Iron Chelator DIBI Suppresses Formation of Ciprofloxacin-Induced Antibiotic Resistance in Staphylococcus aureus
Abstract
:1. Introduction
2. Results
2.1. Staphylococcus aureus Accumulates Excess Medium Iron
2.2. Cellular Iron Status Affects Sensitivity to DIBI but Not to CIP
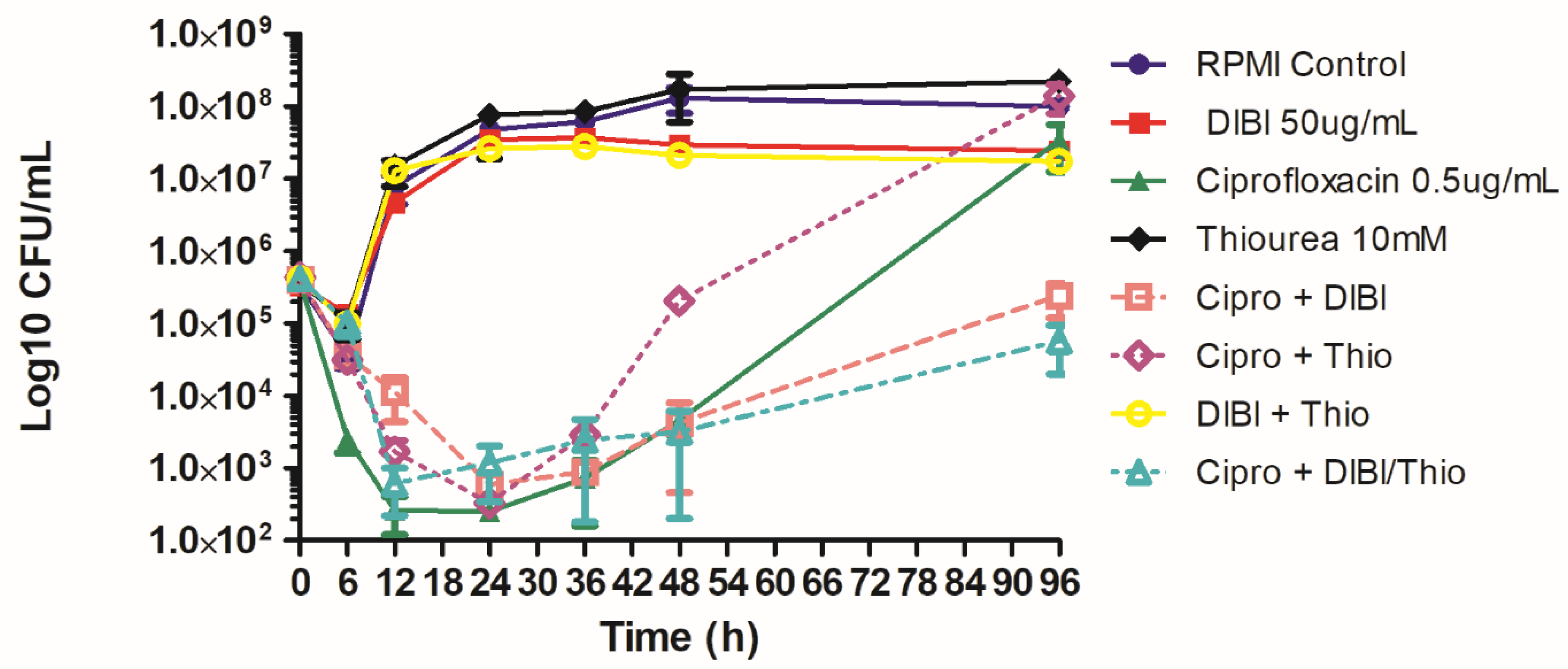
2.3. DIBI Suppresses Recovery Growth of CIP-Exposed Bacteria
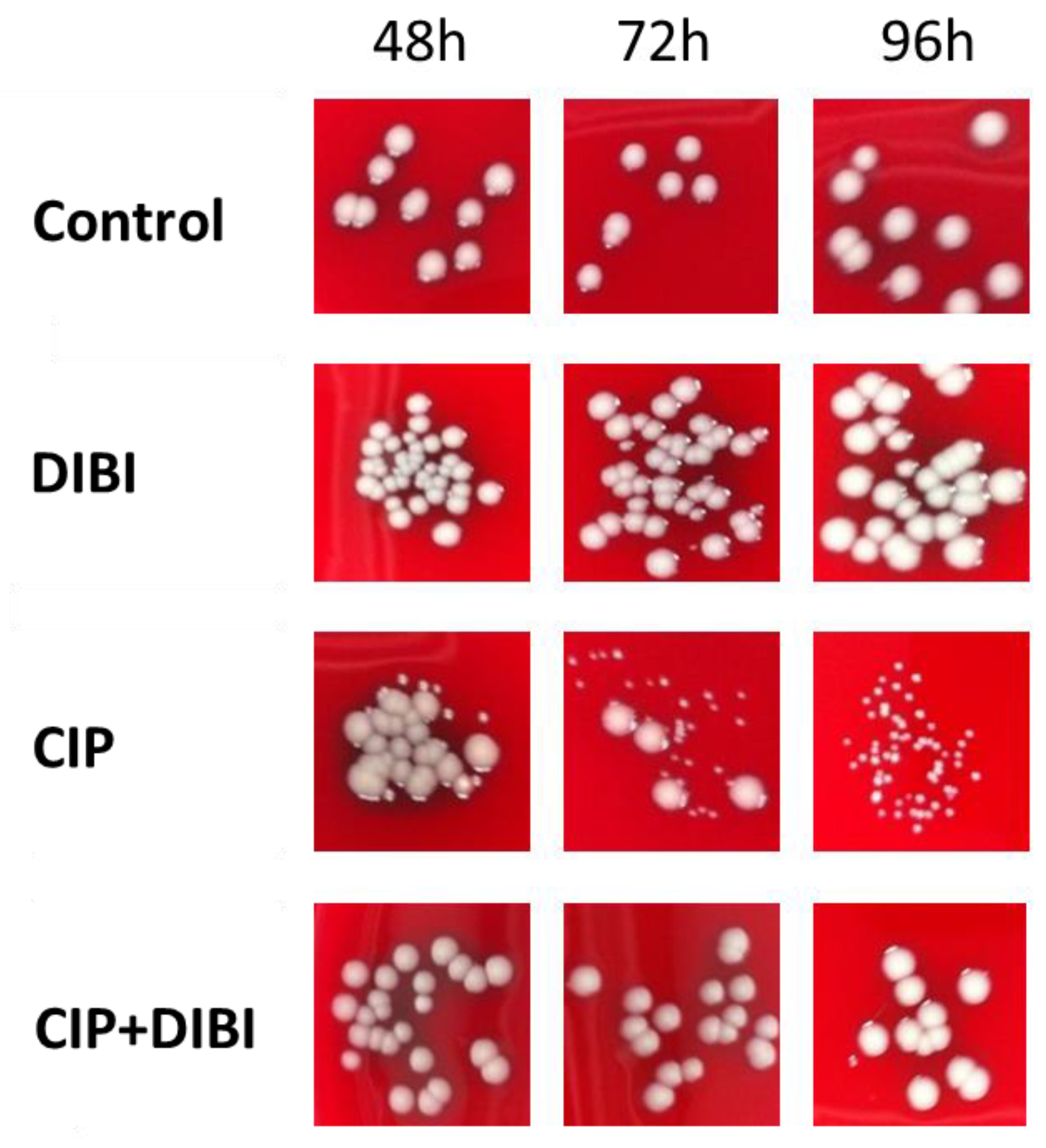
2.4. Prolonged CIP-Exposure Selective for Small Colony Variant Phenotype
2.5. CIP-induced persister SCVs Display Stable Acquired Resistance to Quinolones
3. Discussion
4. Materials and Methods
4.1. Antibiotics, Media, and Bacterial Strains
4.2. Antibiotic Kill and Recovery Growth Testing
4.3. Isolation and Characterization of Treatment Survivors
4.4. Antibiotic Susceptibility
4.5. Trace Element Quantification of Iron, Manganese, and Zinc
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lewis, K. Persister cells. Annu. Rev. Microbiol. 2010, 64, 357–372. [Google Scholar] [CrossRef] [PubMed]
- Michiels, J.; Van den Bergh, B.; Verstraeten, N.; Michiels, J. Molecular mechanisms and clinical implications of bacterial persistence. Drug Resis. Updates 2016, 29, 76–89. [Google Scholar] [CrossRef] [PubMed]
- Schaaff, F.; Bierbaum, G.; Baumert, N.; Bartmann, P.; Sahl, H.G. Mutations are involved in emergence of aminoglycoside-induced small colony variants of Staphylococcus aureus. Int. J. Med. Microbiol. 2003, 293, 427–435. [Google Scholar] [CrossRef]
- Proctor, R.; Kriegeskorte, A.; Kahl, B.; Becker, K.; Löffler, B.; Peters, G. Staphylococcus aureus small colony variants (SCVs): A roadmap for the metabolic pathways involved in persistent infections. Front. Cell. Infect. Microbiol. 2014, 4, 99. [Google Scholar] [CrossRef]
- Proctor, R.; Kahl, B.; von Eiff, C.; Vaudaux, P.; Daniel, P.; Lew, D.; Peters, G. Staphylococcal small colony variants have novel mechanisms for antibiotic resistance. Clin. Infect. Dis. 1998, 27, S68–S74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, L.; Lemaire, S.; Kahl, B.; Becker, K.; Proctor, R.; Denis, O.; Tulkens, P.; Van Bambeke, F. Antibiotic activity against small colony variants of Staphylococcus aureus: Review of in vitro, animal, and clinical data. J. Antimicrob. Chemother. 2013, 68, 1455–1465. [Google Scholar] [CrossRef] [Green Version]
- Richter, K.; Thomas, N.; Zhang, G.; Prestidge, C.; Coenye, T.; Wormald, P.; Vreugde, S. Deferiprone and gallium-protoporphyrin have the capacity to potentiate the activity of antibiotics in Staphylococcus aureus small colony variants. Front. Cell. Infect. Microbiol. 2017, 7, 280. [Google Scholar] [CrossRef] [Green Version]
- Torres-Barcelo, C.; Kojadinovic, M.; Moxon, R.; MacLean, R.C. The SOS response increases bacterial fitness, but not evolvability, under a sublethal dose of antibiotic. Proc. Biol. Sci. 2015, 282, 20150885. [Google Scholar] [CrossRef] [Green Version]
- Maslowska, K.H.; Makiela-Dzbenska, K.; Fijalkowska, I.J. The SOS system: A complex and tightly regulated response to DNA damage. Environ. Mol. Mutagen. 2019, 60, 368–384. [Google Scholar] [CrossRef] [Green Version]
- Holbein, B.E.; Ang, M.T.C.; Allan, D.S.; Chen, W.; Lehmann, C. Iron-withdrawing anti-infectives for new host-directed therapies based on iron dependence, the Achilles’ heel of antibiotic-resistant microbes. Environ. Chem. Lett. 2021, 19, 2789–2808. [Google Scholar] [CrossRef]
- Kohanski, M.; Dwyer, D.; Hayete, B.; Lawrence, C.; Collins, J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 2007, 130, 797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, P.; Briales, A.; Brochmann, R.; Wang, H.; Kragh, K.; Kolpen, M.; Hempel, C.; Bjarnsholt, T.; Høiby, N.; Ciofu, O. Formation of hydroxyl radicals contributes to the bactericidal activity of ciprofloxacin against Pseudomonas aeruginosa biofilms. Pathog. Dis. 2014, 70, 440–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Méhi, O.; Bogos, B.; Csörgő, B.; Pál, F.; Nyerges, A.; Papp, B.; Pál, C. Perturbation of iron homeostasis promotes the evolution of antibiotic resistance. Mol. Biol. Evol. 2014, 31, 2793–2804. [Google Scholar] [CrossRef] [Green Version]
- Kara, M.; Hasinoff, B.; McKay, D.N.; Campbell, N. Clinical and chemical interactions between iron preparations and ciprofloxacin. Br. J. Pharmacol. 1991, 31, 257–261. [Google Scholar] [CrossRef]
- Heeb, S.; Fletcher, M.; Chhabra, S.; Diggle, S.; Williams, P.; Camara, M. Quinolones: From antibiotics to autoinducers. FEMS Microbiol. Rev. 2010, 35, 247–274. [Google Scholar] [CrossRef] [Green Version]
- Cirz, R.; Jones, M.; Gingles, N.; Minogue, T.; Jarrahi, B.; Peterson, S.; Romesberg, F. Complete and SOS-mediated response of Staphylococcus aureus to the antibiotic ciprofloxacin. J. Bacteriol. 2007, 189, 531–539. [Google Scholar] [CrossRef] [Green Version]
- Schroder, W.; Goerke, S.; Wolz, C. Opposing effects of aminocoumarins and fluoroquinolones on the SOS response and adaptability in Staphylococcus aureus. J. Antimicrob. Chemother. 2013, 68, 529–538. [Google Scholar] [CrossRef] [Green Version]
- Yeeles, J.; Cammack, R.; Dillingham, M. An iron-sulfur cluster is essential for the binding of broken DNA by AddAB-type helicase-nucleases. J. Biol. Chem. 2009, 284, 7746–7755. [Google Scholar] [CrossRef] [Green Version]
- Ambrose, P.G.; VanScoy, B.D.; Luna, B.M.; Yan, J.; Ulhaq, A.; Nielsen, T.B.; Rudin, S.; Hujer, K.; Bonomo, R.A.; Actis, L.; et al. Apotransferrin in combination with ciprofloxacin slows bacterial replication, prevents resistance amplification, and increases antimicrobial regimen effect. Antimicrob. Agents Chemother. 2019, 63, e00112-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ang, M.T.C.; Gumbau-Brisa, R.; Allan, D.S.; McDonald, R.; Ferguson, M.J.; Holbein, B.E.; Bierenstiel, M. DIBI, a 3-hydroxypyridin-4-one chelator iron-binding polymer with enhanced antimicrobial activity. Med. Chem. Commun. 2018, 9, 1206–1212. [Google Scholar] [CrossRef]
- Parquet, M.D.C.; Savage, K.A.; Allan, D.S.; Davidson, R.J.; Holbein, B.E. Novel iron-chelator DIBI inhibits Staphylococcus aureus growth, suppresses experimental MRSA infection in mice and enhances the activities of diverse antibiotics in vitro. Front Microbiol. 2018, 9, 1811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horsburgh, M.J.; Clements, M.O.; Crossley, H.; Ingham, E.; Foster, S.J. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 2001, 69, 3744–3754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, M.G.; Corey, B.W.; Si, Y.; Craft, D.W.; Zurawski, D. Antibacterial activities of iron chelators against common nosocomial pathogens. Antimicrob. Agents Chemother. 2012, 56, 5419–5421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, Y.M.; Barwinska-Sendra, A.; Tarrant, E.; Skaar, E.P.; Waldron, K.J.; Kehl-Fie, T.E. Superoxide dismutase capable of functioning with iron or manganese promotes the resistance of Staphylococcus aureus to calprotectin and nutritional immunity. PLoS Pathog. 2017, 13, e1006125. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhao, X.; Malik, M.; Drlica, K. Contribution of reactive oxygen species to pathways of quinolone-mediated bacterial cell death. J. Antimicrob. Chemother. 2010, 65, 520–524. [Google Scholar] [CrossRef] [Green Version]
- Hong, Y.; Li, Q.; Gao, Q.; Xie, J.; Huang, H.; Drlica, K.; Zhao, X. Reactive oxygen species play a dominant role in all pathways of rapid quinolone-mediated killing. J. Antimicrob. Chemother. 2020, 75, 576–585. [Google Scholar] [CrossRef] [Green Version]
- Van Acker, H.; Coenye, T. The role of reactive oxygen species in antibiotic-mediated killing of bacteria. Trends Microbiol. 2017, 25, 456–466. [Google Scholar] [CrossRef]
- Roberts, C.A.; Al-Tameemi, H.M.; Mashruwala, A.A.; Rosario-Cruz, Z.; Chauhan, U.; Sause, W.E.; Torres, V.J.; Belden, W.J.; Boyd, J.M. The Suf iron-sulfur cluster biosynthetic system is essential in Staphylococcus aureus, and decreased Suf function results in global metabolic defects and reduced survival in human neutrophils. Infect. Immun. 2017, 85, e00100-17. [Google Scholar] [CrossRef] [Green Version]
- Lim, C.S.Q.; Ha, K.P.; Clarke, R.S.; Gavin, L.A.; Cook, D.T.; Hutton, J.A.; Sutherell, C.L.; Edwards, A.M.; Evans, L.E.; Tate, E.W.; et al. Identification of a potent small-molecule inhibitor of bacterial DNA repair that potentiates quinolone antibiotic activity in methicillin-resistant Staphylococcus aureus. Bioorg. Med. Chem. 2019, 27, 114962. [Google Scholar] [CrossRef] [PubMed]
- Kuehl, R.; Morata, L.; Meylan, S.; Mensa, J.; Soriano, A. When antibiotics fail: A clinical and microbiological perspective on antibiotic tolerance and persistence of Staphylococcus aureus. J. Antimicrob. Chemother. 2020, 75, 1071–1086. [Google Scholar] [CrossRef]
- Peng, Q.; Zhou, S.; Yao, F.; Hou, B.; Huang, Y.; Hua, D.; Zheng, Y.; Qian, Y. Baicalein suppresses the SOS response system of Staphylococcus aureus induced by ciprofloxacin. Cell. Physiol. Biochem. 2011, 28, 1045–1050. [Google Scholar] [CrossRef] [PubMed]
- Perez, C.; Wei, Y.; Guo, M. Iron-binding and anti-Fenton properties of baicalein and baicalin. J. Inorg. Biochem. 2009, 103, 326–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minear, S.; O’Donnell, A.F.; Ballew, A.; Giaever, G.; Nislow, C.; Stearns, T.; Cyert, M.S. Curcumin inhibits growth of Saccharomyces cerevisiae through iron chelation. Eukaryot. Cell 2011, 10, 1574–1581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azad, G.K.; Singh, V.; Golla, U.; Tomar, R.S. Depletion of cellular iron by curcumin leads to alteration in histone acetylation and degradation of Sml1p in Saccharomyces cerevisiae. PLoS ONE 2013, 8, e59003. [Google Scholar] [CrossRef]
- Dai, C.; Lin, J.; Li, H.; Shen, Z.; Wang, Y.; Velkov, T.; Shen, J. The natural product curcumin as an antibacterial agent: Current achievements and problems. Antioxidants 2022, 11, 459. [Google Scholar] [CrossRef]
- Marathe, S.A.; Sen, M.; Dasgupta, I.; Chakravortty, D. Differential modulation of intracellular survival of cytosolic and vacuolar pathogens by curcumin. Antimicrob. Agents Chemother. 2012, 56, 5555–5567. [Google Scholar] [CrossRef] [Green Version]
- Marathe, S.A.; Kumar, R.; Ajitkumar, P.; Nagaraja, V.; Chakravortty, D. Curcumin reduces the antimicrobial activity of ciprofloxacin against Salmonella Typhimurium and Salmonella Typhi. J. Antimicrob. Chemother. 2013, 68, 139–152. [Google Scholar] [CrossRef] [Green Version]
- Campion, J.J.; McNamara, P.J.; Evans, M.E. Evolution of ciprofloxacin-resistant Staphylococcus aureus in in vitro pharmacokinetic environments. Antimicrob. Agents Chemother. 2004, 48, 4733–4744. [Google Scholar] [CrossRef] [Green Version]
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 12.0. 2022. Available online: http://www.eucast.org (accessed on 1 October 2022).
- Allan, D.S.; Parquet, M.D.C.; Savage, K.A.; Holbein, B.E. Iron sequestrant DIBI, a potential alternative for nares decolonization of methicillin-resistant Staphylococcus aureus, is anti-infective and inhibitory for mupirocin-resistant isolates. Antimicrob. Agents and Chemother. 2020, 64, e02353-19. [Google Scholar] [CrossRef]
- Parquet, M.D.C.; Savage, K.A.; Allan, D.S.; Ang, M.T.C.; Chen, W.; Logan, S.M.; Holbein, B.E. Antibiotic-resistant Acinetobacter baumannii is susceptible to the novel iron-sequestering anti-infective DIBI in vitro and in experimental pneumonia in mice. Antimicrob. Agents Chemother. 2019, 63, e00855-19. [Google Scholar] [CrossRef]
- Holbein, B.E.; Mira de Orduña, R. Effect of trace iron levels and iron withdrawal by chelation on the growth of Candida albicans and Candida vini. FEMS Microbiol. Lett. 2010, 307, 19–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphries, R.M.; Ambler, J.; Mitchell, S.L.; Castanheira, M.; Dingle, T.; Hindler, J.A.; Koeth, L.; Sei, K. CLSI methods development and standardization working group best practices for evaluation of antimicrobial susceptibility tests. J. Clin. Microbiol. 2018, 56, e01934-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chillappagari, S.; Seubert, A.; Trip, H.; Kuipers, O.P.; Marahiel, M.A.; Miethke, M. Copper stress affects iron homeostasis by destabilizing iron-sulfur cluster formation in Bacillus subtilis. J. Bacteriol. 2010, 192, 2512–2524. [Google Scholar] [CrossRef] [PubMed]


| Growth Medium | Medium Metal µM | S. aureus Cellular Metal µM | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ATCC 43300 | ATCC 25923 | ATCC 6538 | ||||||||||
| Fe | Mn | Zn | Fe | Mn | Zn | Fe | Mn | Zn | Fe | Mn | Zn | |
| MHB | 6.420 | 0.300 | 11.067 | 3.700 | 0.460 | 0.828 | 4.730 | 0.440 | 0.953 | 7.310 | 0.690 | 1.279 |
| FECMHB | 0.250 | 0.004 | 2.634 | 0.810 | 0.020 | 1.012 | 1.014 | 0.020 | 0.909 | 1.140 | 0.030 | 1.087 |
| RPMI | 0.080 | 0.025 | 0.559 | 0.390 | 0.040 | 0.959 | 0.610 | 0.030 | 0.605 | 0.570 | 0.040 | 1.057 |
| Cultivation Medium a | ATCC 25923 | ATCC 43300 | ||
|---|---|---|---|---|
| CIP MIC µg/mL (µM) | DIBI MIC µg/mL (µM) | CIP MIC µg/mL (µM) | DIBI MIC µg/mL (µM) | |
| MHB | 0.5 (1.5) | 2000 (222) | 0.5 (1.5) | 8000 (888) |
| FECMHB | ND | 1 (0.11) | ND | 2 (0.22) |
| RPMI | 0.25 (0.75) | 2 (0.22) | 0.25 (0.75) | 2 (0.22) |
| Culture Medium | CFU/mL | |
|---|---|---|
| 0 h | 24 h | |
| MHB | 1 × 105 | 3 × 107 |
| MHB+2 × MIC CIP | 1 × 105 | 2 × 101 |
| FECMHB | 7 × 104 | 3 × 107 |
| FECMHB +2 × MIC CIP | 7 × 104 | 6 × 101 |
| Treatment | DIBI a µg/mL (µM) | CIP a µg/mL (µM) | MOX b µg/mL (µM) | MUP b µg/mL (µM) | GEN c µg/mL (µM) | VAN c µg/mL (µM) |
|---|---|---|---|---|---|---|
| Control | 4 (0.4) | 0.125 (0.38) | 0.062 (0.15) | 0.062 (0.123) | 0.062 (0.13) | 0.5 (0.345) |
| DIBI | 4 (0.4) | 0.125 (0.38) | 0.062 (0.15) | 0.062 (0.123) | 0.062 (0.13) | 0.5 (0.345) |
| CIP | 4 (0.4) | 1.00 (3.0) | 0.25 (0.62) | 0.062 (0.123) | 0.062 (0.13) | 0.5 (0.345) |
| CIP + DIBI | 4 (0.4) | 0.125 (0.38) | 0.062 (0.15) | 0.062 (0.123) | 0.031 (0.065) | 0.5 (0.345) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Allan, D.S.; Holbein, B.E. Iron Chelator DIBI Suppresses Formation of Ciprofloxacin-Induced Antibiotic Resistance in Staphylococcus aureus. Antibiotics 2022, 11, 1642. https://doi.org/10.3390/antibiotics11111642
Allan DS, Holbein BE. Iron Chelator DIBI Suppresses Formation of Ciprofloxacin-Induced Antibiotic Resistance in Staphylococcus aureus. Antibiotics. 2022; 11(11):1642. https://doi.org/10.3390/antibiotics11111642
Chicago/Turabian StyleAllan, David S., and Bruce E. Holbein. 2022. "Iron Chelator DIBI Suppresses Formation of Ciprofloxacin-Induced Antibiotic Resistance in Staphylococcus aureus" Antibiotics 11, no. 11: 1642. https://doi.org/10.3390/antibiotics11111642
APA StyleAllan, D. S., & Holbein, B. E. (2022). Iron Chelator DIBI Suppresses Formation of Ciprofloxacin-Induced Antibiotic Resistance in Staphylococcus aureus. Antibiotics, 11(11), 1642. https://doi.org/10.3390/antibiotics11111642





