Recurrent E. coli Urinary Tract Infections in Nursing Homes: Insight in Sequence Types and Antibiotic Resistance Patterns
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Study Population
2.2. Bacterial Isolates and Definitions
2.3. Genome Sequencing, Assembly and Typing
2.4. Analysis of Strain Recurrence by SNP Analysis (Same-Strain rUTI)
2.5. Antibiotic Resistance Rates
2.6. Descriptive Analysis
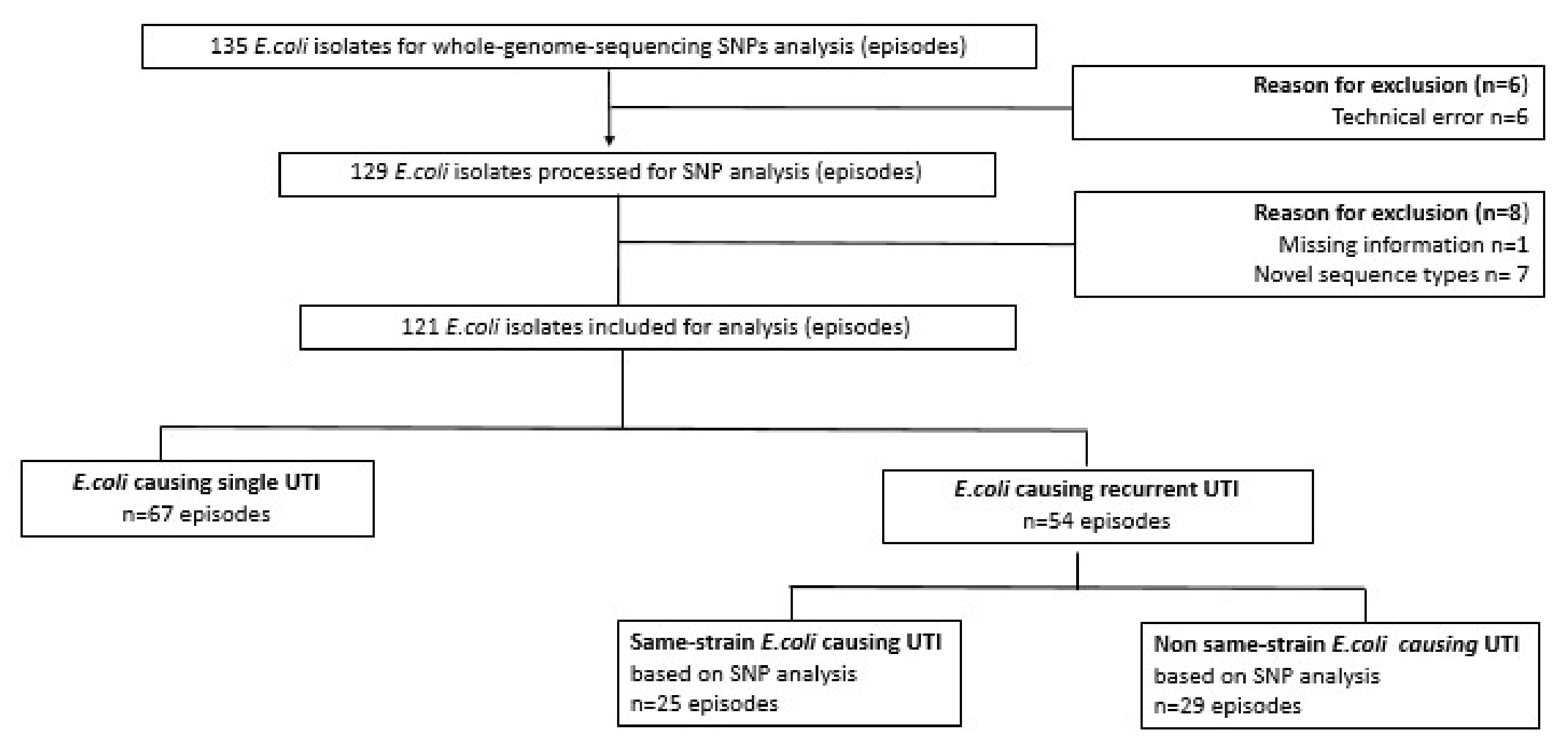
3. Results
3.1. E. coli Sequence Types in Nursing Home Residents with Single Versus Recurrent UTI
3.2. Relatedness of E. coli Isolates Based on SNP Analysis (Same-Strain Recurrence) from Seven Nursing Home Residents
3.3. Antibiotic Resistance in E. coli Causing rUTI
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rowe, T.A.; Juthani-Mehta, M. Diagnosis and management of urinary tract infection in older adults. Infect. Dis. Clin. North Am. 2014, 28, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Stamm, W.E.; Norrby, S.R. Urinary tract infections: Disease panorama and challenges. J. Infect. Dis. 2001, 183 (Suppl. S1), S1–S4. [Google Scholar] [CrossRef] [PubMed]
- Wagenlehner, F.; Wullt, B.; Ballarini, S.; Zingg, D.; Naber, K.G. Social and economic burden of recurrent urinary tract infections and quality of life: A patient web-based study (GESPRIT). Expert. Rev. Pharm. Outcomes Res. 2018, 18, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Terlizzi, M.E.; Gribaudo, G.; Maffei, M.E. UroPathogenic Escherichia coli (UPEC) Infections: Virulence Factors, Bladder Responses, Antibiotic, and Non-antibiotic Antimicrobial Strategies. Front. Microbiol. 2017, 8, 1566. [Google Scholar] [CrossRef] [PubMed]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Riley, L.W. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin. Microbiol. Infect. 2014, 20, 380–390. [Google Scholar] [CrossRef]
- Mody, L.; Juthani-Mehta, M. Urinary tract infections in older women: A clinical review. JAMA 2014, 311, 844–854. [Google Scholar] [CrossRef]
- Raz, R.; Gennesin, Y.; Wasser, J.; Stoler, Z.; Rosenfeld, S.; Rottensterich, E.; Stamm, W.E. Recurrent urinary tract infections in postmenopausal women. Clin. Infect. Dis. 2000, 30, 152–156. [Google Scholar] [CrossRef]
- Ikähelmo, R.; Siitonen, A.; Heiskanen, T.; Kärkkäinen, U.; Kuosmanen, P.; Lipponen, P.; Mäkelä, P.H. Recurrence of urinary tract infection in a primary care setting: Analysis of a 1-year follow-up of 179 women. Clin. Infect. Dis. 1996, 22, 91–99. [Google Scholar] [CrossRef]
- Dwyer, P.L.; O’Reilly, M. Recurrent urinary tract infection in the female. Curr. Opin. Obstet. Gynecol. 2002, 14, 537–543. [Google Scholar] [CrossRef]
- Nielsen, K.; Stegger, M.; Kiil, K.; Lilje, B.; Ejrnæs, K.; Leihof, R.; Skjøt-Rasmussen, L.; Godfrey, P.; Monsen, T.; Ferry, S.; et al. Escherichia coli Causing Recurrent Urinary Tract Infections: Comparison to Non-Recurrent Isolates and Genomic Adaptation in Recurrent Infections. Microorganisms 2021, 9, 1416. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.L.; Wu, M.; Henderson, J.P.; Hooton, T.M.; Hibbing, M.E.; Hultgren, S.J.; Gordon, J.I. Genomic diversity and fitness of E. coli strains recovered from the intestinal and urinary tracts of women with recurrent urinary tract infection. Sci. Transl. Med. 2013, 5, 184ra60. [Google Scholar] [CrossRef] [PubMed]
- Thänert, R.; Reske, K.A.; Hink, T.; Wallace, M.A.; Wang, B.; Schwartz, D.; Seiler, S.; Cass, C.; Burnham, C.-A.D.; Dubberke, E.R.; et al. Comparative Genomics of Antibiotic-Resistant Uropathogens Implicates Three Routes for Recurrence of Urinary Tract Infections. mBio 2019, 10, e01977-19. [Google Scholar] [CrossRef] [PubMed]
- Valentine-King, M.T.; Zoorob, R.; Germanos, G.; Salemi, J.; Gupta, K.; Grigoryan, L. Prior cultures predict subsequent susceptibility in patients with recurrent urinary tract infections. Antimicrob. Steward. Heal. Epidemiol. 2022, 2, s67. [Google Scholar] [CrossRef]
- Worby, C.J.; Olson, B.S.; Dodson, K.W.; Earl, A.M.; Hultgren, S.J. Establishing the role of the gut microbiota in susceptibility to recurrent urinary tract infections. J. Clin. Investig. 2022, 132. [Google Scholar] [CrossRef]
- Russo, T.A.; Stapleton, A.; Wenderoth, S.; Hooton, T.M.; Stamm, W.E. Chromosomal restriction fragment length polymorphism analysis of Escherichia coli strains causing recurrent urinary tract infections in young women. J. Infect. Dis. 1995, 172, 440–445. [Google Scholar] [CrossRef]
- Forde, B.M.; Roberts, L.W.; Phan, M.-D.; Peters, K.M.; Fleming, B.A.; Russell, C.W.; Lenherr, S.M.; Myers, J.B.; Barker, A.P.; Fisher, M.A.; et al. Population dynamics of an Escherichia coli ST131 lineage during recurrent urinary tract infection. Nat. Commun. 2019, 10, 3643. [Google Scholar] [CrossRef]
- Lindblom, A.; Kiszakiewicz, C.; Kristiansson, E.; Yazdanshenas, S.; Kamenska, N.; Karami, N.; Åhrén, C. The impact of the ST131 clone on recurrent ESBL-producing E. coli urinary tract infection: A prospective comparative study. Sci. Rep. 2022, 12, 10048. [Google Scholar] [CrossRef]
- Li, D.; Reid, C.J.; Kudinha, T.; Jarocki, V.M.; Djordjevic, S.P. Genomic analysis of trimethoprim-resistant extraintestinal pathogenic Escherichia coli and recurrent urinary tract infections. Microb. Genom. 2020, 6, e000475. [Google Scholar] [CrossRef]
- Kuil, S.D.; Hidad, S.; Fischer, J.C.; Harting, J.; Hertogh, C.M.P.M.; Prins, J.M.; de Jong, M.D.; van Leth, F.; Schneeberger, C. Sensitivity of C-Reactive Protein and Procalcitonin Measured by Point-of-Care Tests to Diagnose Urinary Tract Infections in Nursing Home Residents: A Cross-Sectional Study. Clin. Infect. Dis. 2021, 73, e3867–e3875. [Google Scholar] [CrossRef]
- Kuil, S.D.; Hidad, S.; Fischer, J.C.; Harting, J.; Hertogh, C.M.; Prins, J.M.; van Leth, F.; de Jong, M.D.; Schneeberger, C. Sensitivity of point-of-care testing C reactive protein and procalcitonin to diagnose urinary tract infections in Dutch nursing homes: PROGRESS study protocol. BMJ Open 2019, 9, e031269. [Google Scholar] [CrossRef] [PubMed]
- Aspevall, O.; Hallander, H.; Gant, V.; Kouri, T. European guidelines for urinalysis: A collaborative document produced by European clinical microbiologists and clinical chemists under ECLM in collaboration with ESCMID. Clin. Microbiol. Infect. 2001, 7, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Roer, L.; Johannesen, T.B.; Hansen, F.; Stegger, M.; Tchesnokova, V.; Sokurenko, E.; Garibay, N.; Allesøe, R.; Thomsen, M.C.F.; Lund, O.; et al. CHTyper, a Web Tool for Subtyping of Extraintestinal Pathogenic Escherichia coli Based on the fumC and fimH Alleles. J. Clin. Microbiol. 2018, 56, e00063-18. [Google Scholar] [CrossRef]
- Bessonov, K.; Laing, C.; Robertson, J.; Yong, I.; Ziebell, K.; Gannon, V.P.J.; Nichani, A.; Arya, G.; Nash, J.H.E.; Christianson, S. ECTyper: In silico Escherichia coli serotype and species prediction from raw and assembled whole-genome sequence data. Microb. Genom. 2021, 7, 000728. [Google Scholar] [CrossRef]
- Waters, N.R.; Abram, F.; Brennan, F.; Holmes, A.; Pritchard, L. Easy phylotyping of Escherichia coli via the EzClermont web app and command-line tool. Access Microbiol. 2020, 2, acmi000143. [Google Scholar] [CrossRef]
- Schwengers, O.; Hain, T.; Chakraborty, T.; Goesmann, A. ReferenceSeeker: Rapid determination of appropriate reference genomes. J. Open Source Softw. 2020, 5, 1994. [Google Scholar] [CrossRef]
- Gorrie, C.L.; Da Silva, A.G.; Ingle, D.J.; Higgs, C.; Seemann, T.; Stinear, T.P.; Williamson, D.A.; Kwong, J.C.; Grayson, M.L.; Sherry, N.L.; et al. Key parameters for genomics-based real-time detection and tracking of multidrug-resistant bacteria: A systematic analysis. Lancet Microbe. 2021, 2, e575–e583. [Google Scholar] [CrossRef]
- StataCorp. Stata Statistical Software: Release 17; StatCorp LLC.: College Station, TX, USA, 2021. [Google Scholar]
- Microsoft Corporation. Microsoft Excel [Internet]. 2016. Available online: https://office.microsoft.com/excel (accessed on 29 June 2022).
- CANVA. Gratis Ontwerptool. Available online: www.canva.nl (accessed on 20 August 2022).
- Verenso. Richtlijn Urineweginfecties Bij Kwetsbare Ouderen, Dutch National Guideline Urinary Tract Infection in Frail Older Adults 2018. Available online: https://www.verenso.nl/richtlijnen-en-praktijkvoering/richtlijnendatabase/urineweginfecties (accessed on 3 October 2022).
- Brauner, A.; Jacobson, S.H.; Kühn, I. Urinary Escherichia coli causing recurrent infections--a prospective follow-up of biochemical phenotypes. Clin. Nephrol. 1992, 38, 318–323. [Google Scholar]
- Foxman, B. Recurring urinary tract infection: Incidence and risk factors. Am. J. Public Health 1990, 80, 331–333. [Google Scholar] [CrossRef] [PubMed]
- Manges, A.R.; Geum, H.M.; Guo, A.; Edens, T.J.; Fibke, C.D.; Pitout, J.D.D. Global Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages. Clin. Microbiol. Rev. 2019, 32, e00135-18. [Google Scholar] [CrossRef] [PubMed]
- Haaijman, J.; Stobberingh, E.E.; van Buul, L.W.; Hertogh, C.; Horninge, H. Urine cultures in a long-term care facility (LTCF): Time for improvement. BMC Geriatr. 2018, 18, 221. [Google Scholar] [CrossRef] [PubMed]
- Hisano, M.; Bruschini, H.; Nicodemo, A.C.; Gomes, C.M.; Lucon, M.; Srougi, M. The Bacterial Spectrum and Antimicrobial Susceptibility in Female Recurrent Urinary Tract Infection: How Different They Are From Sporadic Single Episodes? Urology 2015, 86, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Stegger, M.; Leihof, R.F.; Baig, S.; Sieber, R.N.; Thingholm, K.R.; Marvig, R.L.; Frimodt-Møller, N.; Nielsen, K.L. A snapshot of diversity: Intraclonal variation of Escherichia coli clones as commensals and pathogens. Int. J. Med. Microbiol. 2020, 310, 151401. [Google Scholar] [CrossRef]
- Lorenzo-Gómez, M.F.; Padilla-Fernández, B.; Flores-Fraile, J.; Valverde-Martínez, S.; González-Casado, I.; Hernández, J.-M.D.D.; Sánchez-Escudero, A.; Arroyo, M.-J.V.; Martínez-Huélamo, M.; Criado, F.H.; et al. Impact of whole-cell bacterial immunoprophylaxis in the management of recurrent urinary tract infections in the frail elderly. Vaccine 2021, 39, 6308–6314. [Google Scholar] [CrossRef]


| Sequence Type | Overall (n = 121) | Recurrent UTI (n = 54) | Non-Recurrent UTI (n = 67) |
|---|---|---|---|
| Number of episodes (%) | |||
| 131 | 20 (16.5) | 10 (18.5) | 10 (14.9) |
| 73 | 16 (13.2) | 10 (18.5) | 6 (9.0) |
| 69 | 12 (9.9) | 1 (1.9) | 11 (16.4) |
| 10 | 6 (5.0) | 2 (3.7) | 4 (6.0) |
| 12 | 5 (4.1) | 5 (9.3) | 0 (0.0) |
| 38 | 5 (4.1) | 2 (3.7) | 3 (4.5) |
| 362 | 3 (2.5) | 0 (0.0) | 3 (4.5) |
| 405 | 3 (2.5) | 0 (0.0) | 3 (4.5) |
| 453 | 3 (2.5) | 3 (5.6) | 0 (0.0) |
| 538 | 3 (2.5) | 2 (3.7) | 1 (1.5) |
| Other * | 45 (37.2) | 19 (35.1) | 26 (38.7) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hidad, S.; van der Putten, B.; van Houdt, R.; Schneeberger, C.; Kuil, S.D. Recurrent E. coli Urinary Tract Infections in Nursing Homes: Insight in Sequence Types and Antibiotic Resistance Patterns. Antibiotics 2022, 11, 1638. https://doi.org/10.3390/antibiotics11111638
Hidad S, van der Putten B, van Houdt R, Schneeberger C, Kuil SD. Recurrent E. coli Urinary Tract Infections in Nursing Homes: Insight in Sequence Types and Antibiotic Resistance Patterns. Antibiotics. 2022; 11(11):1638. https://doi.org/10.3390/antibiotics11111638
Chicago/Turabian StyleHidad, Soemeja, Boas van der Putten, Robin van Houdt, Caroline Schneeberger, and Sacha Daniëlle Kuil. 2022. "Recurrent E. coli Urinary Tract Infections in Nursing Homes: Insight in Sequence Types and Antibiotic Resistance Patterns" Antibiotics 11, no. 11: 1638. https://doi.org/10.3390/antibiotics11111638
APA StyleHidad, S., van der Putten, B., van Houdt, R., Schneeberger, C., & Kuil, S. D. (2022). Recurrent E. coli Urinary Tract Infections in Nursing Homes: Insight in Sequence Types and Antibiotic Resistance Patterns. Antibiotics, 11(11), 1638. https://doi.org/10.3390/antibiotics11111638







