Genome Mining and Metabolic Profiling Reveal Cytotoxic Cyclodipeptides in Streptomyces hygrospinosus var. Beijingensis
Abstract
:1. Introduction
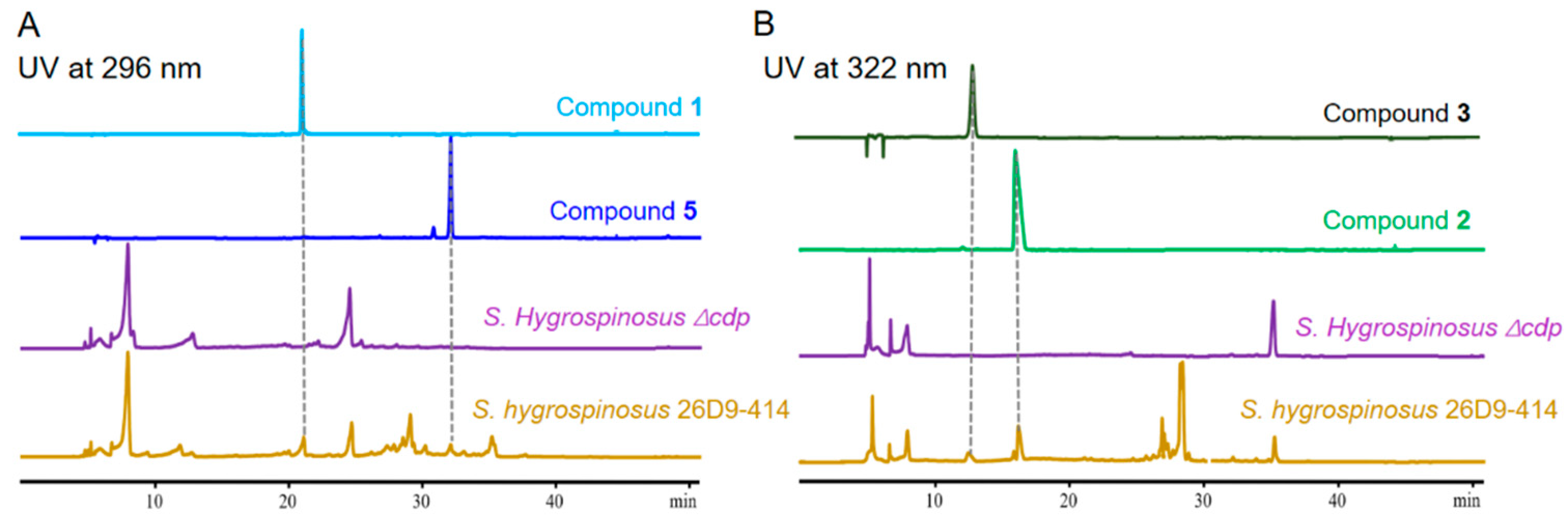
2. Results and Discussion
3. Conclusions
4. Material and Methods
4.1. General Experimental Procedures
4.2. Bacterial Strains, Plasmids, Primers and Culture Conditions
4.3. Construction of S. hygrospinosus Δcdp Mutant
4.4. Strain Fermentation and Chemical Analysis
4.5. Fermentation and Isolation
4.6. ECD Calculations
4.7. Cytotoxicity Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Scherlach, K.; Hertweck, C. Mining and unearthing hidden biosynthetic potential. Nat. Commun. 2021, 12, 3864. [Google Scholar] [CrossRef] [PubMed]
- Shabuer, G.; Ishida, K.; Pidot, S.J.; Roth, M.; Dahse, H.M.; Hertweck, C. Plant pathogenic anaerobic bacteria use aromatic polyketides to access aerobic territory. Science 2015, 350, 670–674. [Google Scholar] [CrossRef] [PubMed]
- Dunbar, K.L.; Dell, M.; Gude, F.; Hertweck, C. Reconstitution of polythioamide antibiotic backbone formation reveals unusual thiotemplated assembly strategy. Proc. Natl. Acad. Sci. USA 2020, 117, 8850–8858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niehs, S.P.; Dose, B.; Richter, S.; Pidot, S.J.; Dahse, H.M.; Stinear, T.P.; Hertweck, C. Mining symbionts of a spider-transmitted fungus lluminates uncharted biosynthetic pathways to cytotoxic benzolactones. Angew. Chem. Int. Ed. Engl. 2020, 59, 7766–7771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, Y.M.; Ahuja, M.; Oakley, C.E.; Entwistle, R.; Asokan, A.; Zutz, C.; Wang, C.C.; Oakley, B.R. Development of genetic dereplication strains in Aspergillus nidulans results in the discovery of aspercryptin. Angew. Chem. Int. Ed. Engl. 2016, 55, 1662–1665. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.; Ye, Y.; Zhang, Y. Large-scale culture as a complementary and practical method for discovering natural products with novel skeletons. Nat. Prod. Rep. 2021, 38, 1775–1793. [Google Scholar] [CrossRef]
- Li, F.; Lin, S.; Zhang, S.; Hao, X.; Li, X.N.; Yang, B.; Liu, J.; Wang, J.; Hu, Z.; Zhang, Y. Alterbrassinoids A-D: Fusicoccane-derived diterpenoid dimers featuring different carbon skeletons from Alternaria brassicicola. Org. Lett. 2019, 21, 8353–8357. [Google Scholar] [CrossRef]
- Wang, F.; Sarotti, A.M.; Jiang, G.; Huguet-Tapia, J.C.; Zheng, S.L.; Wu, X.; Li, C.; Ding, Y.; Cao, S. Waikikiamides A-C: Complex diketopiperazine dimer and diketopiperazine-polyketide hybrids from a Hawaiian marine fungal strain Aspergillus sp. FM242. Org. Lett. 2020, 22, 4408–4412. [Google Scholar] [CrossRef]
- Kersten, R.D.; Weng, J.K. Gene-guided discovery and engineering of branched cyclic peptides in plants. Proc. Natl. Acad. Sci. USA 2018, 115, e10961–e10969. [Google Scholar] [CrossRef] [Green Version]
- Bhushan, A.; Egli, P.J.; Peters, E.E.; Freeman, M.F.; Piel, J. Genome mining- and synthetic biology-enabled production of hypermodified peptides. Nat. Chem. 2019, 11, 931–939. [Google Scholar] [CrossRef]
- Gondry, M.; Jacques, I.B.; Thai, R.; Babin, M.; Canu, N.; Seguin, J.; Belin, P.; Pernodet, J.L.; Moutiez, M. A comprehensive overview of the cyclodipeptide synthase family enriched with the characterization of 32 new enzymes. Front. Microbiol. 2018, 9, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belin, P.; Moutiez, M.; Lautru, S.; Seguin, J.; Pernodet, J.L.; Gondry, M. The nonribosomal synthesis of diketopiperazines in tRNA-dependent cyclodipeptide synthase pathways. Nat. Prod. Rep. 2012, 29, 961–979. [Google Scholar] [CrossRef]
- Stark, T.; Hofmann, T. Structures, sensory activity, and dose/response functions of 2,5-diketopiperazines in roasted cocoa nibs (Theobroma cacao). J. Agric. Food. Chem. 2005, 53, 7222–7231. [Google Scholar] [CrossRef] [PubMed]
- Harken, L.; Li, S.M. Modifications of diketopiperazines assembled by cyclodipeptide synthases with cytochrome P(450) enzymes. Appl. Microbiol. Biotechnol. 2021, 105, 2277–2285. [Google Scholar] [CrossRef]
- Canu, N.; Moutiez, M.; Belin, P.; Gondry, M. Cyclodipeptide synthases: A promising biotechnological tool for the synthesis of diverse 2,5-diketopiperazines. Nat. Prod. Rep. 2020, 37, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Yao, F.; Zheng, X.; Cui, D.; Shao, Y.; Zhu, C.; Deng, Z.; You, D. Genome mining of the biosynthetic gene cluster of the polyene macrolide antibiotic tetramycin and characterization of a P450 monooxygenase involved in the hydroxylation of the tetramycin B polyol segment. Chembiochem 2012, 13, 2234–2242. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Cheng, Q.; Yao, F.; Wang, X.; Kong, L.; Cao, B.; Xu, M.; Lin, S.; Deng, Z.; Chooi, Y.H.; et al. Biosynthesis of the pyrrolidine protein synthesis inhibitor anisomycin involves novel gene ensemble and cryptic biosynthetic steps. Proc. Natl. Acad. Sci. USA 2017, 114, 4135–4140. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Kong, L.; Qiao, Y.; Zhang, D.; Deng, Z.; You, D. Genome mining guided discovery and identification of nystatin and toyocamycin in Streptomyces hygrospinosus var. beijingensis. Acta Microbiol. Sin. 2021, 61, 3076–3086. [Google Scholar]
- Xi, Y.-K.; Zhang, H.; Li, R.-X.; Kang, S.-Y.; Li, J.; Li, Y. Total synthesis of spirotryprostatins through organomediated intramolecular umpolung cyclization. Chemistry 2019, 25, 3005–3009. [Google Scholar] [CrossRef] [PubMed]
- Tatsuta, K.; Fujimoto, K.; Yamashita, M.; Tsuchiya, T.; Umezawa, S. Argvalin, a new microbial metabolite: Isolation and structure. J. Antibiot. 1973, 26, 606–608. [Google Scholar] [CrossRef]
- Lautru, S.; Gondry, M.; Genet, R.; Pernodet, J.L. The albonoursin gene Cluster of S. noursei biosynthesis of diketopiperazine metabolites independent of nonribosomal peptide synthetases. Chem. Biol. 2002, 9, 1355–1364. [Google Scholar] [CrossRef]
- Brown, R.; Kelley, C.; Wiberley, S.E. The production of 3-benzylidene-6-isobutylidene-2, 5-dioxopiperazine, 3, 6-dibenzylidene-2, 5-dioxopiperazine, 3-benzyl-6-benzylidene-2, 5-dioxopiperazine, and 3, 6-dibenzyl-2, 5-dioxopiperazine by a variant of Streptomyces noursei. J. Org. Chem. 1965, 30, 277–280. [Google Scholar] [CrossRef]
- Yang, L.M.; Wu, R.Y.; McPhail, A.T.; Yokoi, T.; Lee, K.H. Neihumicin, a new cytotoxic antibiotic from Micromonospora neihuensis. II. structural determination and total synthesis. J. Antibiot. 1988, 41, 488–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Li, S.; Chen, Y.; Tian, X.; Zhang, H.; Zhang, G.; Zhu, Y.; Zhang, S.; Zhang, W.; Zhang, C. New diketopiperazine derivatives from a deep-sea-derived Nocardiopsis alba SCSIO 03039. J. Antibiot. 2013, 66, 31–36. [Google Scholar] [CrossRef] [Green Version]
- Kawahara, N.; Nozawa, K.; Nakajima, S.; Yamazaki, M.; Kawai, K. Sulfur-containing dioxopiperazine derivatives from Emericella heterothallica. Heterocycles 1989, 29, 397–402. [Google Scholar]
- Achkar, I.W.; Abdulrahman, N.; Al-Sulaiti, H.; Joseph, J.M.; Uddin, S.; Mraiche, F. Cisplatin based therapy: The role of the mitogen activated protein kinase signaling pathway. J. Transl. Med. 2018, 16, 96. [Google Scholar] [CrossRef] [Green Version]
- Cain, C.C.; Lee, D.; Waldo, R.H., 3rd; Henry, A.T.; Casida, E.J., Jr.; Wani, M.C.; Wall, M.E.; Oberlies, N.H.; Falkinham, J.O., 3rd. Synergistic antimicrobial activity of metabolites produced by a nonobligate bacterial predator. Antimicrob. Agents Chemother. 2003, 47, 2113–2117. [Google Scholar] [CrossRef] [Green Version]
- Ström, K.; Sjögren, J.; Broberg, A.; Schnürer, J. Lactobacillus plantarum MiLAB 393 produces the antifungal cyclic dipeptides cyclo(L-Phe-L-Pro) and cyclo(L-Phe-trans-4-OH-L-Pro) and 3-phenyllactic acid. Appl. Env. Microbiol. 2002, 68, 4322–4327. [Google Scholar] [CrossRef] [Green Version]
- Gondry, M.; Sauguet, L.; Belin, P.; Thai, R.; Amouroux, R.; Tellier, C.; Tuphile, K.; Jacquet, M.; Braud, S.; Courçon, M.; et al. Cyclodipeptide synthases are a family of tRNA-dependent peptide bond-forming enzymes. Nat. Chem. Biol. 2009, 5, 414–420. [Google Scholar] [CrossRef]
- Jacques, I.B.; Moutiez, M.; Witwinowski, J.; Darbon, E.; Martel, C.; Seguin, J.; Favry, E.; Thai, R.; Lecoq, A.; Dubois, S.; et al. Analysis of 51 cyclodipeptide synthases reveals the basis for substrate specificity. Nat. Chem. Biol. 2015, 11, 721–727. [Google Scholar] [CrossRef]
- Le Chevalier, F.; Correia, I.; Matheron, L.; Babin, M.; Moutiez, M.; Canu, N.; Gondry, M.; Lequin, O.; Belin, P. In vivo characterization of the activities of novel cyclodipeptide oxidases: New tools for increasing chemical diversity of bioproduced 2,5-diketopiperazines in Escherichia coli. Microb. Cell Fact. 2020, 19, 178. [Google Scholar] [CrossRef] [PubMed]
- Mikulski, L.; Schäfer, J.; Brockmeyer, K.; Kraut, R.; Li, S.M. Comparative studies on similarities and differences of cyclodipeptide oxidases for installation of C-C double bonds at the diketopiperazine ring. Appl. Microbiol. Biotechnol. 2020, 104, 2523–2536. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kong, L.; Shen, J.; Wang, Q.; Liu, Q.; Yang, W.; Deng, Z.; You, D. Characterization of the positive SARP family regulator PieR for improving piericidin A1 production in Streptomyces piomogeues var. Hangzhouwanensis. Synth. Syst. Biotechnol. 2019, 4, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, W.; Chooi, Y.H.; Wang, L.; Cao, B.; Deng, Z.; Chu, Y.; You, D. A multifunctional monooxygenase XanO4 catalyzes xanthone formation in xantholipin biosynthesis via a cryptic demethoxylation. Cell Chem. Biol. 2016, 23, 508–516. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Kong, L.; Zheng, X.; Shen, J.; Wang, J.; Zhang, D.; Qiao, Y.; Wang, J.; Deng, Z.; You, D. Acyltransferase AniI, a tailoring enzyme with broad substrate tolerance for high-level production of anisomycin. Appl. Env. Microbiol. 2021, 87, e0017221. [Google Scholar] [CrossRef]
- Shen, J.; Kong, L.; Li, Y.; Zheng, X.; Wang, Q.; Yang, W.; Deng, Z.; You, D. A LuxR family transcriptional regulator AniF promotes the production of anisomycin and its derivatives in Streptomyces hygrospinosus var. beijingensis. Synth. Syst. Biotechnol. 2019, 4, 40–48. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, X.; Al Anbari, W.H.; Zhang, M.; Chen, X.; Luo, Z.; Li, X.N.; Chen, C.; Liu, J.; Wang, J.; et al. Amiaspochalasins A-H, undescribed aspochalasins with a C-21 ester carbonyl from Aspergillus micronesiensis. J. Org. Chem. 2019, 84, 5483–5491. [Google Scholar] [CrossRef]
- Paget, M.S.; Chamberlin, L.; Atrih, A.; Foster, S.J.; Buttner, M.J. Evidence that the extracytoplasmic function sigma factor sigmaE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 1999, 181, 204–211. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Wang, Z.; Bai, L.; Liang, J.; Zhou, X.; Deng, Z. Two pHZ1358-derivative vectors for efficient gene knockout in Streptomyces. J. Microbiol. Biotechnol. 2010, 20, 678–682. [Google Scholar] [CrossRef] [Green Version]
- Gust, B.; Challis, G.L.; Fowler, K.; Kieser, T.; Chater, K.F. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 2003, 100, 1541–1546. [Google Scholar] [CrossRef] [Green Version]
- Tan, G.Y.; Deng, K.; Liu, X.; Tao, H.; Chang, Y.; Chen, J.; Chen, K.; Sheng, Z.; Deng, Z.; Liu, T. Heterologous biosynthesis of spinosad: An Omics-guided large polyketide synthase gene cluster reconstitution in Streptomyces. ACS Synth. Biol. 2017, 6, 995–1005. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Wang, G.; Xiao, H.; Jiang, J.; Xiao, D.; Xing, B.; Li, A.; Zhang, Y.; Sun, K.; Xu, Y.; et al. Strepimidazoles A-G from the plant endophytic Streptomyces sp. PKU-EA00015 with inhibitory activities against a plant pathogenic fungus. J. Nat. Prod. 2020, 83, 2246–2254. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Liu, Z.; Zhu, X.; Fan, Z.; Huang, X.; Wu, Q.; Zheng, X.; Qin, X.; Zhang, T.; Zhang, H.; et al. Antitubercular ilamycins from marine-derived Streptomyces atratus SCSIO ZH16 ΔilaR. J. Nat. Prod. 2020, 83, 1646–1657. [Google Scholar] [CrossRef] [PubMed]
- Perlatti, B.; Lan, N.; Earp, C.E.; AghaAmiri, S.; Vargas, S.H.; Azhdarinia, A.; Bills, G.F.; Gloer, J.B. Arenicolins: C-glycosylated depsides from Penicillium arenicola. J. Nat. Prod. 2020, 83, 668–674. [Google Scholar] [CrossRef] [PubMed]
- Huo, C.; Zheng, Z.; Xu, Y.; Ding, Y.; Zheng, H.; Mu, Y.; Niu, Y.; Gao, J.; Lu, X. Naphthacemycins from a Streptomyces sp. as protein-tyrosine phosphatase inhibitors. J. Nat. Prod. 2020, 83, 1394–1399. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, L.; Kong, L.; Wang, T.; Chu, Y.; Deng, Z.; You, D. Unveiling the post-PKS redox tailoring steps in biosynthesis of the type II polyketide antitumor antibiotic xantholipin. Chem. Biol. 2012, 19, 422–432. [Google Scholar] [CrossRef]
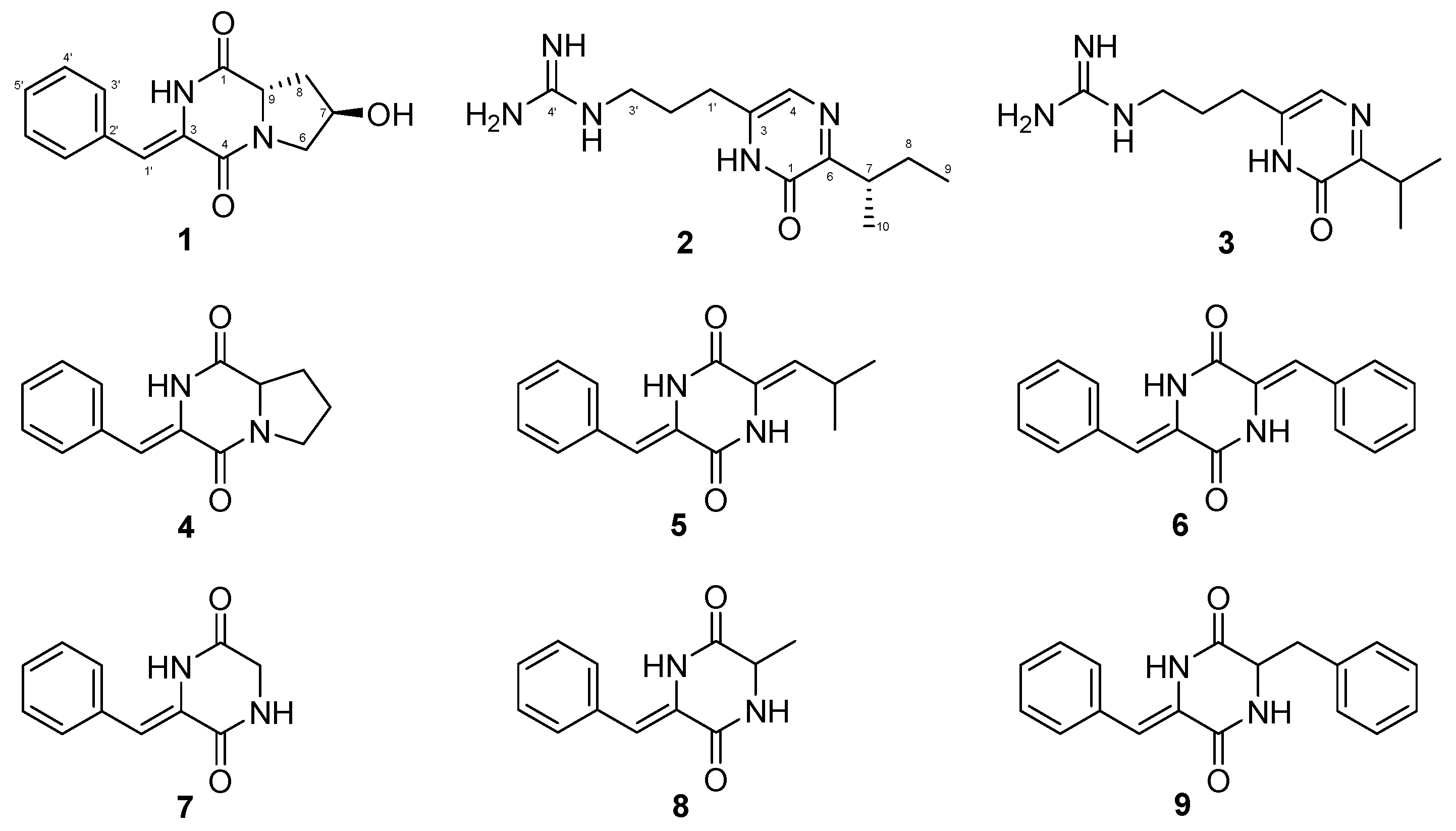
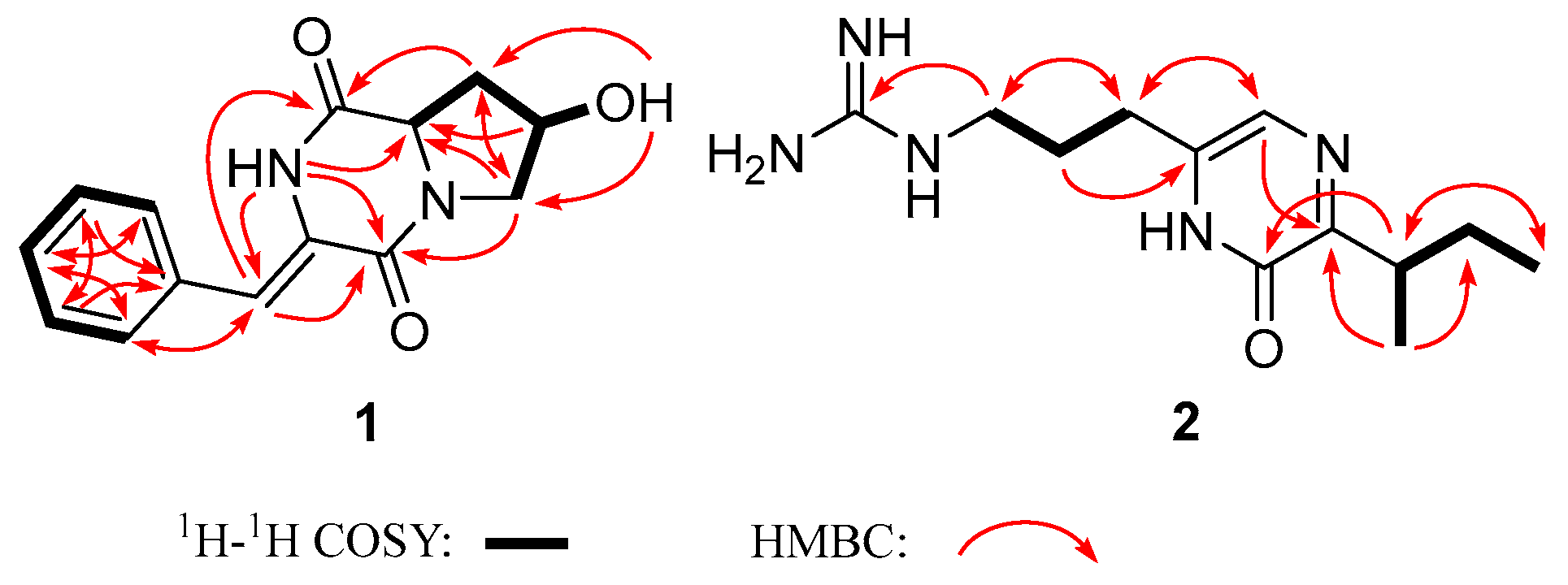
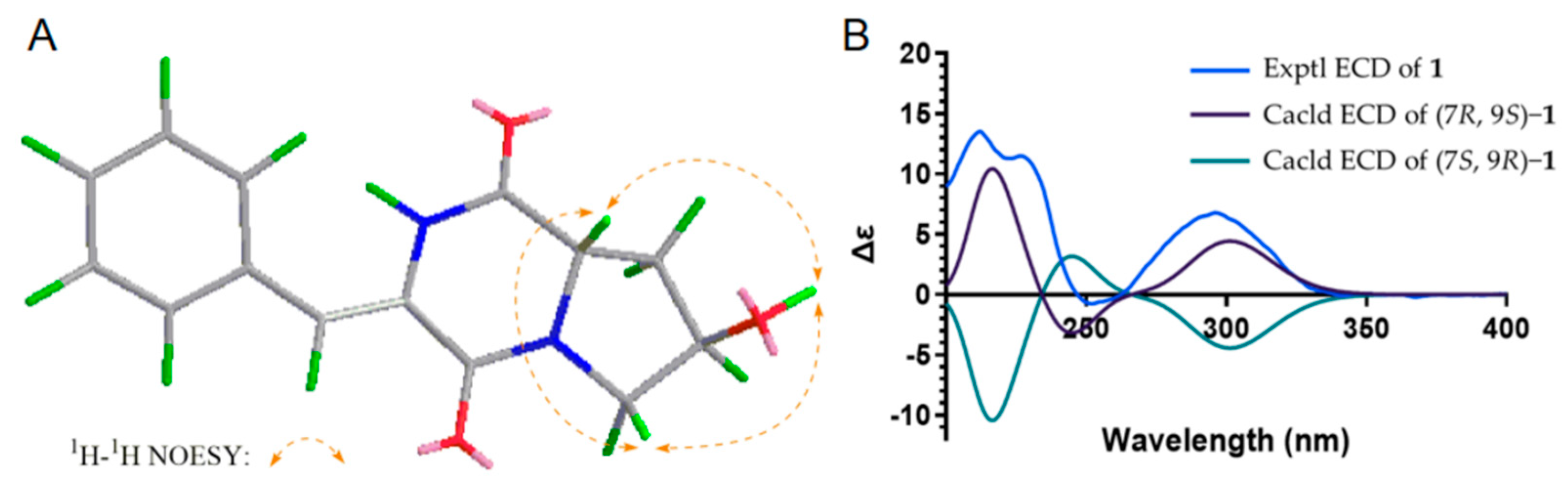
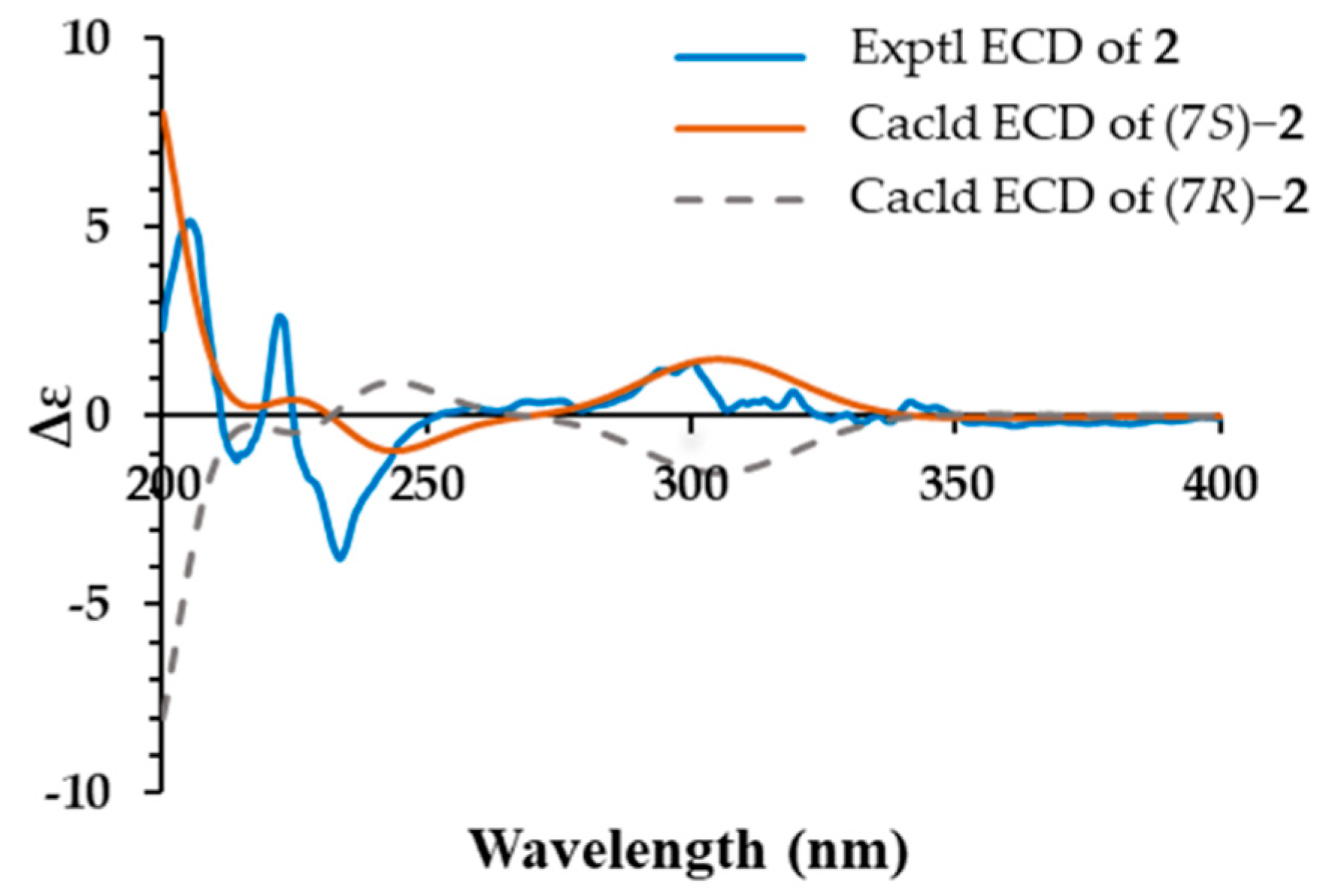
| No. | 1 | 2 | ||
|---|---|---|---|---|
| δH, Mult (J in Hz) | δC, Type | δH, Mult (J in Hz) | δC, Type | |
| 1 | 167.5, C | 158.8, C | ||
| 2 | 10.03, s | 12.10, s | ||
| 3 | 128.6, C | 137.5, C | ||
| 4 | 158.7, C | 7.84, s | 120.1, C | |
| 5 | ||||
| 6 | 3.71, dd (12.7, 4.7) | 54.5, CH2 | 156.0, C | |
| 3.32, d (12.7) | ||||
| 7 | 4.34, m | 66.6, CH | 3.08, m | 35.7, CH |
| 8 | 2.12, dd (12.6, 6.4) | 37.3, CH2 | 1.68, m | 27.1, CH2 |
| 2.01, td (12.6, 4.4) | 1.41, m | |||
| 9 | 4.58, dd (12.6, 6.4) | 56.8, CH | 0.79, t (7.4) | 11.9, CH3 |
| 10 | 1.07, d (6.9) | 17.8, CH3 | ||
| 1′ | 6.67, s | 114.8, CH | 2.43, t (7.5) | 26.7, CH2 |
| 2′ | 133.6, C | 1.78, m | 27.2, CH2 | |
| 3′ | 7.54, d (7.6) | 129.4, CH | 3.10, t (6.4) | 40.1, CH2 |
| 4′ | 7.40, t (7.6) | 128.6, CH | 156.9, C | |
| 5′ | 7.30, t (7.6) | 128.0, CH | ||
| 7-OH | 5.17, d (2.9) | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, D.; Wang, J.; Qiao, Y.; Lin, B.; Deng, Z.; Kong, L.; You, D. Genome Mining and Metabolic Profiling Reveal Cytotoxic Cyclodipeptides in Streptomyces hygrospinosus var. Beijingensis. Antibiotics 2022, 11, 1463. https://doi.org/10.3390/antibiotics11111463
Zhang D, Wang J, Qiao Y, Lin B, Deng Z, Kong L, You D. Genome Mining and Metabolic Profiling Reveal Cytotoxic Cyclodipeptides in Streptomyces hygrospinosus var. Beijingensis. Antibiotics. 2022; 11(11):1463. https://doi.org/10.3390/antibiotics11111463
Chicago/Turabian StyleZhang, Dashan, Junbo Wang, Yongjian Qiao, Baixin Lin, Zixin Deng, Lingxin Kong, and Delin You. 2022. "Genome Mining and Metabolic Profiling Reveal Cytotoxic Cyclodipeptides in Streptomyces hygrospinosus var. Beijingensis" Antibiotics 11, no. 11: 1463. https://doi.org/10.3390/antibiotics11111463
APA StyleZhang, D., Wang, J., Qiao, Y., Lin, B., Deng, Z., Kong, L., & You, D. (2022). Genome Mining and Metabolic Profiling Reveal Cytotoxic Cyclodipeptides in Streptomyces hygrospinosus var. Beijingensis. Antibiotics, 11(11), 1463. https://doi.org/10.3390/antibiotics11111463







