotrophomonas maltophilia with inherent resistance to carbapenems collected from the shed environment, the remaining isolates were all identified from milking environment samples and demonstrated to be blaNDM-positive by PCR, with three A. portensis strains positive for tet(X) as well.
The results of antimicrobial susceptibility testing show that all the carbapenem-resistant isolates conferred resistance to meropenem, ceftiofur and amoxicillin but were susceptible to enrofloxacin, tigecycline and colistin. Five isolates showed resistance to chloramphenicol (38.46%) and seven isolates showed resistance to tetracycline (53.85%). Due to inherent drug resistance in Stenotrophomonas maltophilia, we only incorporated this strain into antimicrobial susceptibility testing, but no more in-depth explorations were conducted.
Three A. portensis isolates exhibited highly similar antimicrobial susceptibility profiles, being resistant to meropenem, ceftiofur, tetracycline and amoxicillin but susceptible to other antibiotics tested (Table 1). Acinetobacter spp. carrying tet(X) but susceptible to tigecycline was reported previously [9,18]. This phenomenon confirmed it was always possible to detect tet(X) in strains without a tigecycline resistance phenotype, which highlighted that the traditional methods of bacterial isolation based on selective media supplemented with antibiotics may impair the recovery of bacteria harboring critical precursors of resistance genes, although they may not confer the resistance phenotype directly. For some reasons to be investigated, one possibility was the inhibition of tet(X) function within certain species, suggesting tet(X) still could express its resistance by transmitting to other hosts [9,18]. In a manner of speaking, the existence of this silent dissemination was even more dangerous.

Table 1.
Antimicrobial susceptibility testing of carbapenem-resistant isolates collected from the dairy farm and their corresponding transconjugants.
All of the Shewanella spp. isolates were sensitive to colistin, tigecycline and enrofloxacin but resistant to meropenem, ceftiofur and amoxicillin. Five strains (55.6%) and four strains (44.4%) were resistant to chloramphenicol and tetracycline, respectively. The minimum inhibitory concentrations of Shewanella spp. isolates to meropenem were in the range from 16 to 128 μg/mL (Table 1). As previously reported, in Shewanella, blaOXA could lead to a low-level resistance (8–16 μg/mL) to meropenem or was incapable of reducing susceptibility to carbapenems in different occasions [19,20,21]. By contrast, the production of NDM was more concerned in terms of meropenem resistance in these bacteria.
2.2. Transfer Ability of MDR Plasmids
Conjugation experiments by broth mating were conducted on twelve blaNDM-positive strains. The results show that the blaNDM-1 of seven Shewanella strains (77.8%) could be transferred to the recipient strain E. coli C600. Antimicrobial susceptibility testing demonstrated that the corresponding transconjugants were resistant to amoxicillin, meropenem and ceftiofur, and MICs of meropenem ranged from 16 to 32 μg/mL (Table 1). The fact that the carbapenem resistance gene blaNDM-1 could spread among different species of bacteria suggested Shewanella spp. was likely to be the reservoir of antibiotic resistance genes, including blaNDM-1.
For three A. portensis, both tet(X3) and blaNDM-1 genes could not be transferred to three different recipient strains by broth mating. Subsequently, we performed electroporation experiments on these strains but also failed after three repeats. Recently, isolates bearing tet(X) without corresponding tigecycline resistance have been reported [9,18]. Plasmidscarrying tet(X3) could be transferred to Acinetobacter baumannii from Acinetobacter towneri susceptible to tigecycline, making a 128-fold increase in the MIC of tigecycline in transconjugant. This species-dependent peculiarity was likely to explain the different resistance phenotypes conferred by the same gene [18]. However, plasmids carrying tet(X3) and blaNDM-1 in the same Acinetobacter species were often non-conjugative in lab experiment conditions [9].
2.3. Characteristics of Nine Shewanella spp. Isolates
For better characterization, the genomic DNA of nine Shewanella spp. isolates were extracted then subjected to short-read and long-read sequencing. Species identification by whole-genome sequences based on PubMLST displayed that the nine Shewanella strains had the highest match with Shewanella putrefaciens (52~60%). Considering the matching degrees were not high enough, we thought conservatively that these nine Shewanella strains belonged to other subspecies rather than Shewanella putrefaciens. There were only small SNPs among the core genomes of JNE2, JNE17, JNE4-2 and JNE8 (Figure 1a) and highly similar antibiotic resistance gene distribution (Figure 1b), indicating these four strains from different samples had a close phylogenetic relationship and may derive from the same ancestor. Likewise, there were no obvious SNPs differences among JNE4-1, JNE9-1, JNE10-2, JNE3-1 and JNE7 (Figure 1a), which means the five strains were clonally related. However, significant differences lay between these two groups of strains (SNPs > 14,500) (Figure 1a). Not only did this suggest that these two groups of strains are clearly not from the same clone, but their subspecies were also likely to be different. Based on this, we tended to classify these nine strains into two new distinct subspecies of Shewanella.
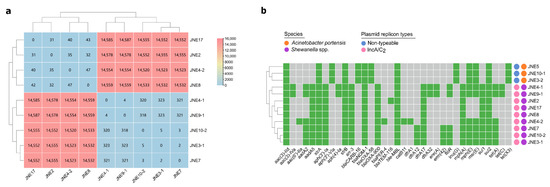
Figure 1.
Characteristics of thirteen carbapenem-resistant isolates. (a) A heatmap of core genome SNPs analysis among nine Shewanella spp. strains carrying blaNDM-1. (b) A heatmap of antibiotic resistance genes, species and plasmid replicon types for thirteen carbapenem-resistant isolates. Resistance genes are marked positive by green and negative by gray. The species and plasmid replicon types are showed by different colored circles.
There were at least 14 resistance genes in every Shewanella strain, and a multicopy of sul1 existed in these strains (Figure 1b). This further implied Shewanella spp. may well be the reservoir of significant antibiotic resistance genes, facilitating the wide spread of drug-resistant bacteria. Replicon analysis found that all of the Shewanella isolates carried plasmids of IncA/C2 type (Figure 1b). Over the years, the spread of blaNDM has been found associated with IncA/C plasmids [22,23].
In line with SNPs analysis, JNE7, JNE2, JNE4-2 and JNE10-2 were chosen as the representative strains to be sequenced with the MinION long-read platform. Results show that blaNDM-1 in four strains was located on IncA/C2 plasmids, pJNE7-NDM (137,224 bp), pJNE2-NDM (152,348 bp), pJNE4-2-NDM (152,348 bp) and pJNE10-2-NDM (166,090 bp), respectively. Based on the WGS analysis, it was found that clonal dissemination existed among 9 Shewanella isolates, and it could be concluded that blaNDM-1 located on IncA/C2 plasmids was prevalent in nine Shewanella isolates. The four plasmids showed high similarity, and pJNE2-NDM showed 100% sequence identity to the plasmid p1540-2 (94% coverage, CP019053) in E. coli and 99.98% sequence identity to the plasmid P2-NDM-1 (94% coverage, CP087671) in K. pneumoniae (Figure 2a), suggesting the dissemination of blaNDM-1 in milking environments was mediated by highly similar IncA/C2 plasmids and highlighting the mobile nature of those resistance genes. We subsequently analyzed the detailed genetic contexts of blaNDM-1 by mapping the assembled sequences to the plasmid pSA70-3 in Shewanella putrefaciens, p1540-2 in E. coli, pCf75 in Citrobacter freundii and P2-NDM-1 in K. pneumoniae from the NCBI database (Figure 2b). Limited by the short-read data, complete accurate genetic structures were difficult to obtain. However, it could still be observed clearly that the genetic contexts of blaNDM-1 among strains in this study and those from the database were strikingly similar, revealing the wide spread of blaNDM-1 among different bacteria.
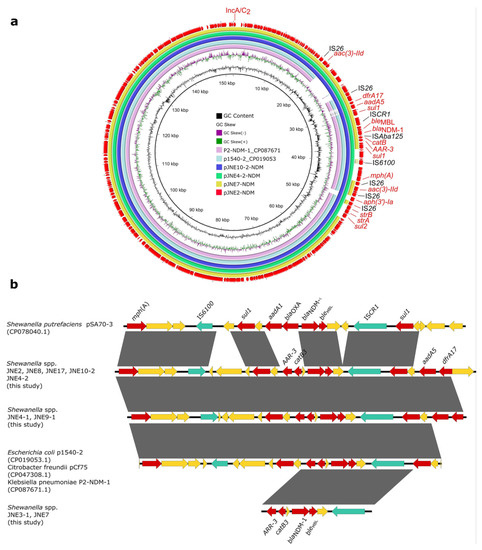
Figure 2.
Genetic features of blaNDM-1 in Shewanella strains. (a) Circular comparison of blaNDM-1-carrying plasmids pJNE2-NDM, pJNE7-NDM, pJNE4-2-NDM and pJNE10-2-NDM. (b) Linear sequence comparison of genetic context of blaNDM-1 in the nine Shewanella spp. isolates in this study with pSA70-3 (CP078040.1), p1540-2 (CP019053.1), pCf75 (CP047308.1) and P2-NDM-1 (CP087671.1) from the NCBI database. Regions with >99% sequence identity are marked by gray shading, and red arrows denote antibiotic resistance genes. Green arrows denote insertion sequences, and yellow arrows represent other genes.
2.4. Characteristics of Three Acinetobacter Portensis Isolates
Three A. portensis strains co-carrying tet(X3) and blaNDM-1 were isolated from different milking environment samples, which were designated as JNE5, JNE3-2 and JNE10-1, respectively. The phylogenetic analysis based on SNPs of core genomes showed that three A. portensis strains had a high degree of similarity (Figure 3), and there were no more than 22 SNPs among core genomes of these strains, implying clonal dissemination was likely to occur in the dairy farm.
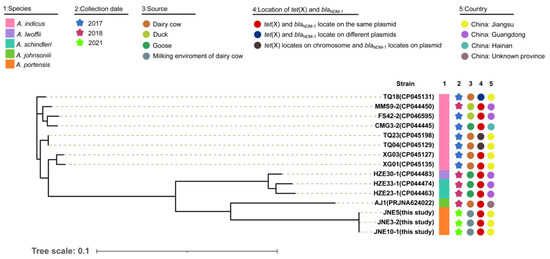
Figure 3.
Phylogenetic analysis and genetic characteristics of Acinetobacter isolates co-harboring tet(X) and blaNDM-1 from this study and the NCBI database. The strain species, collection date, source, country and location of tet(X) and blaNDM-1 in the genomes are showed by subsequent color blocks.
For better comparison, we gathered information of Acinetobacter isolates co-harboring tet(X3) and blaNDM-1 in recent years from an online database (Figure 3). According to the information collected, including the data of this study, Acinetobacter indicus carrying both tet(X3) and blaNDM-1 had been isolated as early as 2017 in China. However, the locations of tet(X3) and blaNDM-1 were not conserved, either on the same plasmid, on different plasmids, or on chromosome and plasmid separately (Figure 3). Such co-existing isolates were concentrated in China and sporadically distributed; they were diverse in species, with the majority of them belonging to A. indicus (Figure 3). Additionally, these isolates came from a wide range of sources, including dairy cows and their environments, ducks and gooses. A. portensis was a newly isolated species of Acinetobacter that harbored both tet(X3) and blaNDM-1; this was the first time that such drug-resistant bacteria had been collected from the milking environment of dairy cows. In the A. portensis in this study, tet(X3) and blaNDM-1 were located on the same plasmid, similar to the majority of such co-existing stains collected in recent years (Figure 3). Considering the diverse sources and wide distribution of strains in China carrying both tet(X3) and blaNDM-1, and the situation that there was probably clonal dissemination in this study, measures must be implemented to avoid their further dissemination and contamination.
Interestingly, each A. portensis isolate carried 13 identically acquired antibiotic resistance genes (Figure 1b), conferring resistance to aminoglycosides (aac(3)-IId, aph(3′)-Via, strA, strB), sulphonamide (sul2), glycopeptides (bleMBL), macrolide (mph(E), msr(E)), lincosamides (lnu(G)), carbapenems (blaNDM-1, blaCARB-16 and blaOXA-58) and tetracyclines (tet(X3)), further supporting the aforementioned clonal dissemination.
2.5. Genetic Contexts of blaNDM-1 and tet(X3)
In the same way, we performed short-read sequencing of three A. portensis. According to the linear comparison of sequences, it was obvious that the genetic contexts of blaNDM-1 and tet(X3) in JNE5, JNE3-2 and JNE10-1 were identical (Figure 4). On account of the clonal dissemination among three A. portensis strains revealed by phylogenetic analysis, we chose the JNE5 strain as the representative strain to be sequenced with the MinION long-read platform to obtain the complete circular genome sequences. Analysis displayed that the genome of JNE5 was comprised of a chromosome (2,568,515 bp) and five non-typeable plasmids, pJNE5-X3_NDM-1 (333,311 bp), pJNE5-64k (64,629 bp), pJNE5-OXA-58 (59,583 bp), pJNE5-3k (3617 bp) and pJNE5-1k (1416 bp). Most of the drug resistance genes were located on plasmid pJNE5-X3_NDM-1. In addition, it should be noted that tet(X3) had two copies, one was located on the chromosome, and the other one was located on the plasmid pJNE5-X3_NDM-1 with blaNDM-1. Moreover, there was one blaOXA-58 existing in pJNE5-OXA-58.
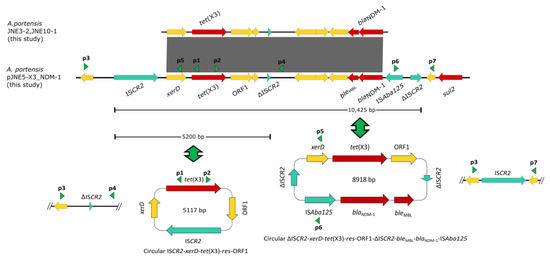
Figure 4.
Linear sequence comparison of genetic context of tet(X3) and blaNDM-1 in three Acinetobacter portensis isolates. Regions of >99% homology are marked by gray shading, red arrows denote antibiotic resistance genes and arrows indicate the direction of transcription of the genes. Green arrowheads p1, p2, p3 and p4 indicate the positions of primers used for amplification of the ISCR2-xerD-tet(X3)-res-ORF1 circular intermediate, arrowheads p3, p5, p6 and p7 are used for amplification of the ΔISCR2-xerD-tet(X3)-res-ORF1-ΔISCR2-bleMBL-blaNDM-1-ISAba125 circular intermediate.
The genetic context of chromosomal tet(X) had high coverage with that of the A. indicus TQ23 chromosome (CP045198.1). Similarly, genetic contexts of blaOXA-58 on plasmid pJNE5-OXA-58 and A. indicus p18TQ-X3 (CP045132.1) separately also had a large number of consistencies (Figure 5a,b). Plasmid comparison revealed that the plasmid pJNE5-X3_NDM-1 shared a low degree of genetic identity with two previously reported plasmids co-harboring tet(X3) and blaNDM-1 (pXG01-X3 and pHZE30-1-1). Although pOXA58_010030 showed the highest sequence identity to pJNE5-X3_NDM-1, it lacked the most important multidrug resistance region (Figure 6). The typical genetic context, ISAba125-blaNDM-1-bleMBL-trpF-dsbC, appeared in plasmid pJNE5-X3_NDM-1 [24]. Additionally, tet(X3) was located within a 5200 bp region with the gene arrangement ISCR2-xerD-tet(X3)-res-ORF1-ΔISCR2, with one intact ISCR2 element located on the upstream of tet(X3) and another ΔISCR2 (83 bp) with the same orientation lay in the downstream (Figure 4).
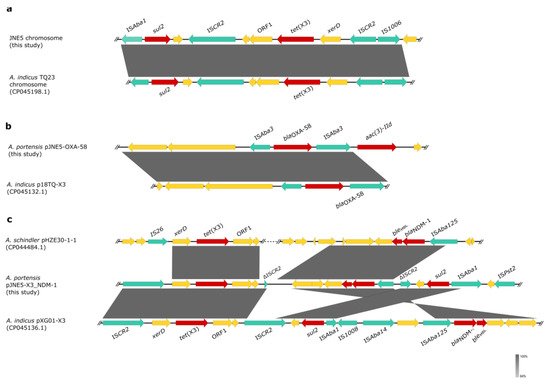
Figure 5.
Genetic features of blaNDM-1, tet(X) and blaOXA-48 in JNE5 strain. (a) Linear alignment of the tet(X3)-carrying chromosome in JNE5 with chromosome from A. indicus TQ23 (CP045198.1). (b) Linear alignment of the blaOXA-48-positive plasmid pJNE5-OXA-58 with p18TQ-X3. (c), Linear sequence comparison of genetic context of blaNDM-1 and tet(X3) in plasmid pJNE5-X3_NDM-1 with that of pHZE30-1 (CP044484.1) and pXG01-X3 (CP045136.1). Regions of homology are marked by shading; red arrows denote antibiotic resistance genes. Green arrows denote insertion sequences, and yellow arrows represent other genes.
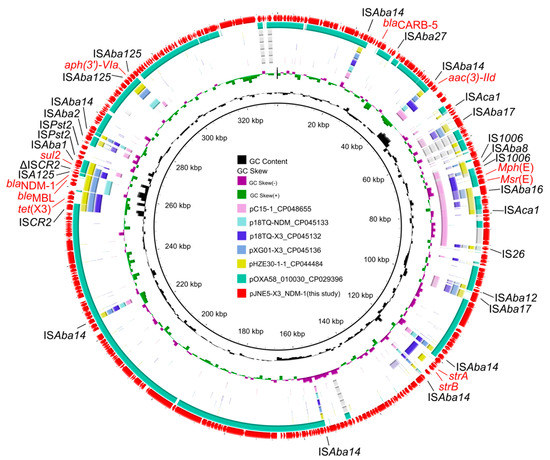
Figure 6.
Circular comparison of the plasmid pJNE5-X3_NDM-1 co-harbouring tet(X3) and blaNDM-1 in Acinetobacter portensis JNE5 strain with similar plasmids from the NCBI database. The outmost circle denotes the reference plasmid pJNE5-X3_NDM-1.
Inverse PCR was carried out to examine whether the minicircle ISCR2-tet(X3) could take shape. Finally, we obtained a fragment of 4245 bp in length, and sequence analysis revealed that it included only one intact copy of ISCR2 element (ISCR2-xerD-tet(X3)-res-orf1) (Figure 4), which was the same as previously reported [9]. To reconfirm the above situation, we used inward-facing primers and then obtained a 1664 bp PCR amplicon, which included only one ΔISCR2 (83 bp) element (Figure 4). Compared with other reported plasmids co-harboring tet(X3) and blaNDM-1, the significant difference in plasmid pJNE5-X3_NDM-1 was that tet(X3) and blaNDM-1 stood exceptionally close to each other (Figure 5c). What was noteworthy was that one ΔISCR2 (389 bp) element lay in the downstream of blaNDM-1; since it was in the same direction as the intact ISCR2 element located on the upstream of tet(X3), outward-facing primers were designed to examine whether this MDR region could also be excised and mobilizable. A 1875 bp amplicon was generated and sequence analysis revealed that it included only one copy of the ΔISCR2 (389 bp) element. The results of PCR using inward-facing primers showed a 2933 bp amplicon, including one intact copy of the ISCR2 element, which verified the instability of circular intermediate ΔISCR2-tet(X3)-blaNDM-1 by excision (Figure 4). The generation of circular intermediate containing tet(X3) and blaNDM-1 was a warning that the two important antibiotic resistance genes were likely to transmit together, which warranted further investigations. Although conjugation experiments failed, the potential hazard of the MDR plasmids still could not be neglected.
3. Materials and Methods
3.1. Bacterial Isolation and Identification
In total, 72 non-duplicated samples were collected from a dairy farm in Xuzhou, Jiangsu Province in May 2021, including 14 milk samples from cows with mastitis, 31 stool samples, 15 milking environment samples and 12 shed environment samples. To enrich the microbiota, solid and liquid samples, as well as cotton swabs (surface samples), were incubated in a 5 mL LB broth without antimicrobials for 6 h. These enrichment broth suspensions were streaked onto MacConkey agar plates containing meropenem (2 mg/L) using a sterile loop to isolate carbapenem-resistant colonies. The boiling method was used to extract bacterial genome DNA, which was subsequently screened for carbapenem resistance genes (blaNDM, blaVIM, blaKPC, blaIMP, blaSIM, blaDIM, blaAIM, blaBIC, blaSPM, blaOXA and blaGIM) using the previously reported primers [25]. The carbapenem-resistant bacteria were screened again for tet(X) genes using primers described earlier [6]. 16S rRNA gene sequencing was conducted to further identify their species using the forward primer 5′-AGAGTTTGATCATGGCTCAG-3′ and the reverse primer 5′-GTGTGACGGGCGGTGTGTAC-3′.
3.2. Antimicrobial Susceptibility Testing
Fresh CAMH broth was prepared for antimicrobial susceptibility testing, and the minimal inhibitory concentrations (MICs) of chloramphenicol, colistin, meropenem, tigecycline, enrofloxacin, ceftiofur, tetracycline and amoxicillin were determined by the broth microdilution method. The results are interpreted according to the guidelines of the Clinical and Laboratory Standards Institute [26] and the European Committee on Antimicrobial Susceptibility Testing (EUCAST, v12.0) (http://www.eucast.org/clinical_breakpoints/ (accessed on 1 January 2022)) with E. coli ATCC 25922 as a quality control strain.
3.3. Conjugation and Electroporation Experiments
Conjugation experiments by broth mating were carried out using blaNDM-positive strains as donors, with strains E. coli C600 (rifampicin resistant), E. coli J53 (sodium azide resistant) and A. baumannii ATCC19606 (chloramphenicol resistant) as recipients. Only E. coli C600 was used as the recipient of nine Shewanella spp. isolates, while all recipients mentioned above were used for A. portensis. Transconjugants were separately selected on LB agar plates containing rifampicin (300 mg/L) with meropenem (2 mg/L), sodium azide (300 mg/L) with meropenem (2 mg/L), or chloramphenicol (64 mg/L) together with meropenem (2 mg/L) and then confirmed by PCR. The broth microdilution method was used to determine the MICs of a range of antimicrobials for the transconjugants. For A. portensis isolates that meropenem resistance phenotype failed to transfer by conjugation, electroporation experiment was performed, in which A. baumannii ATCC19606 was prepared as the receipt strain.
3.4. Whole-Genome Sequencing and Bioinformatics Analysis
Genomic DNA of all blaNDM-positive isolates from overnight cultures was extracted using the FastPure® Bacteria DNA Isolation Mini Kit (Vazyme, Nanjing, China). The genomic DNA was subjected to short-read sequencing (2 × 150 bp) with the Illumina HiSeq 2500 platform (Illumina, San Diego, CA, USA). Short-read Illumina raw reads were de novo assembled using SPAdes [27] with default parameters, and contigs less than 200 bp were discarded. To obtain the complete sequences, we selected several representative strains and extracted their genomic DNA to perform Oxford Nanopore Technologies MinION long-read sequencing [28,29]. The complete genome sequences were modified manually and automatically annotated using RAST (http://rast.nmpdr.org/ (accessed on 1 January 2022)). The BRIG tool was used to perform circular comparison of the plasmids in this study and the homologous plasmids available from the NCBI database [30]. A pairwise SNP distance matrix was generated using snp-dists 0.6.3 (https://github.com/tseemann/snp-dists (accessed on 1 January 2022)). Roary [31] and FastTree [32] based on the SNPs of core genomes were used to construct phylogenetic trees, which were visualized by iTOL v5 [33]. Easyfig was used to generate linear comparison in order to visualize the sequence comparison features of genetic contexts [34].
3.5. Identification of Circular Intermediates
To determine whether the recombination of ISCR2 and ΔISCR2 (83 bp) elements could form the tet(X3)-carrying circular intermediate, all plasmids, including the tet(X3)-carrying plasmid pJNE5-X3_NDM-1 from the JNE5 strain, were extracted for inverse PCR assays, using outward-facing primers p1 5′-TCGGTCGTTGTCTCTTTCGT-3′ and p2 5′-TTGATGTCGCCTTTTGCAGG-3′ for detection of minicircle ISCR2-tet(X3). When the band of target fragment was detected through agarose gel electrophoresis, the same primers were used to sequence the amplified minicircle product. Then inward-facing primers p3 5′-CGCAGCGTTTCGTACATCAG-3′ and p4 5′-AGGTCAATCAGACTGGGCGTT-3′ were used to verify the excision result. In the same way, to see if the recombination of ISCR2 and ΔISCR2 (389 bp) could lead to the formation of circular intermediate co-carrying tet(X3) and blaNDM-1, outward-facing primers p5 5′-TGTTCCATTCCCTTGGTGGT-3′ and p6 5′-ATGTGCCTTTTTGCCAGGGT-3′ were used for detecting minicircle ΔISCR2-tet(X3)-blaNDM-1. Inward-facing primers p3 and p7 5′-GACGGTATTCGTGGCAAAGC-3′ were used to confirm the excision result.
4. Conclusions
In this work, we detected nine NDM-1-producing Shewanella spp. strains and three A. portensis strains co-harboring tet(X3) and blaNDM-1. Clonal dissemination existed among three A. portensis isolates, in which tet(X3) and blaNDM-1 co-located on a novel non-conjugative plasmid, and there was another tet(X3) located on the chromosome. Additionally, we confirmed that the circular intermediate ΔISCR2-tet(X3)-blaNDM-1 could be generated. The emergence of tet(X3) and blaNDM-1-bearing plasmids in different bacteria among dairy cow farming environments constitutes a potential public health concern. Continuous monitoring and surveillance of critical resistance genes in such environments are necessary to ensure good farming standards.
Author Contributions
Conceptualization, Z.W., H.S. and R.L.; methodology, R.L., K.P. and X.L.; validation, R.L., L.Z. and X.L.; formal analysis, R.L., X.X., Y.L. and L.Z.; data curation, L.Z. and X.L.; writing—original draft preparation, L.Z. and X.L.; writing—review and editing, R.L., L.Z. and X.L.; visualization, R.L., L.Z. and K.P. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31872526 and 32161133005), the China Postdoctoral Science Foundation (no. 2020M671632) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The draft genome sequences were submitted to NCBI with the BioProject number PRJNA828415. Complete genome sequences of four representative strains JNE5 (PRJNA828443), JNE10-2 (PRJNA829047), JNE2 (PRJNA829244) and JNE7 (PRJNA829126) were also deposited. The complete sequence of pJNE4-2-NDM was also submitted for reference (ON391944).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Papp-Wallace, K.M.; Endimiani, A.; Taracila, M.A.; Bonomo, R.A. Carbapenems: Past, Present, and Future. Antimicrob. Agents Chemother. 2011, 55, 4943–4960. [Google Scholar] [CrossRef]
- Wang, H.; Li, X.; Liu, B.T. Occurrence and characterization of KPC-2-producing ST11 Klebsiella pneumoniae isolate and NDM-5-producing Escherichia coli isolate from the same horse of equestrian clubs in China. Transbound. Emerg. Dis. 2021, 68, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Kelly, A.M.; Mathema, B.; Larson, E.L. Carbapenem-resistant Enterobacteriaceae in the community: A scoping review. Int. J. Antimicrob. Agents 2017, 50, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Xu, K.; Zhao, L.; Tong, R.; Xiong, L.; Shi, J. Recent research and development of NDM-1 inhibitors. Eur. J. Med. Chem. 2021, 223, 113667. [Google Scholar] [CrossRef]
- Cui, C.Y.; Chen, C.; Liu, B.T.; He, Q.; Wu, X.T.; Sun, R.Y.; Zhang, Y.; Cui, Z.H.; Guo, W.Y.; Jia, Q.L.; et al. Co-occurrence of Plasmid-Mediated Tigecycline and Carbapenem Resistance in Acinetobacter spp. from Waterfowls and Their Neighboring Environment. Antimicrob. Agents Chemother. 2020, 64, e02502-19. [Google Scholar] [CrossRef]
- He, T.; Wang, R.; Liu, D.; Walsh, T.; Zhang, R.; Lv, Y.; Ke, Y.; Ji, Q.; Wei, R.; Liu, Z.; et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 2019, 4, 1450–1456. [Google Scholar] [CrossRef]
- Sun, J.; Chen, C.; Cui, C.Y.; Zhang, Y.; Liu, Y.H. Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat. Microbiol. 2019, 4, 1457–1464. [Google Scholar] [CrossRef]
- Cao, J.; Wang, J.; Wang, Y.; Wang, L.; Gao, G.F. Tigecycline resistance tet(X3) gene is going wild. Biosaf. Health 2020, 2, 9–11. [Google Scholar] [CrossRef]
- Zhang, R.; Dong, N.; Zeng, Y.; Shen, Z.; Lu, J.; Liu, C.; Huang, Z.A.; Sun, Q.; Cheng, Q.; Shu, L.; et al. Chromosomal and Plasmid-Borne Tigecycline Resistance Genes tet(X3) and tet(X4) in Dairy Cows on a Chinese Farm. Antimicrob. Agents Chemother. 2020, 64, e00674-20. [Google Scholar] [CrossRef]
- Ceccarelli, D.; Essen-Zandbergen, A.v.; Veldman, K.T.; Tafro, N.; Haenen, O.; Mevius, D.J. Chromosome-Based blaOXA-48-Like Variants in Shewanella Species Isolates from Food-Producing Animals, Fish, and the Aquatic Environment. Antimicrob. Agents Chemother. 2017, 61, e01013-16. [Google Scholar] [CrossRef] [PubMed]
- Potter, R.F.; D’Souza, A.W.; Wallace, M.A.; Shupe, A.; Patel, S.; Gul, D.; Kwon, J.H.; Andleeb, S.; Burnham, C.-A.D.; Draft, G.D. Genome Sequence of the blaOXA-436 and blaNDM-1-Harboring Shewanella putrefaciens SA70 Isolate. Genome Announc. 2017, 5, e00644-17. [Google Scholar] [CrossRef] [PubMed]
- Janda, J.M.; Abbott, S.L. The genus Shewanella: From the briny depths below to human pathogen. Crit. Rev. Microbiol. 2014, 40, 293–312. [Google Scholar] [CrossRef]
- Yousfi, K.; Bekal, S.; Usongo, V.; Touati, A. Current trends of human infections and antibiotic resistance of the genus Shewanella. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 1353–1362. [Google Scholar] [CrossRef] [PubMed]
- Yousfi, K.; Touati, A.; Lefebvre, B.; Fournier, É.; Côté, J.; Soualhine, H.; Walker, M.; Bougdour, D.; Tremblay, C.; Bekal, S. A Novel Plasmid, pSx1, Harboring a New Tn1696 Derivative from Extensively Drug-Resistant Shewanella xiamenensis Encoding OXA-416. Microb. Drug Resist. 2017, 23, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Cimmino, T.; Olaitan, A.; Rolain, J. Whole genome sequence to decipher the resistome of Shewanella algae, a multidrug-resistant bacterium responsible for pneumonia, Marseille, France. Expert Rev. Anti-Infect. Ther. 2016, 14, 269–275. [Google Scholar] [CrossRef]
- Carvalheira, A.; Gonzales-Siles, L.; Salvà-Serra, F.; Lindgren, S.; Moore, E.R.B. Acinetobacter portensis sp. nov. and Acinetobacter guerrae sp. nov., isolated from raw meat. Int. J. Syst. Evol. Microbiol. 2020, 70, 4544–4554. [Google Scholar] [CrossRef]
- He, T.; Li, R.; Wei, R.; Liu, D.; Bai, L.; Zhang, L.; Gu, J.; Wang, R.; Wang, Y. Characterization of Acinetobacter indicus co-harbouring tet(X3) and blaNDM-1 of dairy cow origin. J. Antimicrob. Chemother. 2020, 75, 2693–2696. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.-Y.; Liu, Y.; Chen, Y.; Huang, F.-M.; Chen, R.-C.; Xiao, Y.-H.; Zhou, K.; Downing, T.; Cevallos, M.; Siddavattam, D.; et al. Sporadic Dissemination of tet(X3) and tet(X6) Mediated by Highly Diverse Plasmidomes among Livestock-Associated Acinetobacter. Microbiol. Spectr. 2021, 9, e01141-21. [Google Scholar] [CrossRef]
- Tacão, M.; Araújo, S.; Vendas, M.; Alves, A.; Henriques, I. Shewanella species as the origin of bla genes: Insights into gene diversity, associated phenotypes and possible transfer mechanisms. Int. J. Antimicrob. Agents 2018, 51, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Antonelli, A.; Di Palo, D.M.; Galano, A.; Becciani, S.; Montagnani, C.; Pecile, P.; Galli, L.; Rossolini, G.M. Intestinal carriage of Shewanella xiamenensis simulating carriage of OXA-48–producing Enterobacteriaceae. Diagn. Microbiol. Infect. Dis. 2015, 82, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Tacão, M.; Correia, A.; Henriques, I. Environmental Shewanella xiamenensis strains that carry bla OXA-48 or bla OXA-204 genes: Additional proof for bla OXA-48-like gene origin. Antimicrob. Agents Chemother. 2013, 57, 6399–6400. [Google Scholar] [CrossRef] [PubMed]
- Kumarasamy, K.K.; Toleman, M.A.; Walsh, T.R.; Bagaria, J.; Woodford, N. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect. Dis. 2010, 10, 578–579. [Google Scholar] [CrossRef]
- Marta, A.; Manuel, O.; Luísa, G.; Augusto, C.; Patrice, N.; Laurent, P. Occurrence of NDM-1-producing Morganella morganii and Proteus mirabilis in a single patient in Portugal: Probable in vivo transfer by conjugation. J. Antimicrob. Chemother. 2020, 75, 903–906. [Google Scholar]
- Fu, Y.; Du, X.; Ji, J.; Chen, Y.; Jiang, Y.; Yu, Y. Epidemiological characteristics and genetic structure of blaNDM-1 in non-baumannii Acinetobacter spp. in China. J. Antimicrob. Chemother. 2012, 67, 2114–2122. [Google Scholar] [CrossRef]
- Poirel, L.; Walsh, T.R.; Cuvillier, V.; Nordmann, P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 2011, 70, 119–123. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI Supplement M100; CLSI: Wayne, PA, USA, 2020. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Li, R.; Xie, M.; Dong, N.; Lin, D.; Yang, X.; Yin, W.; Wai-Chi, C.E.; Chen, S. Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. Gigascience 2018, 7, gix132. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.F.; Petty, N.K.; Zakour, N.L.B.; Beatson, S.A.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. Bmc Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Price, M.; Dehal, P.; Arkin, A. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).