A Meta-Analysis to Estimate Prevalence of Resistance to Tetracyclines and Third Generation Cephalosporins in Enterobacteriaceae Isolated from Food Crops
Abstract
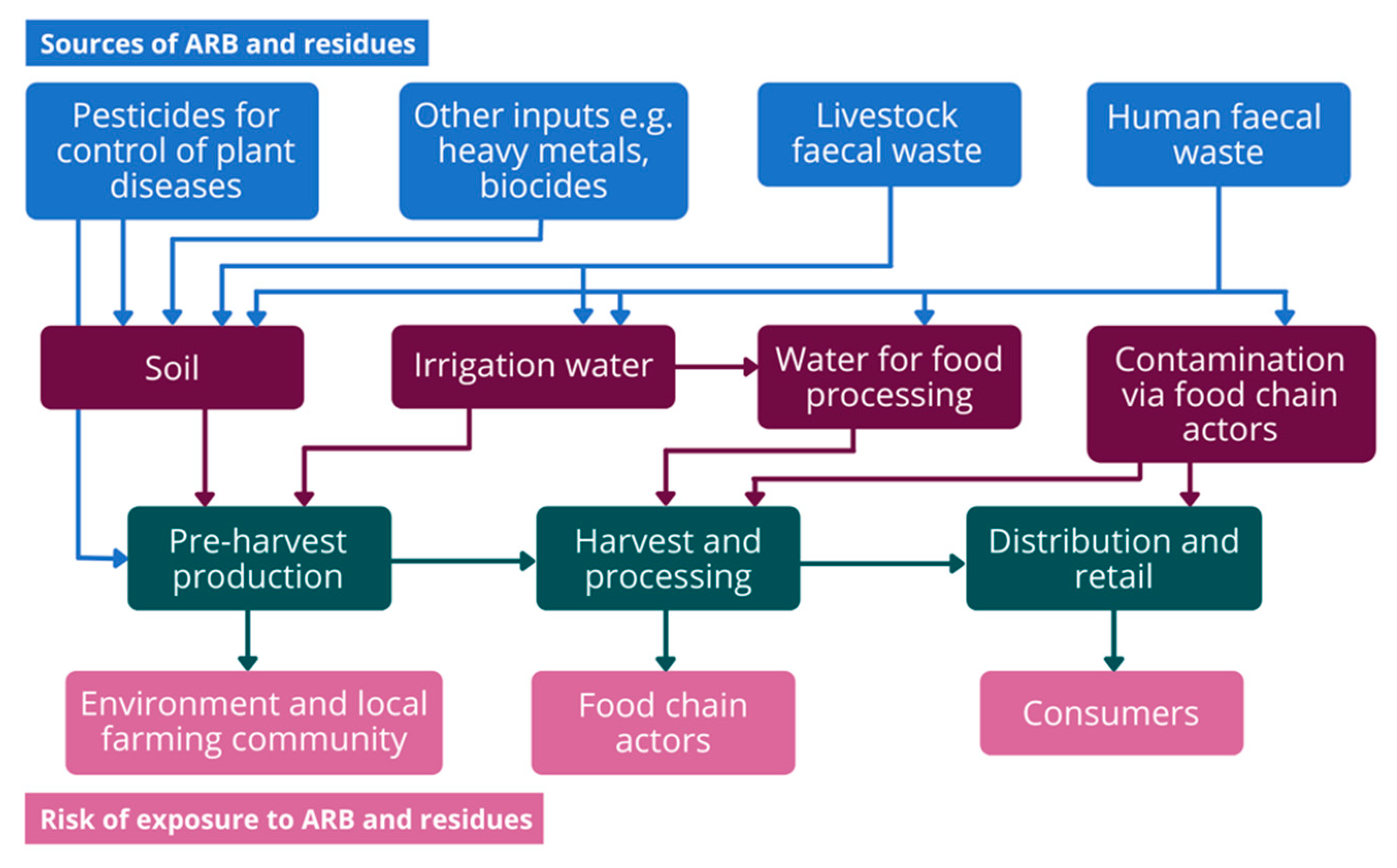
1. Introduction
2. Results
2.1. Risk of Bias Assessment
2.2. Subgroup Meta-Analysis
2.2.1. Enterobacteriaceae Prevalence in Food Crops
2.2.2. Total AMR Prevalence in Produce
2.2.3. AMR Prevalence by Antimicrobial Class
2.2.4. AMR Prevalence by Stage of Value Chain Sampling
2.2.5. AMR Prevalence by Region and Antimicrobial Class Type
2.3. Meta-Regression
2.4. Sensitivity Analysis
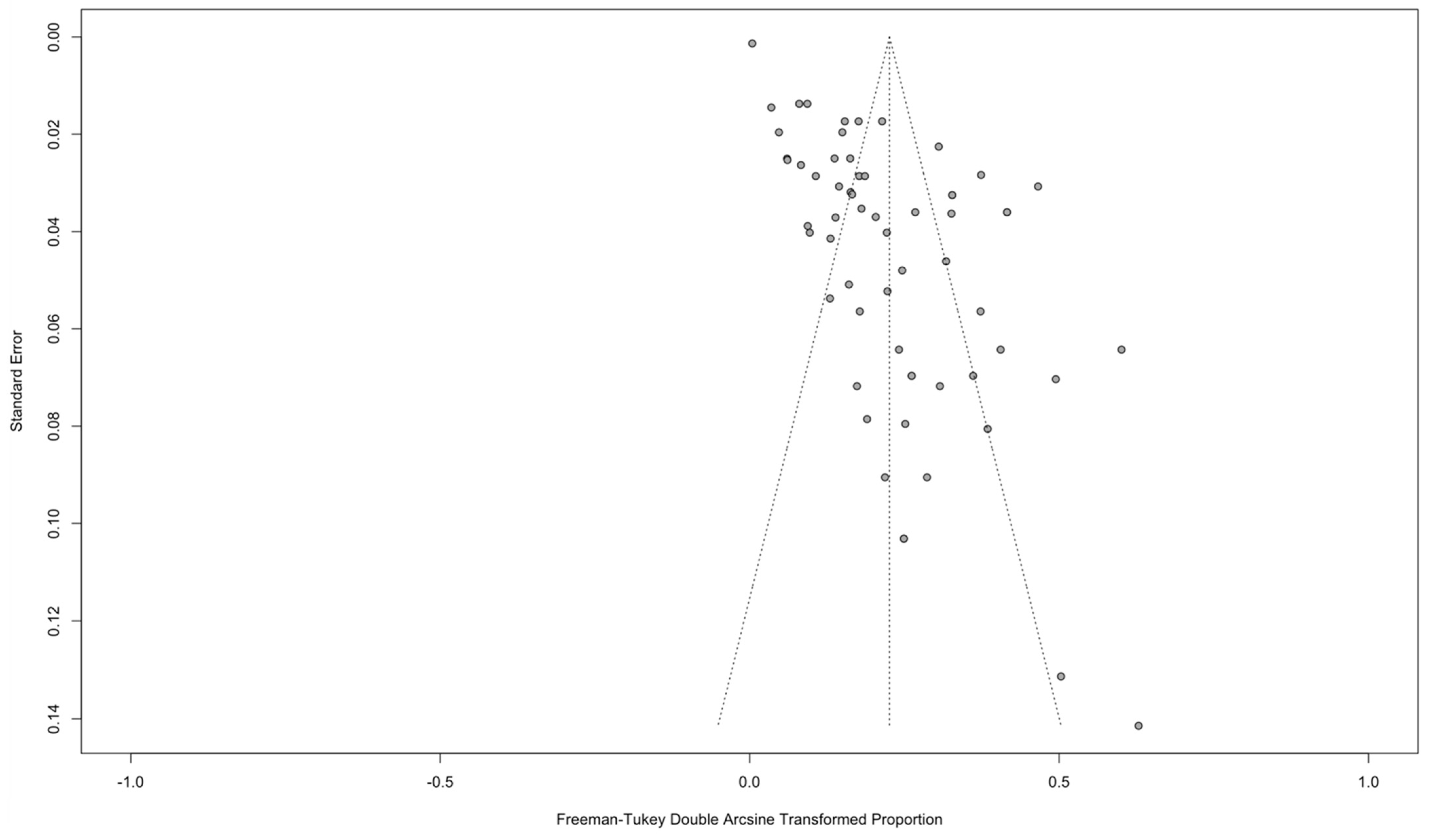
2.5. Publication Bias
2.6. AMR Prevalence Ratio
3. Discussion
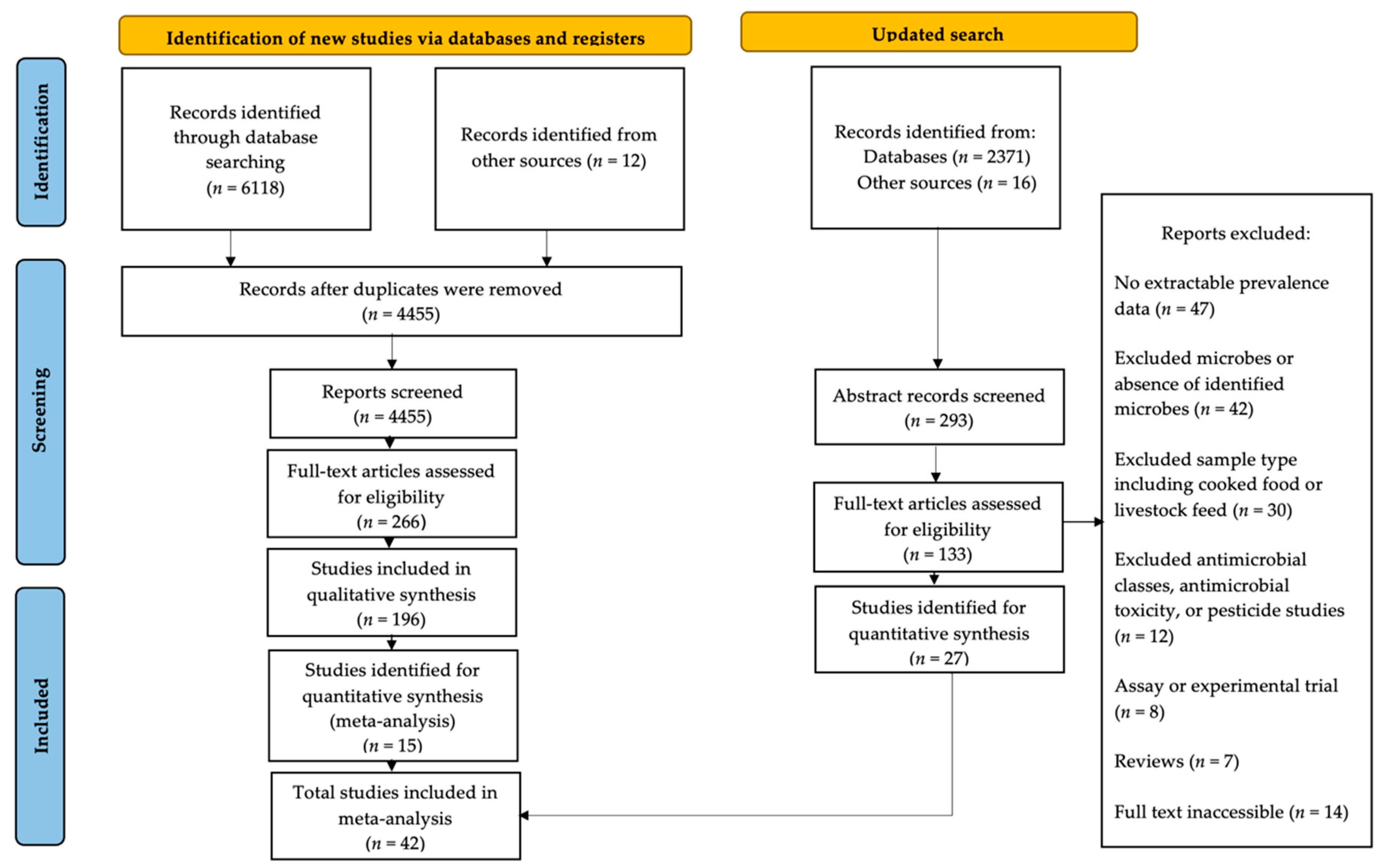
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Sub-Group | Prevalence (%) | Prevalence CI (%) | p-Value | No. of Prevalence Estimates | I2 (%) | I2 CI (%) |
|---|---|---|---|---|---|---|
| 3GC Resistance | ||||||
| ESBL-producers | 4.05 | 1.18, 8.27 | <0.0001 | 11 | 94.8 | 92.5, 96.5 |
| No ESBL-producers | 3.17 | 1.37, 5.54 | <0.0001 | 15 | 89.9 | 85.1, 93.2 |
| TET Resistance | ||||||
| ESBL-producers | 2.85 | 1.06, 5.35 | <0.0001 | 9 | 88.5 | 80.3, 93.2 |
| No ESBL-producers | 5.66 | 2.64, 9.60 | <0.0001 | 21 | 97.4 | 96.8, 97.9 |
References
- Abraham, E.; Chain, E. An Enzyme from Bacteria able to Destroy Penicillin. Nature 1940, 146, 837. [Google Scholar] [CrossRef]
- Hwengwere, K.; Paramel, N.H.; Hughes, K.A.; Peck, L.S.; Clark, M.S.; Walker, C.A. Antimicrobial resistance in Antarctica: Is it still a pristine environment? Microbiome 2021, 10, 71. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. The Review on Antimicrobial Resistance. 2016. Available online: https://amr-review.org/Publications.html (accessed on 30 June 2021).
- Wellcome Trust. The Global Response to AMR: Momentum, Success, and Critical Gaps. 2020. London. Available online: https://wellcome.org/reports/global-response-amr-momentum-success-and-critical-gaps (accessed on 30 June 2021).
- He, Y.; Yuan, Q.; Mathieu, J.; Stadler, L.; Senehi, N.; Sun, R.; Alvarez, P.J. Antibiotic resistance genes from livestock waste: Occurrence, dissemination, and treatment. NPJ Clean Water 2020, 3, 4. [Google Scholar] [CrossRef]
- Rahman, M.; Alam, M.U.; Luies, S.K.; Kamal, A.; Ferdous, S.; Lin, A.; Sharior, F.; Khan, R.; Rahman, Z.; Parvez, S.M.; et al. Contamination of Fresh Produce with Antibiotic-Resistant Bacteria and Associated Risks to Human Health: A Scoping Review. Int. J. Environ. Res. Public Health 2021, 19, 360. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, C.; Lang, L.; Wang, A.; Altendorf, K.; García, F.; Lipski, A. Lettuce for human consumption collected in Costa Rica contains complex communities of culturable oxytetracycline- and gentamicin-resistant bacteria. Appl. Environ. Microbiol. 2006, 72, 5870–5876. [Google Scholar] [CrossRef]
- Food and Agriculture Organization (FAO) and World Health Organization (WHO). Joint FAO/WHO Expert Meeting in collaboration with OIE on Foodborne Antimicrobial Resistance: Role of the Environment, Crops and Biocides. Rome. 2019. Available online: https://www.who.int/publications/i/item/9789241516907 (accessed on 30 June 2021).
- Taylor, P.; Reeder, R. Antibiotic use on crops in low and middle-income countries based on recommendations made by agricultural advisors. CABI Agric. Biosci. 2020, 1, 1–14. [Google Scholar] [CrossRef]
- Sivapalasingam, S.; Friedman, C.R.; Cohen, L.; Tauxe, R.V. Fresh Produce: A Growing Cause of Outbreaks of Foodborne Illness in the United States. J. Food Prot. 2004, 67, 2342–2353. [Google Scholar] [CrossRef] [PubMed]
- Martínez, J.L.; Coque, T.M.; Baquero, F. What is a resistance gene? Ranking risk in resistomes. Nat. Rev. Microbiol. 2015, 13, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Manaia, C.M. Assessing the Risk of Antibiotic Resistance Transmission from the Environment to Humans: Non-Direct Proportionality between Abundance and Risk. Trends Microbiol. 2017, 25, 173–181. [Google Scholar] [CrossRef]
- Jans, C.; Sarno, E.; Collineau, L.; Meile, L.; Stärk, K.D.C.; Stephan, R. Consumer exposure to antimicrobial resistant bacteria from food at Swiss retail level. Front. Microbiol. 2018, 9, 362. [Google Scholar] [CrossRef] [PubMed]
- Reddy, S.P.; Wang, H.; Adams, J.K.; Feng, P.C.H. Prevalence and Characteristics of Salmonella Serotypes Isolated from Fresh Produce Marketed in the United States. J. Food Prot. 2016, 79, 6–16. [Google Scholar] [CrossRef]
- Astill, J.; Dara, R.A.; Campbell, M.; Farber, J.M.; Fraser, E.D.G.; Sharif, S.; Yada, R.Y. Transparency in food supply chains: A review of enabling technology solutions. Trends Food Sci. Technol. 2019, 91, 240–247. [Google Scholar] [CrossRef]
- Brunn, A.; Kadri-Alabi, Z.; Moodley, A.; Guardabassi, L.; Taylor, P.; Mateus, A.; Waage, J. Characteristics and global occurrence of human pathogens harboring antimicrobial resistance in food crops: A scoping review. Front. Sustain. Food Syst. 2022, 6, 824714. [Google Scholar] [CrossRef]
- Caffrey, N.; Invik, J.; Waldner, C.L.; Ramsay, D.; Checkley, S.L. Risk assessments evaluating foodborne antimicrobial resistance in humans: A scoping review. Microbiol. Risk Anal. 2019, 11, 31–46. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Pires, J.; Silvester, R.; Zhao, C.; Song, J.; Criscuolo, N.G.; Gilbert, M.; Bonhoeffer, S.; Laxminarayan, R. Global trends in antimicrobial resistance in animals in low- And middle-income countries. Science 2019, 365, eaaw1944. [Google Scholar] [CrossRef]
- Verraes, C.; Van Boxstael, S.; Van Meervenne, E.; Van Coillie, E.; Butaye, P.; Catry, B.; De Schaetzen, M.-A.; Van Huffel, X.; Imberechts, H.; Dierick, K.; et al. Antimicrobial Resistance in the Food Chain: A Review. Int. J. Environ. Res. Public Health 2013, 10, 2643–2669. [Google Scholar] [CrossRef] [PubMed]
- Hölzel, C.S.; Tetens, J.L.; Schwaiger, K. Unraveling the Role of Vegetables in Spreading Antimicrobial-Resistant Bacteria: A Need for Quantitative Risk Assessment. Foodborne Pathog. Dis. 2018, 15, 671–688. [Google Scholar] [CrossRef] [PubMed]
- Nunes, S.B.; Cadavez, V.; Teixeira, J.A.; Gonzales-Barron, U. Meta-analysis of the incidence of foodborne pathogens in vegetables and fruits from retail establishments in Europe. Curr. Opin. Food Sci. 2017, 18, 21–28. [Google Scholar] [CrossRef]
- Corredor-García, D.; Santiago, G.-P.; Blanco-Lizarazo, C.M. Systematic Review and Meta-analysis: Salmonella spp. prevalence in vegetables and fruits. World J. Microbiol. Biotechnol. 2021, 37, 47. [Google Scholar] [CrossRef]
- Mohammadpour, H.; Berizi, E.; Hosseinzadeh, S.; Majlesi, M.; Zare, M. The prevalence of Campylobacter spp. in vegetables, fruits, and fresh produce: A systematic review and meta-analysis. Gut Pathog. 2018, 10, 41. [Google Scholar] [CrossRef]
- De Oliveira, E.S.; Noronha, T.B.; Tondo, E.C. Salmonella spp. and Escherichia coli O157:H7 prevalence and levels on lettuce: A systematic review and meta-analysis. Food Microbiol. 2019, 84, 103217. [Google Scholar] [CrossRef] [PubMed]
- Iwu, C.D.; Okoh, A.I. Preharvest Transmission Routes of Fresh Produce Associated Bacterial Pathogens with Outbreak Potentials: A Review. Int. J. Environ. Res. Public Health 2019, 16, 4407. [Google Scholar] [CrossRef] [PubMed]
- Niegowska, M.; Wögerbauer, M. Improving the risk assessment of antimicrobial resistance (AMR) along the food/feed chain and from environmental reservoirs using qMRA and probabilistic modelling. EFSA J. Eur. Food Saf. Auth. 2022, 25, e200407. [Google Scholar] [CrossRef]
- Koutsoumanis, K.; Allende, A.; Álvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Herman, L.; Hilbert, F.; et al. Role played by the environment in the emergence and spread of antimicrobial resistance (AMR) through the food chain. EFSA J. Eur. Food Saf. Auth. 2021, 19, e06651. [Google Scholar] [CrossRef]
- Collineau, L.; Boerlin, P.; Carson, C.A.; Chapman, B.; Fazil, A.; Hetman, B.; McEwen, S.A.; Parmley, E.J.; Reid-Smith, R.J.; Taboada, E.N.; et al. Integrating whole-genome sequencing data into quantitative risk assessment of foodborne antimicrobial resistance: A review of opportunities and challenges. Front. Microbiol. 2019, 10, 1107. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Critically Important Antimicrobials for Human Medicine, 6th ed.; World Health Organization: Geneva, Switzerland, 2019. Available online: https://www.who.int/publications/i/item/9789241515528 (accessed on 19 April 2022).
- European Parliament. Committee on the Environment Public Health and Food Safety. Motion for a Resolution on the Draft: Commission Implementing Regulation Designating Antimicrobials or Groups of Antimicrobials Reserved for Treatment of Certain Infections in Humans, in Accordance with Regulation (EU) 2019/6 of the European Parliament and of the Council. 2022. Available online: https://www.europarl.europa.eu/doceo/document/B-9-2022-0327_EN.html (accessed on 7 July 2022).
- Schmidt, C.W. FDA proposes to ban cephalosporins from livestock feed. Environ. Health Perspect. 2012, 120, A106. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Downes, M.J.; Brennan, M.L.; Williams, H.C.; Dean, R.S. Development of a critical appraisal tool to assess the quality of cross-sectional studies. BMJ Open 2016, 6, e011458. [Google Scholar] [CrossRef] [PubMed]
- Aabed, K.; Moubayed, N.; Alzahrani, S. Antimicrobial resistance patterns among different Escherichia coli isolates in the Kingdom of Saudi Arabia. Saudi J. Biol. Sci. 2021, 28, 3776–3782. [Google Scholar] [CrossRef] [PubMed]
- Freitag, C.; Michael, G.B.; Li, J.; Kadlec, K.; Wang, Y.; Hassel, M.; Schwarz, S. Occurrence and characterisation of ESBL-encoding plasmids among Escherichia coli isolates from fresh vegetables. Vet. Microbiol. 2018, 219, 63. [Google Scholar] [CrossRef] [PubMed]
- Hartantyo, S.H.P.; Chau, M.L.; Koh, T.H.; Yap, M.; Yi, T.; Cao, D.Y.H.; Gutiérrez, R.A.; Ng, L.C. Foodborne Klebsiella pneumoniae: Virulence potential, antibiotic resistance, and risks to food safety. J. Food Protect. 2020, 83, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Janalíková, M.; Pleva, P.; Pavlícková, S.; Lecomte, M.; Godillon, T.; Holko, I. Characterization of Escherichia Coli strains isolated from raw vegetables. Potravinarstvo Slovak Journal of Food Sciences 2018, 12, 304–312. [Google Scholar] [CrossRef]
- Kim, H.S.; Chon, J.W.; Kim, Y.J.; Kim, D.H.; Kim, M.S.; Seo, K.H. Prevalence and characterization of extended-spectrum-β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in ready-to-eat vegetables. Int. J. Food Microbiol. 2015, 207, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Lopes, R.; Fuentes-Castillo, D.; Fontana, H.; Rodrigues, L.; Dantas, K.; Cerdeira, L.; Henriques, I.; Lincopan, N. Endophytic Lifestyle of Global Clones of Extended-Spectrum β-Lactamase-Producing Priority Pathogens in Fresh Vegetables: A Trojan Horse Strategy Favoring Human Colonization? MSystems 2021, 6, e01125-20. [Google Scholar] [CrossRef]
- Niyomdecha, N.; Mungkornkaew, N.; Samosornsuk, W. Serotypes and antimicrobial resistance of Salmonella enterica isolated from pork, chicken meat and lettuce, Bangkok and Central Thailand. Southeast Asian J. Trop. Med. Public Health. 2016, 47, 31–39. [Google Scholar]
- Pintor-Cora, A.; Álvaro-Llorente, L.; Otero, A.; Rodríguez-Calleja, J.M.; Santos, J.A.; Allende, A.; Burgess, C. Extended-Spectrum Beta-Lactamase-Producing Enterobacteriaceae in Fresh Produce. Foods. 2021, 10, 2609. [Google Scholar] [CrossRef]
- Somda, N.S.; Bonkoungou, I.J.O.; Sambe-Ba, B.; Drabo, M.S.; Wane, A.A.; Sawadogo-Lingani, H.; Savadogo, A. Diversity and antimicrobial drug resistance of non-typhoidal Salmonella serotypes isolated in lettuce, irrigation water and clinical samples in Burkina Faso. J. Agric. Food Res. 2021, 5, 100167. [Google Scholar] [CrossRef]
- Abakpa, G.O.; Umoh, V.J.; Kamaruzaman, S.; Ibekwe, M. Fingerprints of resistant Escherichia coli O157:H7 from vegetables and environmental samples. J. Sci. Food Agric. 2017, 98, 80–86. [Google Scholar] [CrossRef]
- Al-Kharousi, Z.S.; Guizani, N.; Al-Sadi, A.M.; Al-Bulushi, I.M. Tetracycline Resistance in Enterococci and Escherichia coli Isolated from Fresh Produce and Why it Matters. Int. J. Food Microbiol. 2021, 10, 359–370. [Google Scholar] [CrossRef]
- Ananchaipattana, C.; Hosotani, Y.; Kawasaki, S.; Bari, M.L.; Yamaguchi, K.A.; Inatsu, Y. Serotyping, RAPD grouping and antibiotic susceptibility testing of Salmonella enterica isolated from retail foods in Thailand. Food Sci. Technol. Res. 2014, 20, 905–913. [Google Scholar] [CrossRef]
- Campos, J.; Mourão, J.; Pestana, N.; Peixe, L.; Novais, C.; Antunes, P. Microbiological quality of ready-to-eat salads: An underestimated vehicle of bacteria and clinically relevant antibiotic resistance genes. Int. J. Food Microbiol. 2013, 166, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Chanseyha, C.; Sadiq, M.B.; Cho, T.Z.A.; Anal, A.K. Prevalence and Analysis of Antibiotic Resistant Genes in Escherichia coli and Salmonella Isolates from Green Leaf Lettuce. Chiang Mai J. Sci. 2018, 45, 1–13. [Google Scholar]
- Colosi, I.A.; Baciu, A.M.; Opriș, R.V.; Peca, L.; Gudat, T.; Simon, L.M.; Colosi, H.A.; Costache, C. Prevalence of ESBL, AMPc and carbapenemase-producing enterobacterales isolated from raw vegetables retailed in Romania. Foods. 2020, 9, 1726. [Google Scholar] [CrossRef]
- Gómez-Aldapa, C.A.; Cerna-Cortes, J.F.; Rangel-Vargas, E.; Torres-Vitela, M.R.; Villarruel-López, A.; Gutiérrez-Alcántara, E.J.; Castro-Rosas, J. Presence of Multidrug-Resistant Shiga Toxin-Producing Escherichia coli, Enteropathogenic E. coli and Enterotoxigenic E. coli, on Raw Nopalitos (Opuntia ficus-indica L.) and in Nopalitos Salads from Local Retail Markets in Mexico. Foodborne Pathog. Dis. 2016, 13, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Güran, M.; Şanlıtürk, G.; Hadid, Z.; Hadid, K.; Aksay, B.; Rahhal, M.; Ayman, N.; Alhrasshir, J.; Akçay, N.İ. Comparative analysis of Escherichia Coli contamination on fresh produce at the market: Human handling is a significant parameter of contamination. Prog. Nutr. 2021, 23. [Google Scholar] [CrossRef]
- Kabir, A.; Das, A.K.; Kabir, M.S. Incidence of antibiotic resistant pathogenic bacteria in vegetable items sold by local and super shops in Dhaka city. Stamford J. Microbiol. 2014, 4, 13. [Google Scholar] [CrossRef]
- Kholdi, S.; Motamedifar, M.; Fani, F.; Mohebi, S.; Bazargani, A. Virulence factors, serogroups, and antibiotic resistance of Shiga-toxin producing Escherichia coli from raw beef, chicken meat, and vegetables in Southwest Iran. Iran. J. Vet. Res. 2021, 22, 180–187. [Google Scholar] [CrossRef]
- Kurittu, P.; Khakipoor, B.; Arnio, M.; Nykasenoja, S.; Brouwer, M.; Myllyniemi, A.L.; Vatunen, E.; Heikinheimo, A. Plasmid-borne and chromosomal ESBL/AmpC genes in Esherichia coli and Klebsiella pneumonia in global food products. Front. Microbiol. 2021, 12, 592291. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, M.; Luo, J.; Chen, J.; Wang, Q.; Lu, S.; Ji, H. Antimicrobial resistance of Escherichia coli isolated from retail foods in northern Xinjiang, China. Food Sci. Nutr. 2020, 8, 2035–2051. [Google Scholar] [CrossRef]
- Najwa, M.S.; Rukayadi, Y.; Ubong, A.; Loo, Y.Y.; Chang, W.S.; Lye, Y.L.; Thung, T.Y.; Aimi, S.A.; Malcolm, T.T.H.; Goh, S.G.; et al. Quantification and antibiotic susceptibility of Salmonella spp., Salmonella enteritidis and Salmonella typhimurium in raw vegetables (ulam). Int. Food Res. J. 2015, 22, 1761–1769. [Google Scholar]
- Rasheed, M.U.; Thajuddin, N.; Ahamed, P.; Teklemariam, Z.; Jamil, K. Antimicrobial drug resistance in strains of Escherichia coli isolated from food sources. Rev. Inst. Med. Trop. Sao Paulo. 2014, 56, 341–346. [Google Scholar] [CrossRef]
- Rodrigues, C.; Hauser, K.; Cahill, N.; Ligowska-Marzęta, M.; Centorotola, G.; Cornacchia, A.; Fierro, R.G.; Haenni, M.; Nielsen, E.M.; Piveteau, P.; et al. High Prevalence of Klebsiella pneumoniae in European Food Products: A Multicentric Study Comparing Culture and Molecular Detection Methods. Microbiol. Spectr. 2022, 10, 2376–2397. [Google Scholar] [CrossRef] [PubMed]
- Romyasamit, C.; Sornsenee, P.; Chimplee, S.; Yuwalaksanakun, S.; Wongprot, D.; Saengsuwan, P. Prevalence and characterization of extended-spectrum -lactamase producing Escherichia coli and Klebsiella pneumoniae isolated from raw vegetables retailed in southern Thailand. PeerJ. 2021, 9. [Google Scholar] [CrossRef] [PubMed]
- Skočková, A.; Karpíšková, R.; Koláčková, I.; Cupáková, Š. Characteristics of Escherichia coli from raw vegetables at a retail market in the Czech Republic. Int. J. Food Microbiol. 2013, 167, 196–201. [Google Scholar] [CrossRef] [PubMed]
- Zekar, F.M.; Granier, S.A.; Marault, M.; Yaici, L.; Gassilloud, B.; Manceau, C.; Touati, A.; Millemann, Y. From farms to markets: Gram-negative bacteria resistant to third-generation cephalosporins in fruits and vegetables in a region of North Africa. Front. Microbiol. 2017, 8, 1569. [Google Scholar] [CrossRef]
- Chotinantakul, K.; Woottisin, S.; Okada, S. The emergence of CTX-M-55 in ESBL-producing Escherichia coli from vegetables sold in local markets of northern Thailand. Jpn J Infect Dis. 2022, 75, 296. [Google Scholar] [CrossRef]
- Haile, A.F.; Alonso, S.; Berhe, N.; Bekele Atoma, T.; Boyaka, P.N.; Grace, D. Escherichia coli O157:H7 in Retail Lettuce (Lactuca sativa) in Addis Ababa City: Magnitude of Contamination and Antimicrobial Susceptibility Pattern. Front. Microbiol. 2021, 12, 694506. [Google Scholar] [CrossRef]
- Holvoet, K.; Sampers, I.; Callens, B.; Dewulf, J.; Uyttendaele, M. Moderate prevalence of antimicrobial resistance in escherichia coli isolates from lettuce, irrigation water, and soil. Appl. Environ. Microb. 2013, 79, 6677–6683. [Google Scholar] [CrossRef]
- Kaesbohrer, A.; Bakran-Lebl, K.; Irrgang, A.; Fischer, J.; Kämpf, P.; Schiffmann, A.; Werckenthin, C.; Busch, M.; Kreienbrock, L.; Hille, K. Diversity in prevalence and characteristics of ESBL/pAmpC producing E. coli in food in Germany. Vet. Microbiol. 2019, 233, 52–60. [Google Scholar] [CrossRef]
- Montero, L.; Irazabal, J.; Cardenas, P.; Graham, J.P.; Trueba, G. Extended-spectrum beta-lactamase producing-Escherichia coli isolated from irrigation waters and produce in Ecuador. Front. Microbiol. 2021, 12, 709418. [Google Scholar] [CrossRef]
- Mwanza, F.; Komba, E.V.G.; Kambarage, D.M. Occurrence and Determination of Antimicrobial Resistant Escherichia coli Isolates in Fish and Vegetables as Indicator Organism of Faecal Contamination in Dar es Salaam, Tanzania. Int. J. Microbiol. 2021, 2021, 6633488. [Google Scholar] [CrossRef] [PubMed]
- Priyanka, P.R.M.; Meghwanshi, K.K.; Rana, A.; Singh, A.P. Leafy greens as a potential source of multidrug-resistant diarrhoeagenic Escherichia coli and salmonella. Microbiology 2021, 167, 001059. [Google Scholar] [CrossRef] [PubMed]
- Richter, L.; du Plessis, E.M.; Duvenage, S.; Korsten, L. Occurrence, Phenotypic and Molecular Characterization of Extended-Spectrum- and AmpC- β-Lactamase Producing Enterobacteriaceae Isolated From Selected Commercial Spinach Supply Chains in South Africa. Front. Microbiol. 2020, 11, 638. [Google Scholar] [CrossRef] [PubMed]
- Shahin, K.; Bouzari, M.; Wang, R.; Yazdi, M. Prevalence and molecular characterization of multidrug-resistant Shigella species of food origins and their inactivation by specific lytic bacteriophages. Int. J. Food Microbiol. 2019, 305, 108252. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Oh, S.S.; Kim, J.; Shin, J. Extended-spectrum β-lactamase-producing Escherichia coli isolated from raw vegetables in South Korea. Sci. Rep-UK. 2020, 10, 19721. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Wang, Y.; Ma, S.; Zhang, S.; Liu, D.; Wang, Y.; Wu, C. Surveillance of antimicrobial resistance in Escherichia coli and enterococci from food products at retail in Beijing, China. Food Control. 2021, 119, 107483. [Google Scholar] [CrossRef]
- Thung, T.Y.; Mazlan, N.; Lee, E.; New, C.Y.; Tan, C.W.; Son, R.; Rinai, K.R.; Anua, S.M.; Mastor, N.N. Antimicrobial resistance profile of salmonella present in organic farming in Selangor, Malaysia. Food Res. 2020, 4, 2176–2180. [Google Scholar] [CrossRef]
- van Hoek, A.H.A.M.; Veenman, C.; van Overbeek, W.M.; Lynch, G.; de Roda Husman, A.M.; Blaak, H. Prevalence and characterization of ESBL- and AmpC-producing Enterobacteriaceae on retail vegetables. Int. J. Food Microbiol. 2015, 204, 1–8. [Google Scholar] [CrossRef]
- Zou, H.; Zheng, B.; Sun, M.; Ottoson, J.; Li, Y.; Berglund, B.; Chi, X.; Ji, X.; Li, X.; Stålsby Lundborg, C.; et al. Evaluating Dissemination Mechanisms of Antibiotic-Resistant Bacteria in Rural Environments in China by Using CTX-M-Producing Escherichia coli as an Indicator. Microb. Drug Resist. 2019, 25, 975–984. [Google Scholar] [CrossRef]
- European Medicines Agency. Categorisation of Antibiotics in the European Union. 2019. Available online: https://www.ema.europa.eu/en/documents/report/categorisation-antibiotics-european-union-answer-request-european-commission-updating-scientific_en.pdf (accessed on 8 July 2022).
- World Health Organization. Integrated Global Surveillance on ESBL-Producing E. coli Using a “One Health” Approach: Implementation and Opportunities; World Health Organization: Geneva, Switzerland, 2021.
- Grace, D. Food Safety in Low- and Middle-Income Countries. Int. J. Environ. Res. Public Health 2015, 12, 10490–10507. [Google Scholar] [CrossRef]
- Zhang, H.; Li, X.; Yang, Q.; Sun, L.; Yang, X.; Zhou, M.; Deng, R.; Bi, L. Plant Growth, Antibiotic Uptake, and Prevalence of Antibiotic Resistance in an Endophytic System of Pakchoi under Antibiotic Exposure. Int. J. Environ. Res. Public Health 2017, 14, 1336. [Google Scholar] [CrossRef]
- Borges, M.C.; Stein, C.; Colpani, V.; Barker, T.H.; Munn, Z.; Falavigna, M. How are systematic reviews of prevalence conducted? A methodological study. BMC Med. Res. Methodol. 2020, 20, 96. [Google Scholar] [CrossRef]
- Chandrasekaran, S.; Jiang, S.C. A dose response model for quantifying the infection risk of antibiotic-resistant bacteria. Sci. Rep. 2019, 9, 17093. [Google Scholar] [CrossRef] [PubMed]
- Oniciuc, E.A.; Likotrafiti, E.; Alvarez-Molina, A.; Prieto, M.; Santos, J.A.; Alvarez-Ordóñez, A. The present and future of whole genome sequencing (WGS) and whole metagenome sequencing (WMS) for surveillance of antimicrobial resistant microorganisms and antimicrobial resistance genes across the food chain. Genes 2018, 9, 268. [Google Scholar] [CrossRef]
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.K.; Colquhoun, H.; Levac, D.; Moher, D.; Peters, M.D.J.; Horsley, T.; Weeks, L.; et al. PRISMA extension for scoping reviews (PRISMA-ScR): Checklist and explanation. Ann. Intern. Med. 2018, 169, 7. [Google Scholar] [CrossRef] [PubMed]
- World Organization for Animal Health (WOAH). Criteria Used for Categorisation List of Antimicrobial Agents. Oie List of Antimicrobial Agents of Veterinary Importance. 2018. Available online: https://www.woah.org/app/uploads/2021/03/a-oie-list-antimicrobials-may2018.pdf (accessed on 19 April 2022).
- van der Bij, A.K.; van Dik, K.; Muilwijk, J.; Thijsen, S.F.T.; Notermans, D.W.; de Greeff, S.; van de Sande-Bruinsma, N.; on behalf of the ISIS-AR study group. Clinical breakpoint changes and their impact on surveillance of antimicrobial resistance in Escherichia coli causing bacteraemia. Clin. Microbiol. Infect. 2012, 18, E466–E472. [Google Scholar] [CrossRef] [PubMed]
- Barendregt, J.J.; Doi, S.A.; Lee, Y.Y.; Norman, R.E.; Vos, T. Meta-analysis of prevalence. J. Epidemiol. Community Health 2013, 67, 974–978. [Google Scholar] [CrossRef]
| Risk of Bias | Number of Studies | Citations |
|---|---|---|
| Very High | 9 | Aabed et al. 2021 [34]; Freitag et al. 2018 [35]; Hartentyo et al. 2020 [36]; Janalikova et al. 2018 [37]; Kim et al. 2015 [38]; Lopes et al. 2021 [39]; Niyomdecha et al. 2016 [40]; Pintor-Cora et al. 2021 [41]; Somda et al. 2021 [42] |
| High | 18 | Abakpa et al. 2017 [43]; Al-Kharousi et al. 2021 [44]; Ananchaipattana et al. 2014 [45]; Campos et al. 2013 [46]; Chanseyha et al. 2018 [47]; Colosi et al. 2020 [48]; Gomez-Aldapa et al. 2016 [49]; Güran et al. 2021 [50]; Kabir et al. 2014 [51]; Kholdi et al. 2021 [52]; Kurittu et al. 2021 [53]; Li et al. 2020 [54]; Najwa et al. 2015 [55]; Rasheed et al. 2014 [56]; Rodrigues et al. 2022 [57]; Romyasamit et al. 2021 [58]; Skočková et al. 2013 [59]; Zekar et al. 2017 [60] |
| Moderate | 15 | Chotinantakul et al. 2022 [61]; Haile et al. 2021 [62]; Holvoet et al. 2013 [63]; Kaesbohrer et al. 2019 [64]; Montero et al. 2021 [65]; Mwanza et al. 2021 [66]; Priyanka et al. 2021 [67]; Reddy et al. 2016 [14]; Richter et al. 2020 [68]; Shahin et al. 2019 [69]; Song et al. 2020 [70]; Sun et al. 2021 [71]; Thung et al. 2020 [72]; Van Hoeck et al. 2015 [73]; Zou et al. 2019 [74] |
| Low | 0 | None |
| Total studies | 42 | - |
| Sub-Group | No. of Samples Tested a | No. of Resistant Samples b | Prevalence (%) | Prevalence CI (%) | p-Value | No. of Prevalence Estimates | I2 (%) | I2 CI (%) |
|---|---|---|---|---|---|---|---|---|
| Pathogen Prevalence | ||||||||
| Total Enterobacteriaceae | 149,751 | 950 | 11.45 | 7.96, 15.45 | <0.0001 | 44 | 98.5 | 98.3, 98.7 |
| Pre-Harvest Samples | 1442 | 186 | 13.86 | 5.78, 24.53 | <0.0001 | 8 | 96.0 | 93.9, 97.3 |
| Post-Harvest Samples | 148,309 | 764 | 10.85 | 7.30, 14.97 | <0.0001 | 36 | 98.4 | 98.2, 98.6 |
| AMR Prevalence | ||||||||
| Total AMR | 149,298 | 404 | 4.75 | 2.92, 6.94 | <0.0001 | 45 | 97.3 | 96.8, 97.7 |
| 3GC Resistance | ||||||||
| All Stages | 7841 | 257 | 3.75 | 2.13, 5.74 | <0.0001 | 27 | 93.2 | 91.2, 94.7 |
| Pre-Harvest Samples | 569 | 33 | 4.45 | 1.44, 8.71 | 0.0031 | 5 | 74.9 | 38.2, 89.8 |
| Post-Harvest Samples | 7155 | 213 | 3.44 | 1.64, 5.76 | <0.0001 | 21 | 94.0 | 92.1, 95.5 |
| TET Resistance | ||||||||
| All Stages | 146,037 | 259 | 4.63 | 2.57, 7.18 | <0.0001 | 30 | 97.2 | 96.7, 97.7 |
| Pre-Harvest Samples | 1442 | 64 | 4.55 | 1.83, 8.24 | <0.0001 | 8 | 86.9 | 76.3, 92.7 |
| Post-Harvest Samples | 144,595 | 195 | 4.72 | 2.11, 8.16 | <0.0001 | 22 | 97.4 | 96.7, 97.9 |
| Region | 3GC Resistance | TET Resistance | Combined AMR | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of Samples Tested a | No. of Resistant Samples b | Prevalence (CI) | p-Value | No. of Prevalence Estimates | No. of Samples Tested a | No. of Resistant Samples b | Prevalence (CI) | p-Value | No. of Prevalence Estimates | Prevalence (CI) | |
| Africa | 842 | 81 | 0.0659 (0.0241; 0.1240) | <0.0001 | 6 | 949 | 33 | 0.0431 (0.0087; 0.0981) | <0.0001 | 5 | 0.0544 (0.0254; 0.0924) |
| Americas | * | - | - | - | - | 139,638 | 33 | 0.0248 (0.0000; 0.0882) | <0.0001 | 4 | 0.0420 (0.0074; 0.0988) |
| Eastern Med. | * | - | - | - | - | * | - | - | - | - | 0.0146 (0.0036, 0.0313) |
| Europe | 1977 | 20 | 0.0184 (0.0000; 0.0602) | <0.0001 | 5 | 807 | 32 | 0.0628 (0.0068; 0.1572) | <0.0001 | 6 | 0.0362 (0.0077; 0.0803) |
| South East Asia | 1179 | 33 | 0.0404 (0.0040; 0.1013) | 0.0293 | 4 | 1273 | 43 | 0.0509 (0.0091; 0.1177) | <0.0001 | 5 | 0.0463 (0.0163; 0.0875) |
| Western Pacific | 2018 | 43 | 0.0278 (0.0071; 0.0586) | <0.0001 | 7 | 2281 | 92 | 0.0623 (0.0079; 0.1550) | <0.0001 | 7 | 0.0440 (0.0149; 0.0852) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brunn, A.A.; Roustit, M.; Kadri-Alabi, Z.; Guardabassi, L.; Waage, J. A Meta-Analysis to Estimate Prevalence of Resistance to Tetracyclines and Third Generation Cephalosporins in Enterobacteriaceae Isolated from Food Crops. Antibiotics 2022, 11, 1424. https://doi.org/10.3390/antibiotics11101424
Brunn AA, Roustit M, Kadri-Alabi Z, Guardabassi L, Waage J. A Meta-Analysis to Estimate Prevalence of Resistance to Tetracyclines and Third Generation Cephalosporins in Enterobacteriaceae Isolated from Food Crops. Antibiotics. 2022; 11(10):1424. https://doi.org/10.3390/antibiotics11101424
Chicago/Turabian StyleBrunn, Ariel A., Manon Roustit, Zaharat Kadri-Alabi, Luca Guardabassi, and Jeff Waage. 2022. "A Meta-Analysis to Estimate Prevalence of Resistance to Tetracyclines and Third Generation Cephalosporins in Enterobacteriaceae Isolated from Food Crops" Antibiotics 11, no. 10: 1424. https://doi.org/10.3390/antibiotics11101424
APA StyleBrunn, A. A., Roustit, M., Kadri-Alabi, Z., Guardabassi, L., & Waage, J. (2022). A Meta-Analysis to Estimate Prevalence of Resistance to Tetracyclines and Third Generation Cephalosporins in Enterobacteriaceae Isolated from Food Crops. Antibiotics, 11(10), 1424. https://doi.org/10.3390/antibiotics11101424






