Identification and Characterization of Genes Related to Ampicillin Antibiotic Resistance in Zymomonas mobilis
Abstract
:1. Introduction
2. Results
2.1. In Silico Analysis of the AR Genes of Z. mobilis ZM4
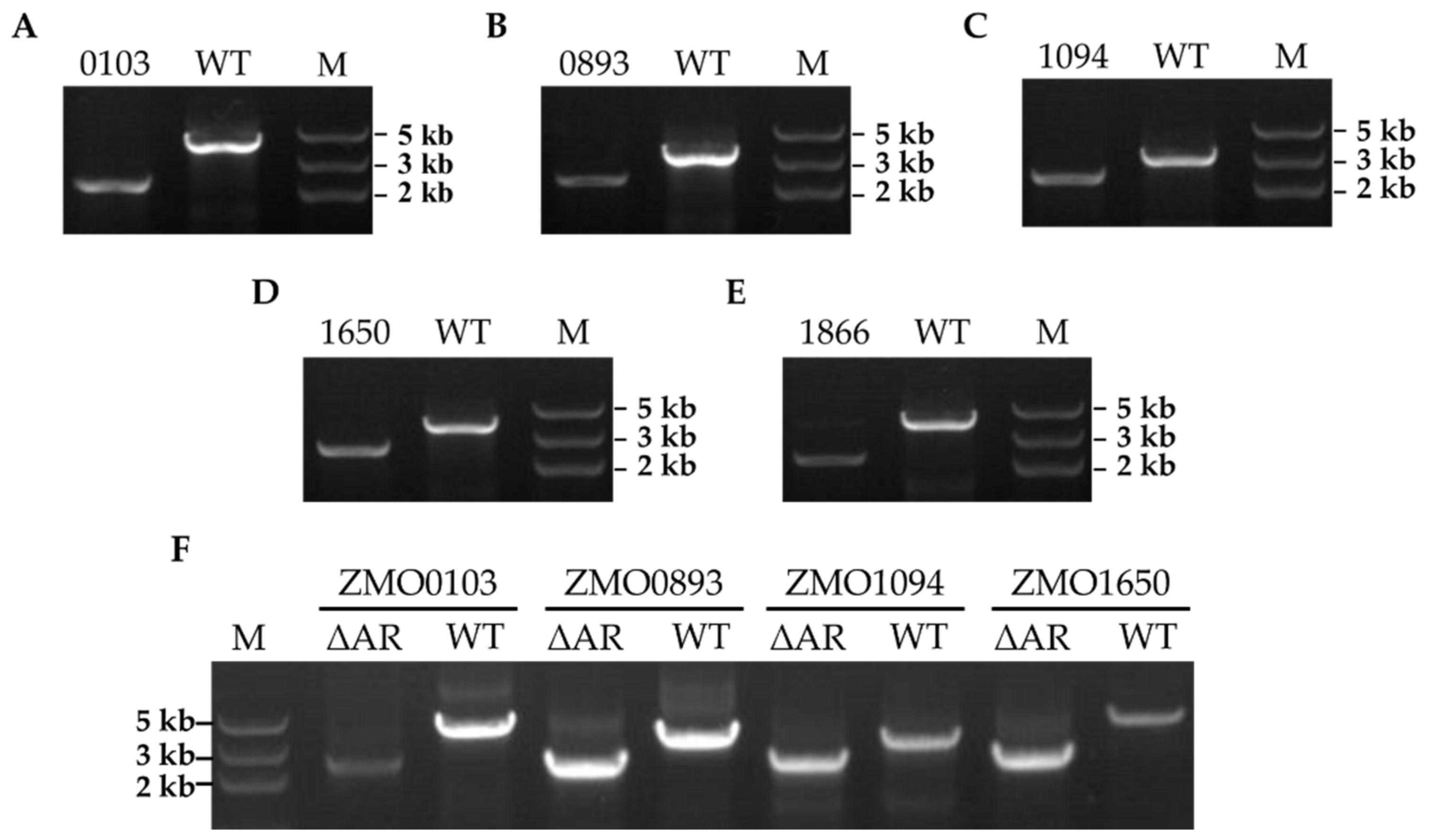
2.2. Ampicillin Resistance−Related Gene Knockout in Z. mobilis ZM4
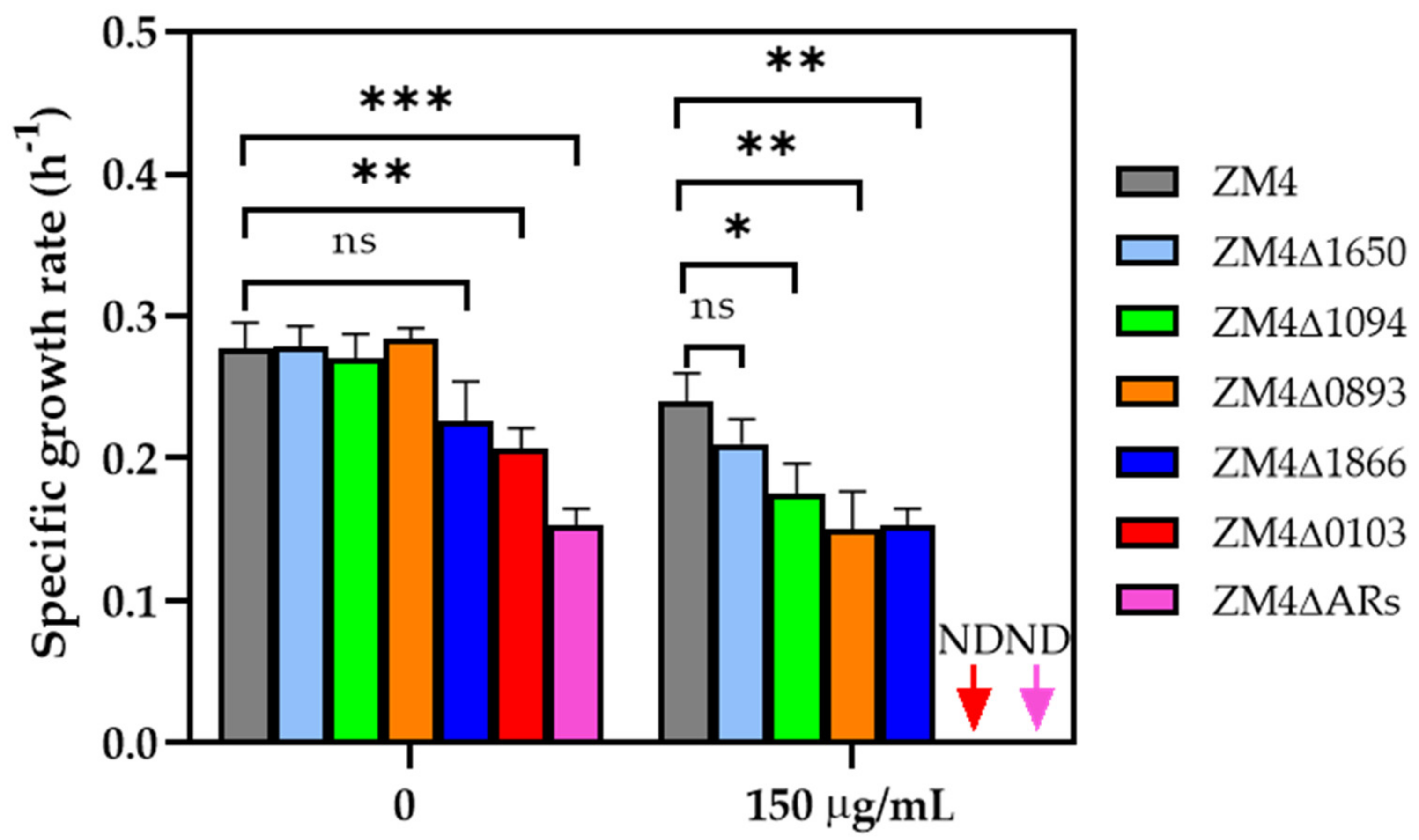
2.3. Antibiotic Tolerance of Ampicillin−Resistant (AR) Gene Knockout Strains
2.4. Cell Morphology of Mutants Treated with Ampicillin
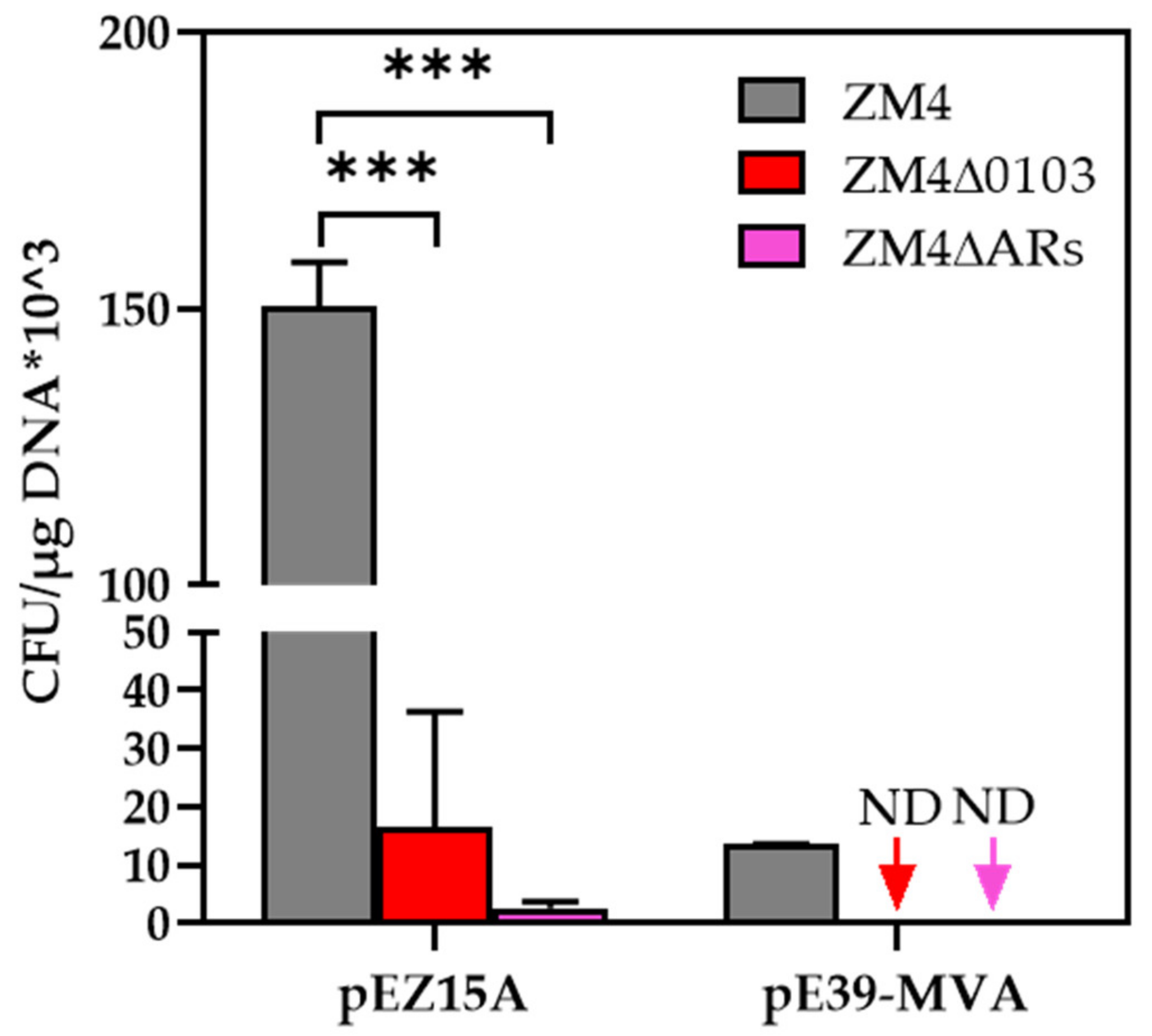
2.5. Effects of Ampicillin−Resistant (AR) Gene Mutagenesis on Genetic Transformation Efficiency
3. Discussion
4. Materials and Methods
4.1. Strains and Cultural Conditions
4.2. In Silico Analysis of the AR Genes of Z. mobilis ZM4
4.3. Construction of Editing Plasmids of Ampicillin−Resistant (AR) Genes in Z. mobilis ZM4
4.4. Curing of Editing Plasmids
4.5. Electroporation of Editing Plasmids to Z. mobilis ZM4
4.6. Construction of ZM4∆ARs by Continuous Gene Editing
4.7. Genetic Transformation Efficiency of β−lactamase Mutants
4.8. Confirmation of Ampicillin−Resistant Gene Deletion in Z. mobilis ZM4
4.9. Growth Studies and Analysis of Ampicillin−Resistant Gene Mutants
4.10. Cell Morphology Observation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Awad, H.; El–Shahed, K.Y.I.; Aziz, R.; Sarmidi, M.R.; El–Enshasy, H.A. Antibiotics as microbial secondary metabolites: Production and application. J. Teknol. 2012, 59, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Parker, D.S.; Armstrong, D.G. Antibiotic feed additives and livestock production. Proc. Nutr. Soc. 1987, 46, 415–421. [Google Scholar] [CrossRef] [Green Version]
- Procopio, R.E.; Silva, I.R.; Martins, M.K.; Azevedo, J.L.; Araujo, J.M. Antibiotics produced by Streptomyces. Braz. J. Infect. Dis. 2012, 16, 466–471. [Google Scholar] [CrossRef] [Green Version]
- Urban−Chmiel, R.; Marek, A.; Stepien−Pysniak, D.; Wieczorek, K.; Dec, M.; Nowaczek, A.; Osek, J. Antibiotic resistance in bacteria−a review. Antibiotics 2022, 11, 1079. [Google Scholar] [CrossRef]
- Singh, S.B.; Young, K.; Silver, L.L. What is an “ideal” antibiotic? Discovery challenges and path forward. Biochem. Pharmacol. 2017, 133, 63–73. [Google Scholar] [CrossRef]
- Ezelarab, H.A.A.; Abbas, S.H.; Hassan, H.A.; Abuo−Rahma, G.E.A. Recent updates of fluoroquinolones as antibacterial agents. Arch Pharm 2018, 351, e1800141. [Google Scholar] [CrossRef]
- Ang, C.W.; Jarrad, A.M.; Cooper, M.A.; Blaskovich, M.A.T. Nitroimidazoles: Molecular fireworks that combat a broad spectrum of infectious diseases. J. Med. Chem. 2017, 60, 7636–7657. [Google Scholar] [CrossRef]
- Vilhena, C.; Bettencourt, A. Daptomycin: A review of properties, clinical use, drug delivery and resistance. Mini Rev. Med. Chem. 2012, 12, 202–209. [Google Scholar] [CrossRef]
- El−Sayed Ahmed, M.A.E.; Zhong, L.L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.B. Colistin and its role in the Era of antibiotic resistance: An extended review (2000–2019). Emerg. Microbes Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef] [Green Version]
- Bush, K.; Bradford, P.A. β−Lactams and β−Lactamase Inhibitors: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a025247. [Google Scholar] [CrossRef]
- Wilson, D.N. Ribosome−targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 2014, 12, 35–48. [Google Scholar] [CrossRef]
- Fernandez, L.; Hancock, R.E. Adaptive and mutational resistance: Role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 2012, 25, 661–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMurry, L.; Petrucci, R.E., Jr.; Levy, S.B. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc. Natl. Acad. Sci. USA 1980, 77, 3974–3977. [Google Scholar] [CrossRef] [Green Version]
- Poole, K. Efflux−mediated antimicrobial resistance. J. Antimicrob. Chemother. 2005, 56, 20–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; He, Q.N.; Yang, Y.F.; Wang, J.W.; Haning, K.; Hu, Y.; Wu, B.; He, M.X.; Zhang, Y.P.; Bao, J.; et al. Advances and prospects in metabolic engineering of Zymomonas mobilis. Metab. Eng. 2018, 50, 57–73. [Google Scholar] [CrossRef]
- Yang, Y.F.; Geng, B.N.; Song, H.Y.; He, Q.N.; He, M.X.; Bao, J.; Bai, F.W.; Yang, S.H. Progress and perspectives on developing Zymomonas mobilis as a chassis cell. Synth. Biol. J. 2021, 2, 59–90. [Google Scholar] [CrossRef]
- Yang, S.H.; Pelletier, D.A.; Lu, T.Y.; Brown, S.D. The Zymomonas mobilis regulator hfq contributes to tolerance against multiple lignocellulosic pretreatment inhibitors. BMC Microbiol. 2010, 10, 135. [Google Scholar] [CrossRef] [Green Version]
- Zou, S.L.; Zhang, K.; Jing, X.; Wang, Z.R.; Zhang, M.H. Studies on selectable marker for genetic engineering of Zymomonas mobilis ZM4 and CP4 strain. Ind. Microbiol. 2012, 42, 72–77. [Google Scholar]
- Rajnish, K.N.; Asraf, S.A.; Manju, N.; Gunasekaran, P. Functional characterization of a putative beta−lactamase gene in the genome of Zymomonas mobilis. Biotechnol. Lett. 2011, 33, 2425–2430. [Google Scholar] [CrossRef]
- Yang, S.; Pappas, K.M.; Hauser, L.J.; Land, M.L.; Chen, G.L.; Hurst, G.B.; Pan, C.; Kouvelis, V.N.; Typas, M.A.; Pelletier, D.A.; et al. Improved genome annotation for Zymomonas mobilis. Nat. Biotechnol. 2009, 27, 893–894. [Google Scholar] [CrossRef]
- Yang, S.; Vera, J.M.; Grass, J.; Savvakis, G.; Moskvin, O.V.; Yang, Y.; McIlwain, S.J.; Lyu, Y.; Zinonos, I.; Hebert, A.S.; et al. Complete genome sequence and the expression pattern of plasmids of the model ethanologen Zymomonas mobilis ZM4 and its xylose−utilizing derivatives 8b and 2032. Biotechnol. Biofuels 2018, 11, 125. [Google Scholar] [CrossRef]
- Seo, J.S.; Chong, H.; Park, H.S.; Yoon, K.O.; Jung, C.; Kim, J.J.; Hong, J.H.; Kim, H.; Kim, J.H.; Kil, J.I.; et al. The genome sequence of the ethanologenic bacterium Zymomonas mobilis ZM4. Nat. Biotechnol. 2005, 23, 63–68. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.L.; Han, J.M.; Wang, B.Y.; Hu, X.Y.; Li, R.X.; Shen, W.; Ma, X.D.; Ma, L.X.; Yi, L.; Yang, S.H.; et al. Characterization and repurposing of the endogenous Type I−F CRISPR−Cas system of Zymomonas mobilis for genome engineering. Nucleic Acids Res. 2019, 47, 11461–11475. [Google Scholar] [CrossRef] [Green Version]
- Shen, W.; Zhang, J.; Geng, B.N.; Qiu, M.Y.; Hu, M.M.; Yang, Q.; Bao, W.W.; Xiao, Y.B.; Zheng, Y.L.; Peng, W.F.; et al. Establishment and application of a CRISPR−Cas12a assisted genome−editing system in Zymomonas mobilis. Microb. Cell Fact. 2019, 18, 162. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.F.; Shen, W.; Huang, J.; Li, R.X.; Xiao, Y.B.; Wei, H.; Chou, Y.C.; Zhang, M.; Himmel, M.E.; Chen, S.W.; et al. Prediction and characterization of promoters and ribosomal binding sites of Zymomonas mobilis in system biology era. Biotechnol. Biofuels 2019, 12, 52. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [Green Version]
- Chan, E.L.; Harris, R.C.; Dalton, H.P. The effect of antibiotics on the cell morphology of Legionella pneumophila. J. Med. Microbiol. 1987, 23, 149–154. [Google Scholar] [CrossRef] [Green Version]
- Rodgers, F.G.; Tzianabos, A.O.; Elliott, T.S. The effect of antibiotics that inhibit cell−wall, protein, and DNA synthesis on the growth and morphology of Legionella pneumophila. J. Med. Microbiol. 1990, 31, 37–44. [Google Scholar] [CrossRef]
- Li, R.X.; Shen, W.; Yang, Y.F.; Du, J.; Li, M.; Yang, S.H. Investigation of the impact of a broad range of temperatures on the physiological and transcriptional profiles of Zymomonas mobilis ZM4 for high−temperature−tolerant recombinant strain development. Biotechnol. Biofuels 2021, 14, 146. [Google Scholar] [CrossRef]
- Mohagheghi, A.; Linger, J.G.; Yang, S.H.; Smith, H.; Dowe, N.; Zhang, M.; Pienkos, P.T. Improving a recombinant Zymomonas mobilis strain 8b through continuous adaptation on dilute acid pretreated corn stover hydrolysate. Biotechnol. Biofuels 2015, 8, 55. [Google Scholar] [CrossRef]
- Lou, J.Y.; Wang, J.W.; Yang, Y.F.; Yang, Q.; Li, R.X.; Hu, M.M.; He, Q.N.; Du, J.; Wang, X.; Li, M.; et al. Development and characterization of efficient xylose utilization strains of Zymomonas mobilis. Biotechnol. Biofuels 2021, 14, 231. [Google Scholar] [CrossRef]
- Fuchino, K.; Bruheim, P. Increased salt tolerance in Zymomonas mobilis strain generated by adaptative evolution. Microb. Cell Fact. 2020, 19, 147. [Google Scholar] [CrossRef]
- Yang, S.; Pan, C.; Hurst, G.B.; Dice, L.; Davison, B.H.; Brown, S.D. Elucidation of Zymomonas mobilis physiology and stress responses by quantitative proteomics and transcriptomics. Front. Microbiol. 2014, 5, 246. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Yang, Y.; Tang, Y.; Wang, X.; Chen, Y.; Shen, W.; Zhan, Y.; Gao, J.; Wu, B.; He, M.; et al. Development and characterization of acidic−pH−tolerant mutants of Zymomonas mobilis through adaptation and next−generation sequencing−based genome resequencing and RNA−Seq. Biotechnol. Biofuels 2020, 13, 144. [Google Scholar] [CrossRef]
- He, M.X.; Wu, B.; Shui, Z.X.; Hu, Q.C.; Wang, W.G.; Tan, F.R.; Tang, X.Y.; Zhu, Q.L.; Pan, K.; Li, Q.; et al. Transcriptome profiling of Zymomonas mobilis under furfural stress. Appl. Microbiol. Biotechnol. 2012, 95, 189–199. [Google Scholar] [CrossRef]
- Mack, K.; Titball, R.W. Transformation of Burkholderia pseudomallei by electroporation. Anal. Biochem. 1996, 242, 73–76. [Google Scholar] [CrossRef]
- Wu, B.; He, M.; Feng, H.; Zhang, Y.; Hu, Q.; Zhang, Y. Construction and characterization of restriction−modification deficient mutants in Zymomonas mobilis ZM4. Chin. J. Appplied Environ. Biol. 2013, 19, 189–197. [Google Scholar] [CrossRef]
- Kerr, A.L.; Jeon, Y.J.; Svenson, C.J.; Rogers, P.L.; Neilan, B.A. DNA restriction−modification systems in the ethanologen Zymomonas mobilis ZM4. Appl. Microbiol. Biotechnol. 2011, 89, 761–769. [Google Scholar] [CrossRef]
- Lal, P.B.; Wells, F.; Myers, K.S.; Banerjee, R.; Guss, A.M.; Kiley, P.J. Improving mobilization of foreign DNA into Zymomonas mobilis strain ZM4 by removal of multiple restriction systems. Appl. Environ. Microbiol. 2021, 87, e0080821. [Google Scholar] [CrossRef]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geng, B.; Huang, X.; Wu, Y.; He, Q.; Yang, S. Identification and Characterization of Genes Related to Ampicillin Antibiotic Resistance in Zymomonas mobilis. Antibiotics 2022, 11, 1476. https://doi.org/10.3390/antibiotics11111476
Geng B, Huang X, Wu Y, He Q, Yang S. Identification and Characterization of Genes Related to Ampicillin Antibiotic Resistance in Zymomonas mobilis. Antibiotics. 2022; 11(11):1476. https://doi.org/10.3390/antibiotics11111476
Chicago/Turabian StyleGeng, Binan, Xingyu Huang, Yalun Wu, Qiaoning He, and Shihui Yang. 2022. "Identification and Characterization of Genes Related to Ampicillin Antibiotic Resistance in Zymomonas mobilis" Antibiotics 11, no. 11: 1476. https://doi.org/10.3390/antibiotics11111476
APA StyleGeng, B., Huang, X., Wu, Y., He, Q., & Yang, S. (2022). Identification and Characterization of Genes Related to Ampicillin Antibiotic Resistance in Zymomonas mobilis. Antibiotics, 11(11), 1476. https://doi.org/10.3390/antibiotics11111476






