Removal of Antibiotic Resistance Genes, Class 1 Integrase Gene and Escherichia coli Indicator Gene in a Microalgae-Based Wastewater Treatment System
Abstract
:1. Introduction
2. Results
2.1. Time Course Changes of Water Quality Parameters in Microalgal Cultures
2.1.1. Total Dissolved Solids
2.1.2. Electric Conductivity
2.1.3. Total Suspended Solids
2.1.4. pH
2.1.5. Temperature
2.1.6. Nitrate Nitrogen
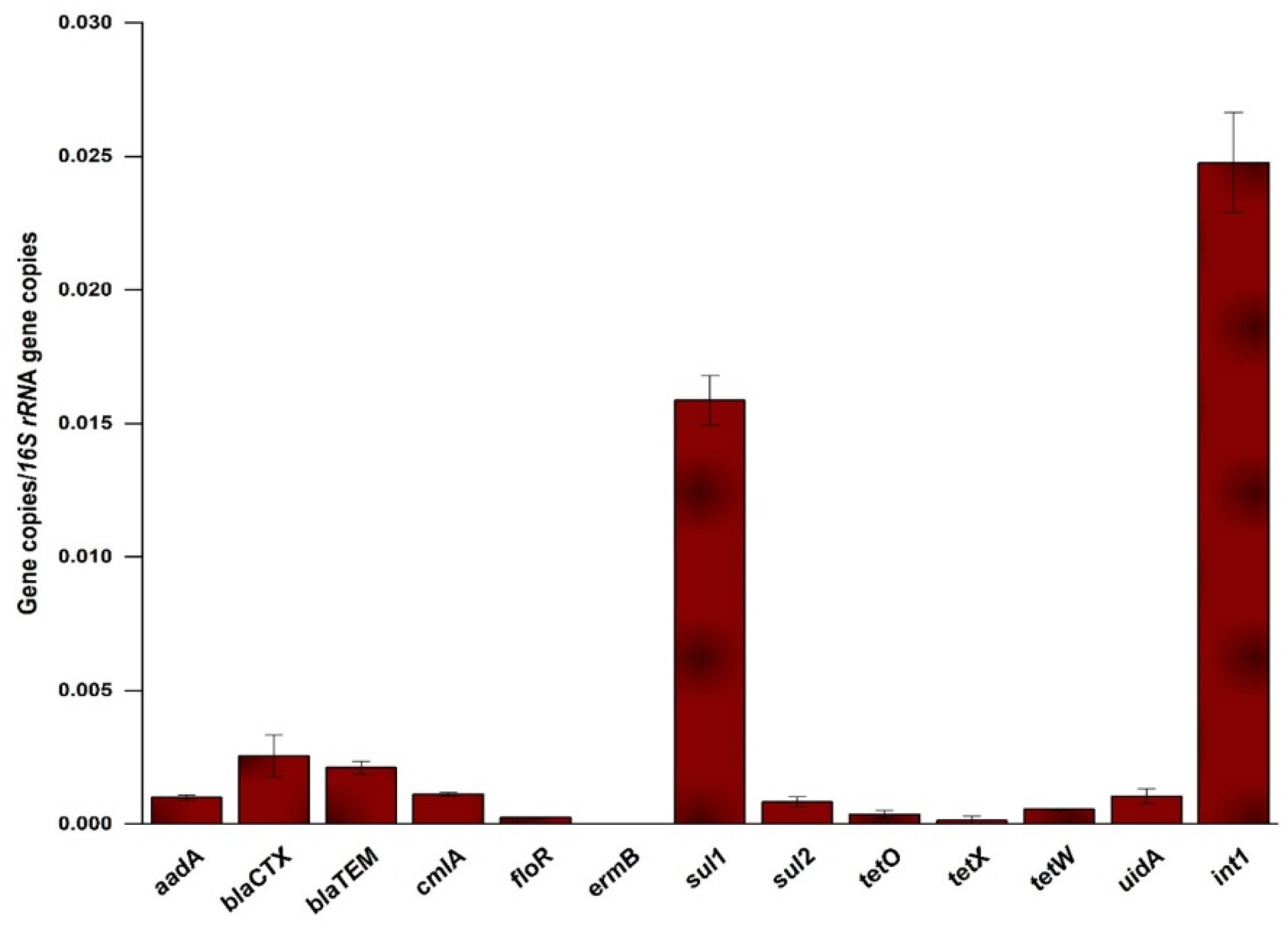
2.2. Occurrence and Relative Abundance of the Genes in the Influent
2.3. The Effect of Treatment on the Relative Abundance of Individual Genes
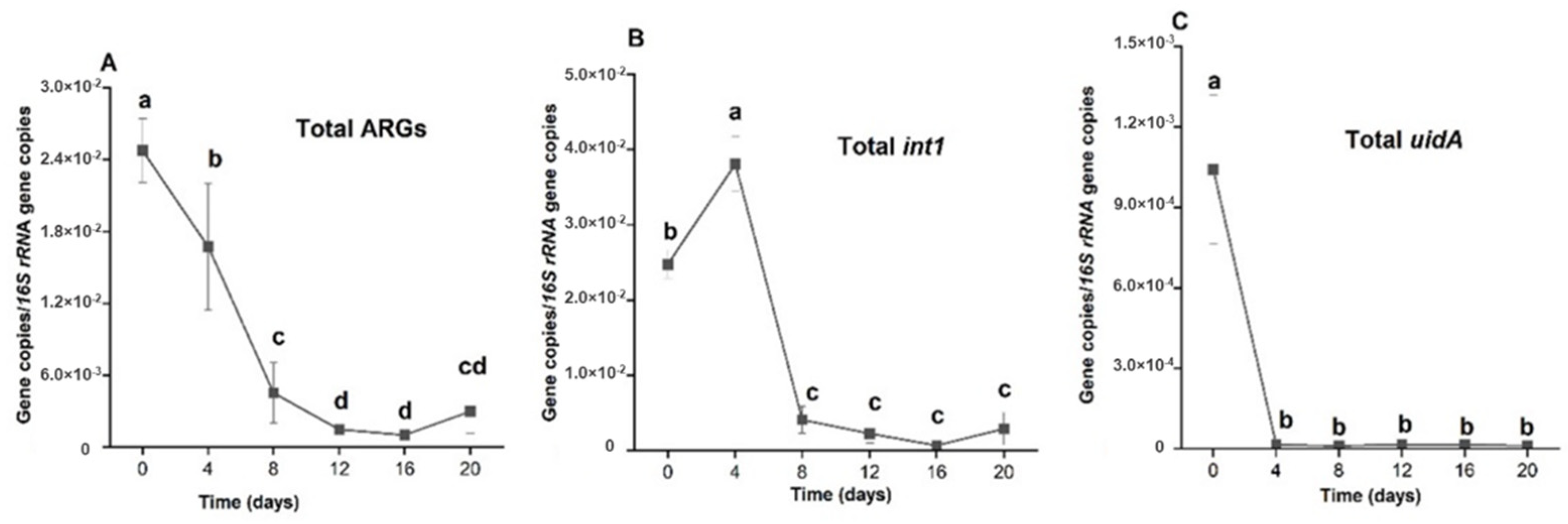
2.4. Comparison of the Efficiencies of Holding Times in Reduction of Relative Abundances of Total ARGs, Total int1 and Total uidA
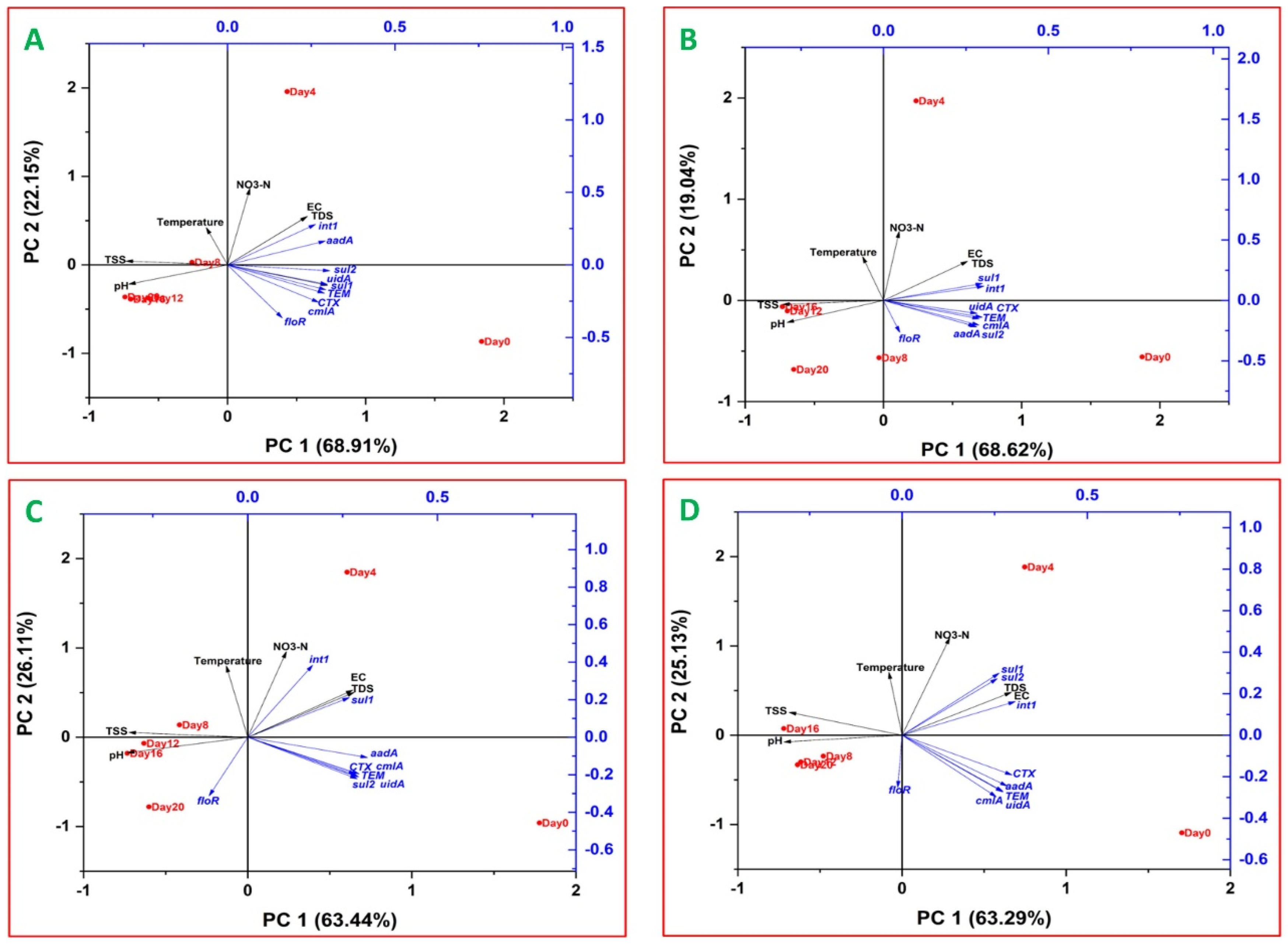
2.5. Relationships between ARGs, Indicator Genes and Water Quality Parameters
3. Discussion
3.1. The Effect of Microalgal Cultivation on Water Quality Indicators
3.2. Occurrence and Abundance of ARGs in the MWW
3.3. The Effect of Treatment on ARGs Relative Abundance
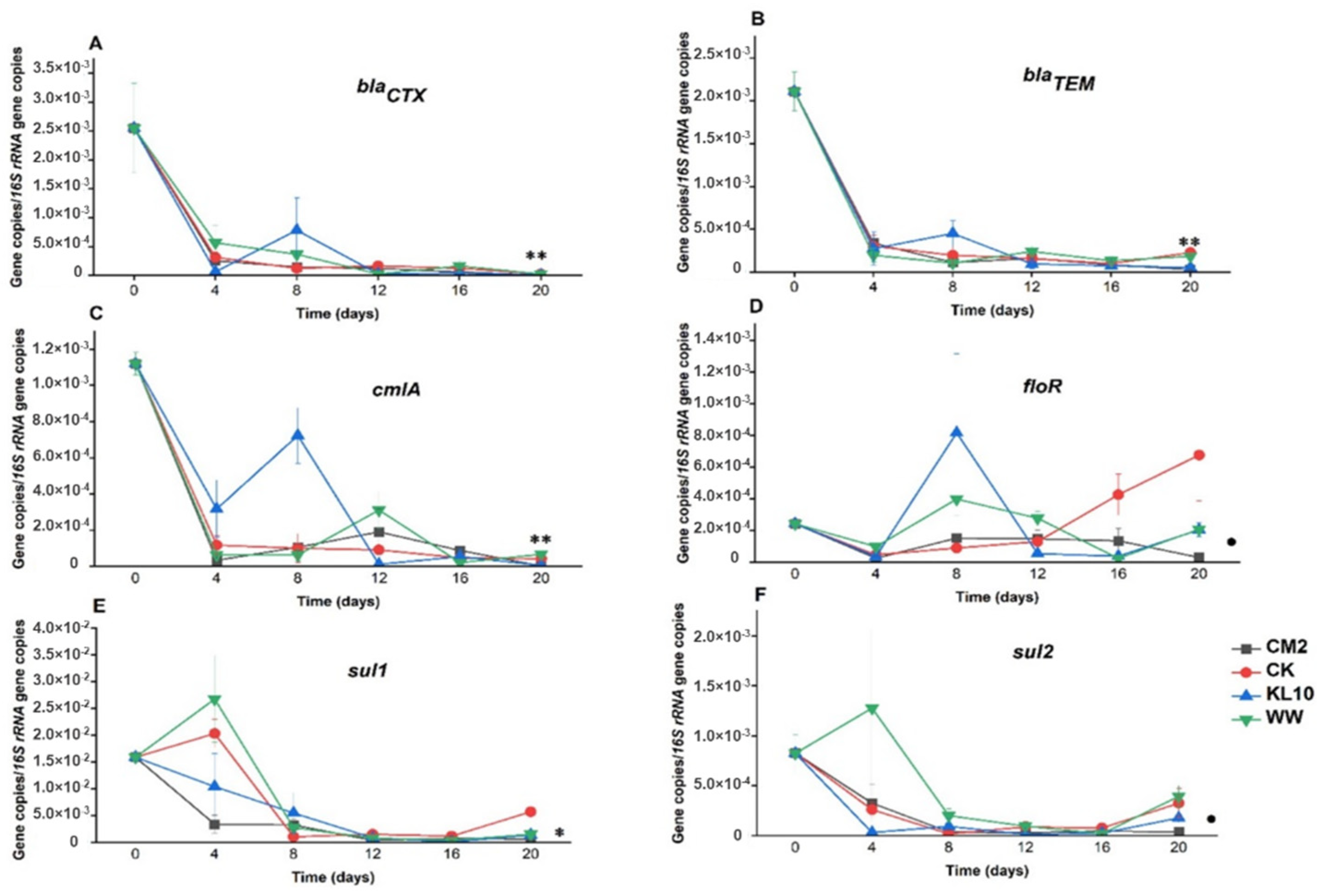
3.3.1. β-lactam Resistance Genes: blaCTX and blaTEM
3.3.2. Sulfonamides Resistance Genes: sul1 and sul2
3.3.3. Phenicol Resistance Genes: cmlA and floR
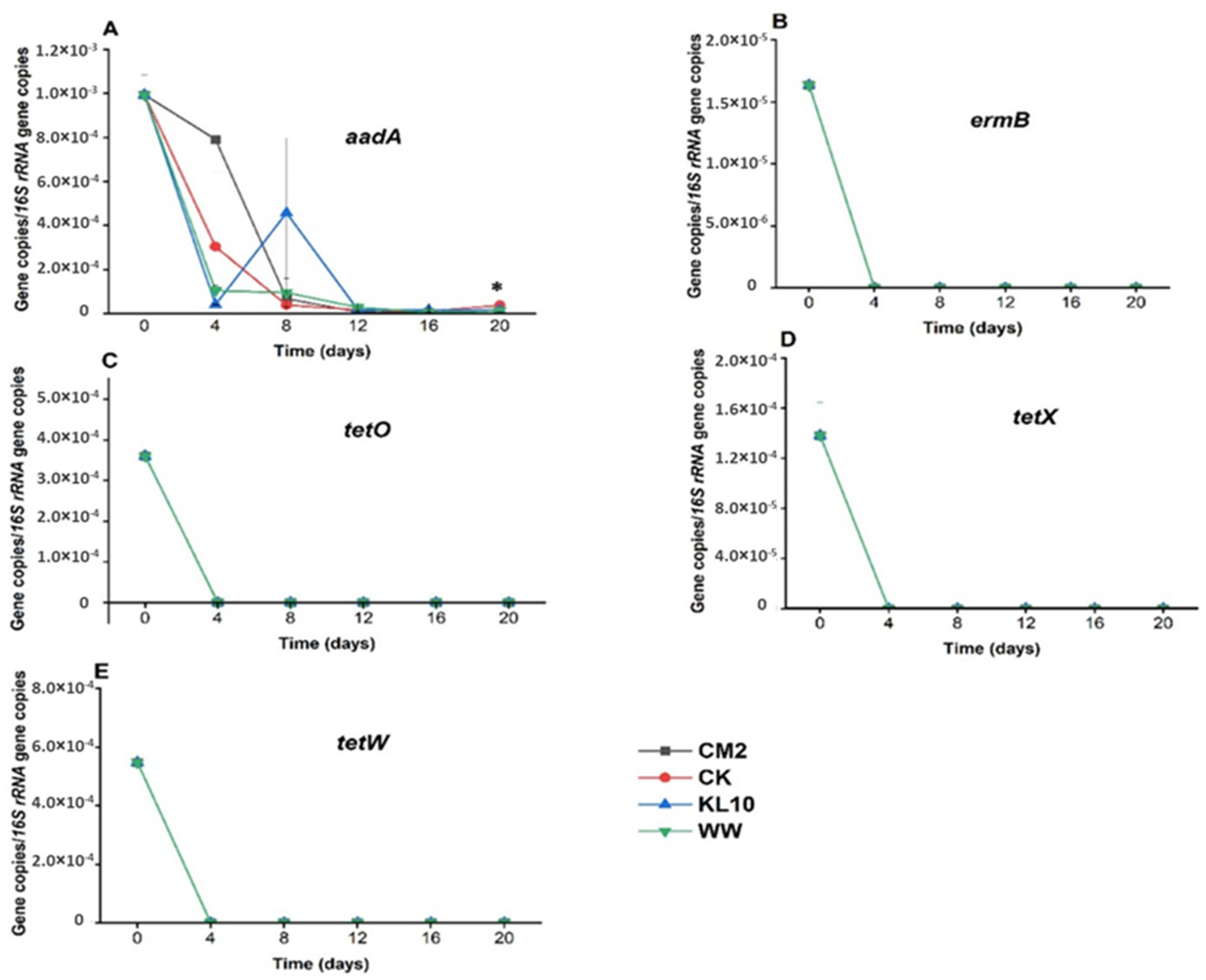
3.3.4. Aminoglycosides Resistance Gene: aadA
3.3.5. Tetracycline Resistance Genes: tetO, tetW and tetX
3.3.6. Macrolide-Lincosamide-Streptogramin Resistance Gene: ermB
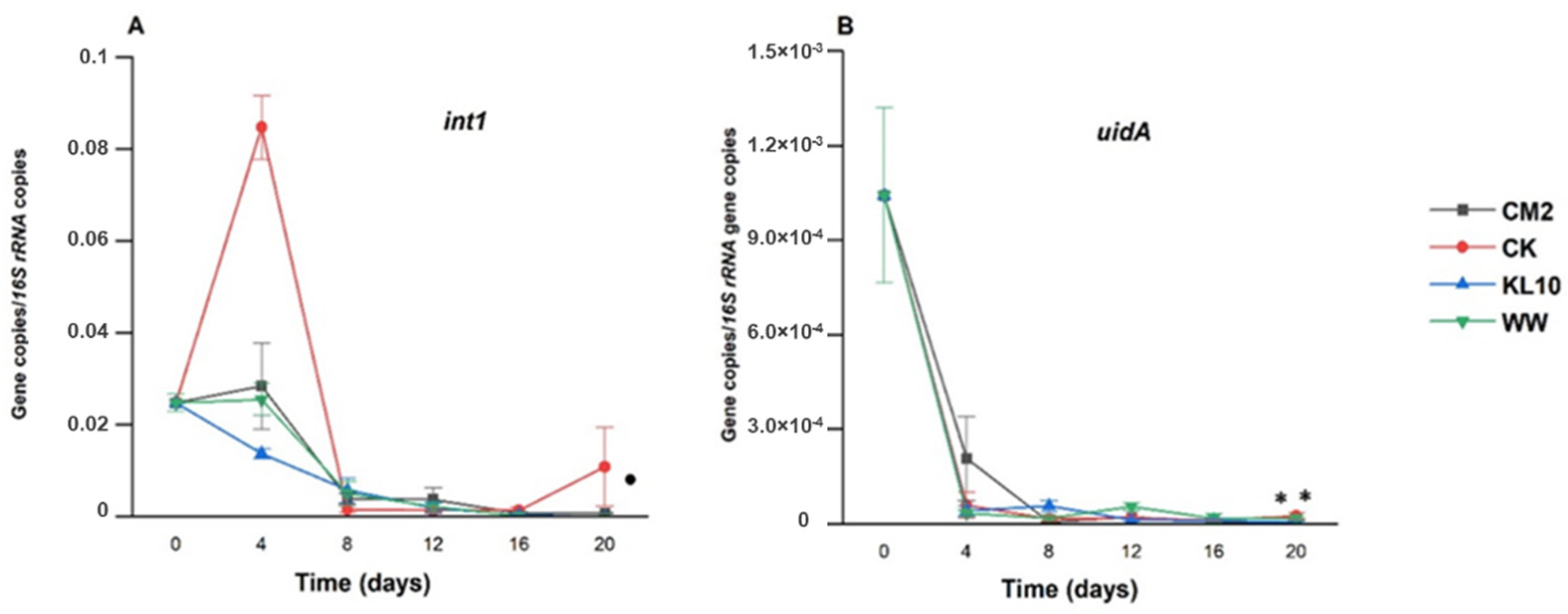
3.3.7. The Effect of Treatment on int1 Relative Abundance
3.3.8. The Effect of the Treatment on uidA Relative Abundance
3.4. Comparison of Genes Reduction Performance between Cultures
3.5. Relationships between ARGs, Indicator Genes and Water Quality Parameters
3.5.1. Relationships between ARGs and Indicator Genes
3.5.2. Relationships between ARGs and Water Quality Parameters
4. Materials and Methods
4.1. Outdoor Cultivation of Microalgae Combinations in MWW
4.2. Determination of Physicochemical Water Quality Parameters
4.3. Antibiotic Resistance Genes Pollution Studies
4.3.1. Genomic DNA Extraction
4.3.2. Qualitative Polymerase Chain Reaction
4.3.3. Real-Time Polymerase Chain Reaction
4.4. Statistical Analyses
5. Conclusions, Future Prospects, Limitations and Recommendations
5.1. Conclusions
- Two indicator genes, int1 and uidA, analyzed in this study proved reliable biomarkers for ARGs pollution in the microalgae-based WWT system. Interestingly, the uidA level could be used to monitor potential fecal and/or ARGs pollution. However, the relationship is most precise with aadA, blaCTX, blaTEM and cmlA. In the same vein, int1 proved to be a reliable biomarker for sul1 and aadA genes.
- The abundance of ARGs in the system can be represented indirectly by the concentrations of TDS, EC and TSS. More specifically, TDS and EC related most with sul1, whereas TSS showed the most solid relationship with blaCTX, blaTEM and aadA.
- Tetracycline resistance genes (tetO, tetW and tetX) and ermB were highly vulnerable to reduction in the microalgae-based WWT system, whereas floR was the most persistent.
- Although all the cultures proved promising in removing the conventional MWW quality parameters, the monocultures were the most efficient in terms of gene reduction from the MWW stream. This proves the superiority of the monocultures over the multi-species cultures, synthetic or natural, both in binary or polyculture combinations.
5.2. Future Prospects, Limitations and Recommendations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guo, N.; Ma, X.; Ren, S.; Wang, S.; Wang, Y. Mechanisms of metabolic performance enhancement during electrically assisted anaerobic treatment of chloramphenicol wastewater. Water Res. 2019, 156, 199–207. [Google Scholar] [CrossRef] [PubMed]
- McConnell, M.M.; Hansen, L.T.; Jamieson, R.C.; Neudorf, K.D.; Yost, C.K.; Tong, A. Removal of antibiotic resistance genes in two tertiary level municipal wastewater treatment plants. Sci. Total Environ. 2018, 643, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, M.; Zhong, H.; Liu, M.; Sui, Q.; Zheng, L.; Tong, J.; Wei, Y. Deciphering the factors influencing the discrepant fate of antibiotic resistance genes in sludge and water phases during municipal wastewater treatment. Bioresour. Technol. 2018, 265, 310–319. [Google Scholar] [CrossRef]
- Barancheshme, F.; Munir, M. Strategies to Combat Antibiotic Resistance in the Wastewater Treatment Plants. Front. Microbiol. 2018, 8, 2603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langbehn, R.K.; Camila, L.; Hugo, M.; Soares, M. Antibiotics in wastewater: From its occurrence to the biological removal by environmentally conscious technologies. Environ. Pollut. 2021, 275, 116603. [Google Scholar] [CrossRef]
- Leng, L.; Wei, L.; Xiong, Q.; Xu, S.; Li, W.; Lv, S.; Lu, Q.; Wan, L.; Wen, Z.; Zhou, W. Use of microalgae based technology for the removal of antibiotics from wastewater: A review. Chemosphere 2019, 238, 124680. [Google Scholar] [CrossRef]
- Liu, S.-S.; Qu, H.-M.; Yang, D.; Hu, H.; Liu, W.-L.; Qiu, Z.-G.; Hou, A.-M.; Guo, J.; Li, J.-W.; Shen, Z.-Q.; et al. Chlorine disinfection increases both intracellular and extracellular antibiotic resistance genes in a full-scale wastewater treatment plant. Water Res. 2018, 136, 131–136. [Google Scholar] [CrossRef] [Green Version]
- An, X.-L.; Su, J.-Q.; Li, B.; Ouyang, W.-Y.; Zhao, Y.; Chen, Q.-L.; Cui, L.; Chen, H.; Gillings, M.R.; Zhang, T.; et al. Tracking antibiotic resistome during wastewater treatment using high throughput quantitative PCR. Environ. Int. 2018, 117, 146–153. [Google Scholar] [CrossRef]
- Noman, S.M.; Shafiq, M.; Bibi, S.; Mittal, B.; Yuan, Y.; Zeng, M.; Li, X.; Olawale, O.A.; Jiao, X.; Irshad, M. Exploring antibiotic resistance genes, mobile gene elements, and virulence gene factors in an urban freshwater samples using metagenomic analysis. Environ. Sci. Pollut. Res. 2022, 1–14. [Google Scholar] [CrossRef]
- Munasinghe-Arachchige, S.P.; Delanka-Pedige, H.M.K.; Henkanatte-Gedera, S.M.; Tchinda, D.; Zhang, Y.; Nirmalakhandan, N. Factors contributing to bacteria inactivation in the Galdieria sulphuraria—Based wastewater treatment system. Algal Res. 2019, 38. [Google Scholar] [CrossRef]
- Nõlvak, H.; Truu, M.; Oopkaup, K.; Kanger, K.; Krustok, I.; Nehrenheim, E.; Truu, J. Reduction of antibiotic resistome and integron-integrase genes in laboratory-scale photobioreactors treating municipal wastewater. Water Res. 2018, 142, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Davies-Colley, R.; Craggs, R.; Nagels, J. Disinfection in a pilot-scale “advanced” pond system (APS) for domestic sewage treatment in New Zealand. Water Sci. Technol. 2003, 48, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Delanka-Pedige, H.M.; Cheng, X.; Munasinghe-Arachchige, S.P.; Abeysiriwardana-Arachchige, I.S.; Xu, J.; Nirmalakhandan, N.; Zhang, Y. Metagenomic insights into virus removal performance of an algal-based wastewater treatment system utilizing Galdieria sulphuraria. Algal Res. 2020, 47, 101865. [Google Scholar] [CrossRef]
- Ferro, L.; Gentili, F.G.; Funk, C. Isolation and characterization of microalgal strains for biomass production and wastewater reclamation in Northern Sweden. Algal Res. 2018, 32, 44–53. [Google Scholar] [CrossRef]
- Ji, X.; Jiang, M.; Zhang, J.; Jiang, X.; Zheng, Z. The interactions of algae-bacteria symbiotic system and its effects on nutrients removal from synthetic wastewater. Bioresour. Technol. 2018, 247, 44–50. [Google Scholar] [CrossRef]
- Piligaev, A.; Sorokina, K.; Shashkov, M.; Parmon, V. Screening and comparative metabolic profiling of high lipid content microalgae strains for application in wastewater treatment. Bioresour. Technol. 2018, 250, 538–547. [Google Scholar] [CrossRef]
- Schumacher, G.; Sekoulov, I. Improving the effluent of small wastewater treatment plants by bacteria reduction and nutrient removal with an algal biofilm. Water Sci. Technol. 2003, 48, 373–380. [Google Scholar] [CrossRef]
- Tohá, J.; Soto, M.A.; Contreras, S. Removal of faecal coliform in high pH ponds using rapid growth alga biomass. Int. J. Environ. Health Res. 1991, 1, 236–239. [Google Scholar] [CrossRef]
- Hendriksen, R.S.; Munk, P.; Njage, P.; Van Bunnik, B.; McNally, L.; Lukjancenko, O.; Röder, T.; Nieuwenhuijse, D.; Pedersen, S.K.; Kjeldgaard, J.; et al. Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat. Commun. 2019, 10, 1124. [Google Scholar] [CrossRef] [Green Version]
- Foladori, P.; Petrini, S.; Andreottola, G. Evolution of real municipal wastewater treatment in photobioreactors and microalgae-bacteria consortia using real-time parameters. Chem. Eng. J. 2018, 345, 507–516. [Google Scholar] [CrossRef]
- Kang, D.; Kim, K.; Jang, Y.; Moon, H.; Ju, D.; Jahng, D. Nutrient removal and community structure of wastewater-borne algal-bacterial consortia grown in raw wastewater with various wavelengths of light. Int. Biodeterior. Biodegrad. 2018, 126, 10–20. [Google Scholar] [CrossRef]
- Newton, A.R.; Pederson, D.O. Revised National Environmental Quality Standards (Neqs). 1996. Available online: https://www.lawinsider.com/dictionary/national-environmental-quality-standards-neqs (accessed on 28 September 2022).
- Cheng, X.; Delanka-Pedige, H.M.K.; Munasinghe-Arachchige, S.P.; Arachchige, I.S.A.A.; Smith, G.B.; Nirmalakhandan, N.; Zhang, Y. Removal of antibiotic resistance genes in an algal-based wastewater treatment system employing Galdieria sulphuraria: A comparative study. Sci. Total Environ. 2019, 711, 134435. [Google Scholar] [CrossRef] [PubMed]
- Nirmalakhandan, N.; Selvaratnam, T.; Henkanatte-Gedera, S.M.; Tchinda, D.; Abeysiriwardana-Arachchige, I.S.A.; Delanka-Pedige, H.M.K.; Munasinghe-Arachchige, S.P.; Zhang, Y.; Holguin, F.O.; Lammers, F.J. Algal wastewater treatment: Photoautotrophic vs. mixotrophic processes. Algal Res. 2019, 41, 101569. [Google Scholar] [CrossRef]
- Chen, J.; Su, Z.; Dai, T.; Huang, B.; Mu, Q.; Zhang, Y.; Wen, D. Occurrence and distribution of antibiotic resistance genes in the sediments of the East China Sea bays. J. Environ. Sci. 2019, 81, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.-P.; Yang, Y.; Lu, D.-P.; Niu, Z.-S.; Feng, J.-N.; Chen, Y.-R.; Tou, F.-Y.; Garner, E.; Xu, J.; Liu, M.; et al. Biofilms as a sink for antibiotic resistance genes (ARGs) in the Yangtze Estuary. Water Res. 2018, 129, 277–286. [Google Scholar] [CrossRef]
- Szekeres, E.; Baricz, A.; Chiriac, C.M.; Farkas, A.; Opris, O.; Soran, M.-L.; Andrei, A.-S.; Rudi, K.; Balcázar, J.L.; Dragos, N.; et al. Abundance of antibiotics, antibiotic resistance genes and bacterial community composition in wastewater effluents from different Romanian hospitals. Environ. Pollut. 2017, 225, 304–315. [Google Scholar] [CrossRef]
- Lee, J.; Jeon, J.H.; Shin, J.; Jang, H.M.; Kim, S.; Song, M.S.; Kim, Y.M. Quantitative and qualitative changes in antibiotic resistance genes after passing through treatment processes in municipal wastewater treatment plants. Sci. Total Environ. 2017, 605–606, 906–914. [Google Scholar] [CrossRef]
- Liu, L.; Liu, C.; Zheng, J.; Huang, X.; Wang, Z.; Liu, Y.; Zhu, G. Elimination of veterinary antibiotics and antibiotic resistance genes from swine wastewater in the vertical flow constructed wetlands. Chemosphere 2013, 91, 1088–1093. [Google Scholar] [CrossRef]
- Puljko, A.; Milaković, M.; Križanović, S.; Kosić-Vukšić, J.; Babić, I.; Petrić, I.; Maravić, A.; Jelić, M.; Udiković-Kolić, N. Prevalence of enteric opportunistic pathogens and extended-spectrum cephalosporin- and carbapenem-resistant coliforms and genes in wastewater from municipal wastewater treatment plants in Croatia. J. Hazard. Mater. 2022, 427. [Google Scholar] [CrossRef]
- Wen, Q.; Yang, L.; Duan, R.; Chen, Z. Monitoring and evaluation of antibiotic resistance genes in four municipal wastewater treatment plants in Harbin, Northeast China. Environ. Pollut. 2016, 212, 34–40. [Google Scholar] [CrossRef]
- Sabri, N.; van Holst, S.; Schmitt, H.; van der Zaan, B.; Gerritsen, H.; Rijnaarts, H.; Langenhoff, A. Fate of antibiotics and antibiotic resistance genes during conventional and additional treatment technologies in wastewater treatment plants. Sci. Total Environ. 2020, 741, 140199. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, H.; Zhao, H. Propagation of antibiotic resistance genes in an industrial recirculating aquaculture system located at northern China. Environ. Pollut. 2020, 261, 114155. [Google Scholar] [CrossRef]
- Liang, C.; Wei, D.; Yan, W.; Zhang, S.; Shi, J.; Liu, L. Fates of intracellular and extracellular antibiotic resistance genes during the cattle farm wastewater treatment process. Bioresour. Technol. 2021, 344, 126272. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Qin, X.; Guo, J.; Jia, W.; Wang, Q.; Zhang, M.; Huang, Y. Evidence of selective enrichment of bacterial assemblages and antibiotic resistant genes by microplastics in urban rivers. Water Res. 2020, 183, 116113. [Google Scholar] [CrossRef]
- Su, H.-C.; Liu, Y.-S.; Pan, C.-G.; Chen, J.; He, L.-Y.; Ying, G.-G. Persistence of antibiotic resistance genes and bacterial community changes in drinking water treatment system: From drinking water source to tap water. Sci. Total Environ. 2018, 616–617, 453–461. [Google Scholar] [CrossRef]
- Binh, C.T.; Heuer, H.; Kaupenjohann, M.; Smalla, K. Diverse aadA gene cassettes on class 1 integrons introduced into soil via spread manure. Res. Microbiol. 2009, 160, 427–433. [Google Scholar] [CrossRef]
- Zhu, Y.-G.; Johnson, T.A.; Su, J.-Q.; Qiao, M.; Guo, G.-X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Wen, Q.; Chen, Z.; Duan, R.; Yang, P. Impacts of advanced treatment processes on elimination of antibiotic resistance genes in a municipal wastewater treatment plant. Front. Environ. Sci. Eng. 2019, 13, 32. [Google Scholar] [CrossRef]
- Hayward, J.L.; Huang, Y.; Yost, C.K.; Hansen, L.T.; Lake, C.; Tong, A.; Jamieson, R.C. Lateral flow sand filters are effective for removal of antibiotic resistance genes from domestic wastewater. Water Res. 2019, 162, 482–491. [Google Scholar] [CrossRef]
- Du, J.; Geng, J.; Ren, H.; Ding, L.; Xu, K.; Zhang, Y. Variation of antibiotic resistance genes in municipal wastewater treatment plant with A2O-MBR system. Environ. Sci. Pollut. Res. 2014, 22, 3715–3726. [Google Scholar] [CrossRef]
- Chen, J.; Wei, X.-D.; Liu, Y.-S.; Ying, G.-G.; Liu, S.-S.; He, L.-Y.; Su, H.-C.; Hu, L.-X.; Chen, F.-R.; Yang, Y.-Q. Removal of antibiotics and antibiotic resistance genes from domestic sewage by constructed wetlands: Optimization of wetland substrates and hydraulic loading. Sci. Total Environ. 2016, 565, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ma, X.; Liu, Y.; Yi, X.; Du, G.; Li, J. Fate of antibiotics, antibiotic-resistant bacteria, and cell-free antibiotic-resistant genes in full-scale membrane bioreactor wastewater treatment plants. Bioresour. Technol. 2020, 302, 122825. [Google Scholar] [CrossRef] [PubMed]
- Di Cesare, A.; Eckert, E.M.; D’Urso, S.; Bertoni, R.; Gillan, D.C.; Wattiez, R.; Corno, G. Co-occurrence of integrase 1, antibiotic and heavy metal resistance genes in municipal wastewater treatment plants. Water Res. 2016, 94, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Narciso-Da-Rocha, C.; Rocha, J.; Vaz-Moreira, I.; Lira, F.; Tamames, J.; Henriques, I.; Martinez, J.L.; Manaia, C.M. Bacterial lineages putatively associated with the dissemination of antibiotic resistance genes in a full-scale urban wastewater treatment plant. Environ. Int. 2018, 118, 179–188. [Google Scholar] [CrossRef]
- Ma, X.; Guo, N.; Ren, S.; Wang, S.; Wang, Y. Response of antibiotic resistance to the co-existence of chloramphenicol and copper during bio-electrochemical treatment of antibiotic-containing wastewater. Environ. Int. 2019, 126, 127–133. [Google Scholar] [CrossRef]
- Osińska, A.; Korzeniewska, E.; Harnisz, M.; Felis, E.; Bajkacz, S.; Jachimowicz, P.; Niestępski, S.; Konopka, I. Small-scale wastewater treatment plants as a source of the dissemination of antibiotic resistance genes in the aquatic environment. J. Hazard. Mater. 2020, 381, 121221. [Google Scholar] [CrossRef]
- Makowska, N.; Koczura, R.; Mokracka, J. Class 1 integrase, sulfonamide and tetracycline resistance genes in wastewater treatment plant and surface water. Chemosphere 2016, 144, 1665–1673. [Google Scholar] [CrossRef]
- Luprano, M.L.; De Sanctis, M.; Del Moro, G.; Di Iaconi, C.; Lopez, A.; Levantesi, C. Antibiotic resistance genes fate and removal by a technological treatment solution for water reuse in agriculture. Sci. Total Environ. 2016, 571, 809–818. [Google Scholar] [CrossRef]
- Seger, M.; Unc, A.; Starkenburg, S.R.; Holguin, F.O.; Lammers, P.J. Nutrient-driven algal-bacterial dynamics in semi-continuous, pilot-scale photobioreactor cultivation of Nannochloropsis salina CCMP1776 with municipal wastewater nutrients. Algal Res. 2019, 39, 101457. [Google Scholar] [CrossRef]
- Shakeri, A.; Soheili, V.; Karimi, M.; Hosseininia, S.A.; Bazzaz, B.S.F. Biological activities of three natural plant pigments and their health benefits. J. Food Meas. Charact. 2017, 12, 356–361. [Google Scholar] [CrossRef]
- Toshkova-Yotova, T.; Georgieva, A.; Iliev, I.; Alexandrov, S.; Ivanova, A.; Pilarski, P.; Toshkova, R. Antitumor and antimicrobial activity of fatty acids from green microalga Coelastrella sp. BGV. South Afr. J. Bot. 2022. [Google Scholar] [CrossRef]
- Jiang, X.; Liu, L.; Chen, J.; Fan, X.; Xie, S.; Huang, J.; Yu, G. Antibiotic resistance genes and mobile genetic elements in a rural river in Southeast China: Occurrence, seasonal variation and association with the antibiotics. Sci. Total Environ. 2021, 778, 146131. [Google Scholar] [CrossRef]
- Wu, D.-L.; Zhang, M.; He, L.-X.; Zou, H.-Y.; Liu, Y.-S.; Li, B.-B.; Yang, Y.-Y.; Liu, C.; He, L.-Y.; Ying, G.-G. Contamination profile of antibiotic resistance genes in ground water in comparison with surface water. Sci. Total Environ. 2020, 715, 136975. [Google Scholar] [CrossRef]
- Adekanmbi, A.O.; Adejoba, A.T.; Banjo, O.A.; Saki, M. Detection of sul1 and sul2 genes in sulfonamide-resistant bacteria (SRB) from sewage, aquaculture sources, animal wastes and hospital wastewater in South-West Nigeria. Gene Rep. 2020, 20, 100742. [Google Scholar] [CrossRef]
- Ma, Q.; Zhu, C.; Yao, M.; Yuan, G.; Sun, Y. Correlation between the sulfamethoxazole-trimethoprim resistance of Shigella flexneri and the sul genes. Medicine 2021, 100, e24970. [Google Scholar] [CrossRef] [PubMed]
- Hoa, P.T.P.; Nonaka, L.; Viet, P.H.; Suzuki, S. Detection of the sul1, sul2, and sul3 genes in sulfonamide-resistant bacteria from wastewater and shrimp ponds of north Vietnam. Sci. Total Environ. 2008, 405, 377–384. [Google Scholar] [CrossRef]
- Li, Z.; Li, T.; Xing, X.; Bi, Z.; Qi, P.; Hu, C.; Xu, G.; Chen, C.; Ma, K.; Chen, J. Inhibiting the increase of antibiotic resistance genes during drinking water distribution by superior microbial interface using Fe modified granular activated carbon. J. Clean. Prod. 2021, 335, 130225. [Google Scholar] [CrossRef]
- Bischoff, K.M.; White, D.G.; McDermott, P.F.; Zhao, S.; Gaines, S.; Maurer, J.J.; Nisbet, D.J. Characterization of Chloramphenicol Resistance in Beta-Hemolytic Escherichia coli Associated with Diarrhea in Neonatal Swine. J. Clin. Microbiol. 2002, 40, 389–394. [Google Scholar] [CrossRef] [Green Version]
- Bischoff, K.M.; White, D.G.; Hume, M.E.; Poole, T.L.; Nisbet, D.J. The chloramphenicol resistance gene cmlAis disseminated on transferable plasmids that confer multiple-drug resistance in swine Escherichia coli. FEMS Microbiol. Lett. 2005, 243, 285–291. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.-Y.; Chang, Y.-H.; Chang, L.-L.; Chang, S.-F.; Lee, T.-M. Characterisation of drug resistance gene cassettes associated with class 1 integrons in clinical isolates of Escherichia coli from Taiwan, ROC. J. Med. Microbiol. 2000, 49, 1097–1102. [Google Scholar] [CrossRef]
- Gil-Molino, M.; Gonçalves, P.; Risco, D.; Martín-Cano, F.E.; García, A.; Rey, J.; Fernández-Llario, P.; Quesada, A. Dissemination of antimicrobial-resistant isolates of Salmonella spp. in wild boars and its relationship with management practices. Transbound. Emerg. Dis. 2022. [Google Scholar] [CrossRef]
- Handrova, L.; Kmet, V. Antibiotic resistance and virulence factors of Escherichia coli from eagles and goshawks. J. Environ. Sci. Health Part B 2019, 54, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Rajasekar, A.; Qiu, M.; Wang, B.; Murava, R.T.; Norgbey, E. Relationship between water quality, heavy metals and antibiotic resistance genes among three freshwater lakes. Environ. Monit. Assess. 2022, 194, 1–10. [Google Scholar] [CrossRef]
- Zhou, R.; Wang, Y.; Hilal, M.G.; Yu, Q.; Feng, T.; Li, H. Temporal succession of water microbiomes and resistomes during carcass decomposition in a fish model. J. Hazard. Mater. 2020, 403, 123795. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Gomes-Silva, G.; Plath, M.; Pereira, B.B.; UeiraVieira, C.; Wang, Z. Shifts in bacterial communities and antibiotic resistance genes in surface water and gut microbiota of guppies (Poecilia reticulata) in the upper Rio Uberabinha, Brazil. Ecotoxicol. Environ. Saf. 2021, 211, 111955. [Google Scholar] [CrossRef] [PubMed]
- Quijano, G.; Arcila, J.S.; Buitrón, G. Microalgal-bacterial aggregates: Applications and perspectives for wastewater treatment. Biotechnol. Adv. 2017, 35, 772–781. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, A.L.; Pires, J.C.M.; Simões, M. A review on the use of microalgal consortia for wastewater treatment. Algal Res. 2017, 24, 403–415. [Google Scholar] [CrossRef]
- Kumar, A.; Ergas, S.; Yuan, X.; Sahu, A.; Zhang, Q.; Dewulf, J.; Malcata, F.X.; van Langenhove, H. Enhanced CO2 fixation and biofuel production via microalgae: Recent developments and future directions. Trends Biotechnol. 2010, 28, 371–380. [Google Scholar] [CrossRef]
- Awuah, E.; Anohene, F.; Asante, K.; Lubberding, H.; Gijzen, H. Environmental conditions and pathogen removal in macrophyte- and algal-based domestic wastewater treatment systems. Water Sci. Technol. 2001, 44, 11–18. [Google Scholar] [CrossRef]
- Parhad, N.M.; Rao, N.U. Effect of pH on survival of Escherichia coli. J. Water Pollut. Control Fed. 1974, 46, 980–986. [Google Scholar]
- Inuwa, A.B.; Iftikhar, Z.; Qaisar, M.; Usman, I.; Muhammad, I.; Farhan, H.; Akhtar, I.; Arshid, P.; Nazir, R. Novel Microalgae Strains from Selected Lower Himalayan Aquatic Habitats as Potential Sources of Green Products. PLoS ONE 2022, 17, e0267788. [Google Scholar] [CrossRef] [PubMed]
- American Public Health Association (APHA); American Water Works Association (AWWA); Water Environment Federation (WEF). Standard Methods for the Examination of Water and Wastewater; American Public Health Association: Washington, DC, USA, 2005. [Google Scholar]
- Looft, T.; Johnson, T.A.; Allen, H.K.; Bayles, D.O.; Alt, D.P.; Stedtfeld, R.D.; Sul, W.J.; Stedtfeld, T.M.; Chai, B.; Cole, J.R.; et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl. Acad. Sci. USA 2012, 109, 1691–1696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Wang, Y.; Zhou, S.; Jiang, X.; Ma, X.; Liu, C. Robust performance of a membrane bioreactor for removing antibiotic resistance genes exposed to antibiotics: Role of membrane foulants. Water Res. 2018, 130, 139–150. [Google Scholar] [CrossRef]
- Le, T.-H.; Ng, C.; Tran, N.H.; Chen, H.; Gin, K.Y.-H. Removal of antibiotic residues, antibiotic resistant bacteria and antibiotic resistance genes in municipal wastewater by membrane bioreactor systems. Water Res. 2018, 145, 498–508. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Inuwa, A.B.; Mahmood, Q.; Iqbal, J.; Widemann, E.; Shafiq, S.; Irshad, M.; Irshad, U.; Iqbal, A.; Hafeez, F.; Nazir, R. Removal of Antibiotic Resistance Genes, Class 1 Integrase Gene and Escherichia coli Indicator Gene in a Microalgae-Based Wastewater Treatment System. Antibiotics 2022, 11, 1531. https://doi.org/10.3390/antibiotics11111531
Inuwa AB, Mahmood Q, Iqbal J, Widemann E, Shafiq S, Irshad M, Irshad U, Iqbal A, Hafeez F, Nazir R. Removal of Antibiotic Resistance Genes, Class 1 Integrase Gene and Escherichia coli Indicator Gene in a Microalgae-Based Wastewater Treatment System. Antibiotics. 2022; 11(11):1531. https://doi.org/10.3390/antibiotics11111531
Chicago/Turabian StyleInuwa, Abdullahi B., Qaisar Mahmood, Jamshed Iqbal, Emilie Widemann, Sarfraz Shafiq, Muhammad Irshad, Usman Irshad, Akhtar Iqbal, Farhan Hafeez, and Rashid Nazir. 2022. "Removal of Antibiotic Resistance Genes, Class 1 Integrase Gene and Escherichia coli Indicator Gene in a Microalgae-Based Wastewater Treatment System" Antibiotics 11, no. 11: 1531. https://doi.org/10.3390/antibiotics11111531
APA StyleInuwa, A. B., Mahmood, Q., Iqbal, J., Widemann, E., Shafiq, S., Irshad, M., Irshad, U., Iqbal, A., Hafeez, F., & Nazir, R. (2022). Removal of Antibiotic Resistance Genes, Class 1 Integrase Gene and Escherichia coli Indicator Gene in a Microalgae-Based Wastewater Treatment System. Antibiotics, 11(11), 1531. https://doi.org/10.3390/antibiotics11111531








