Comparative Molecular Mechanisms of Biosynthesis of Naringenin and Related Chalcones in Actinobacteria and Plants: Relevance for the Obtention of Potent Bioactive Metabolites
Abstract
1. Introduction: Flavonoids and Chalcones, Naringenin and Related Compounds in Nature
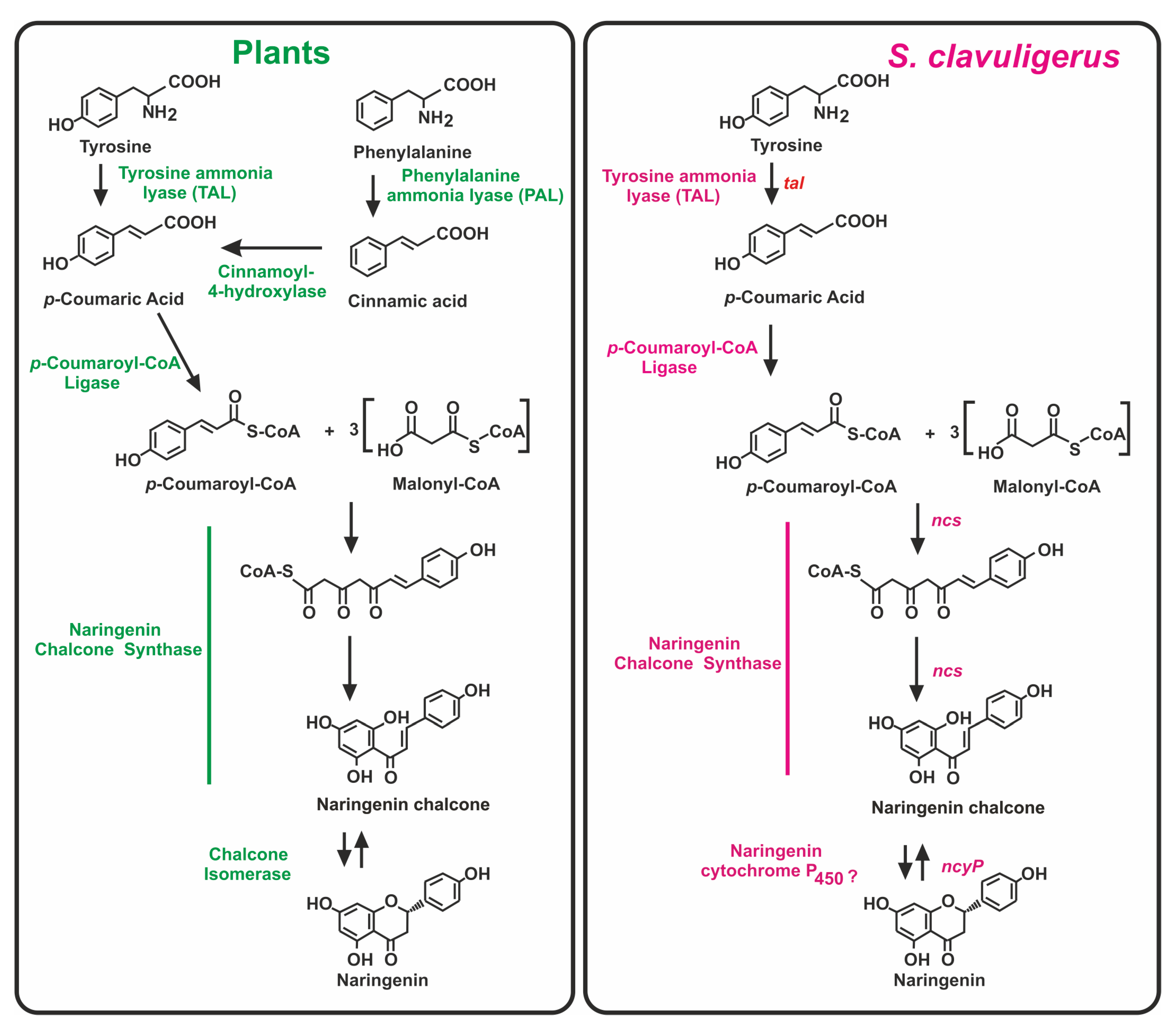
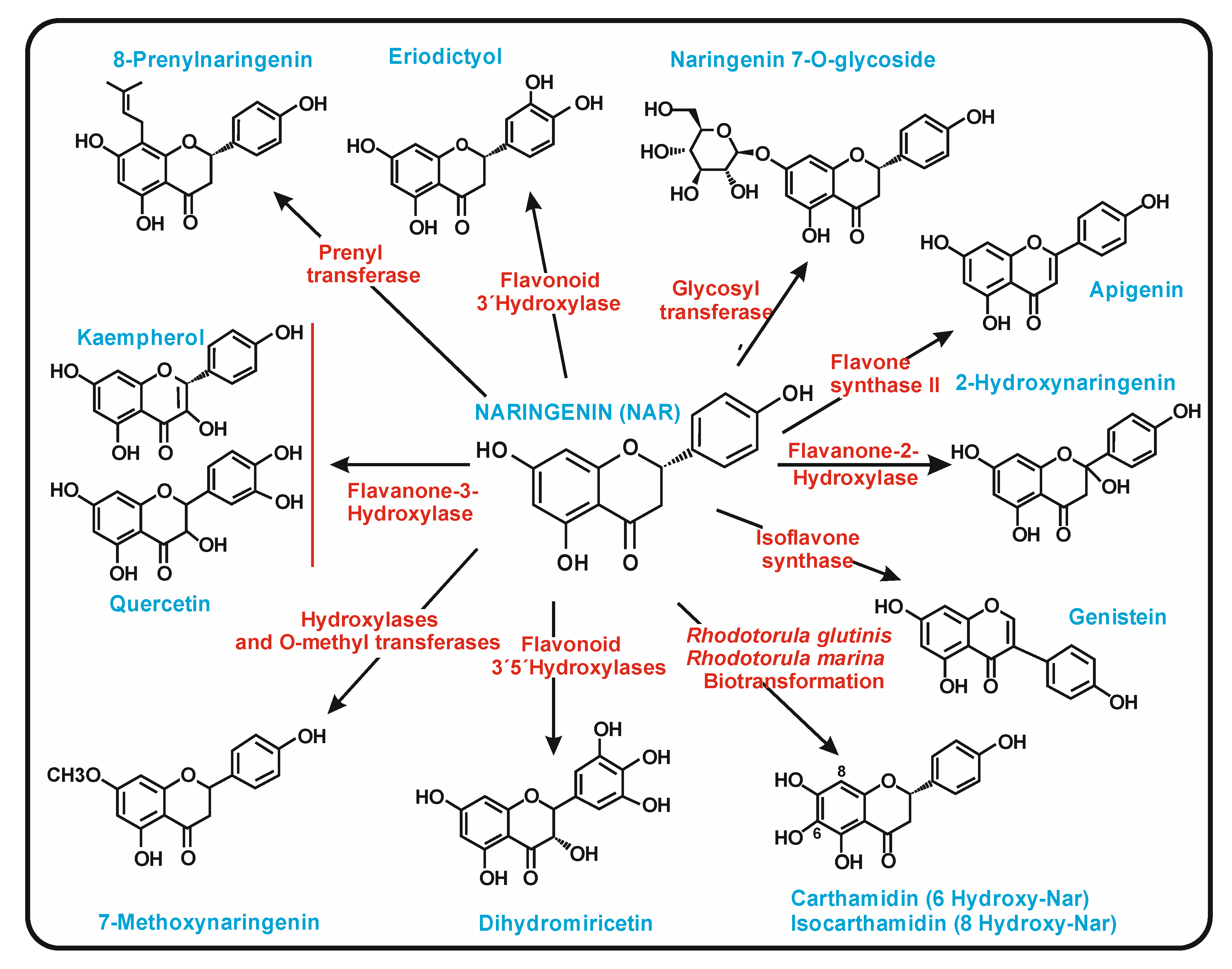
2. Biosynthesis of (2S)-Naringenin in Plants
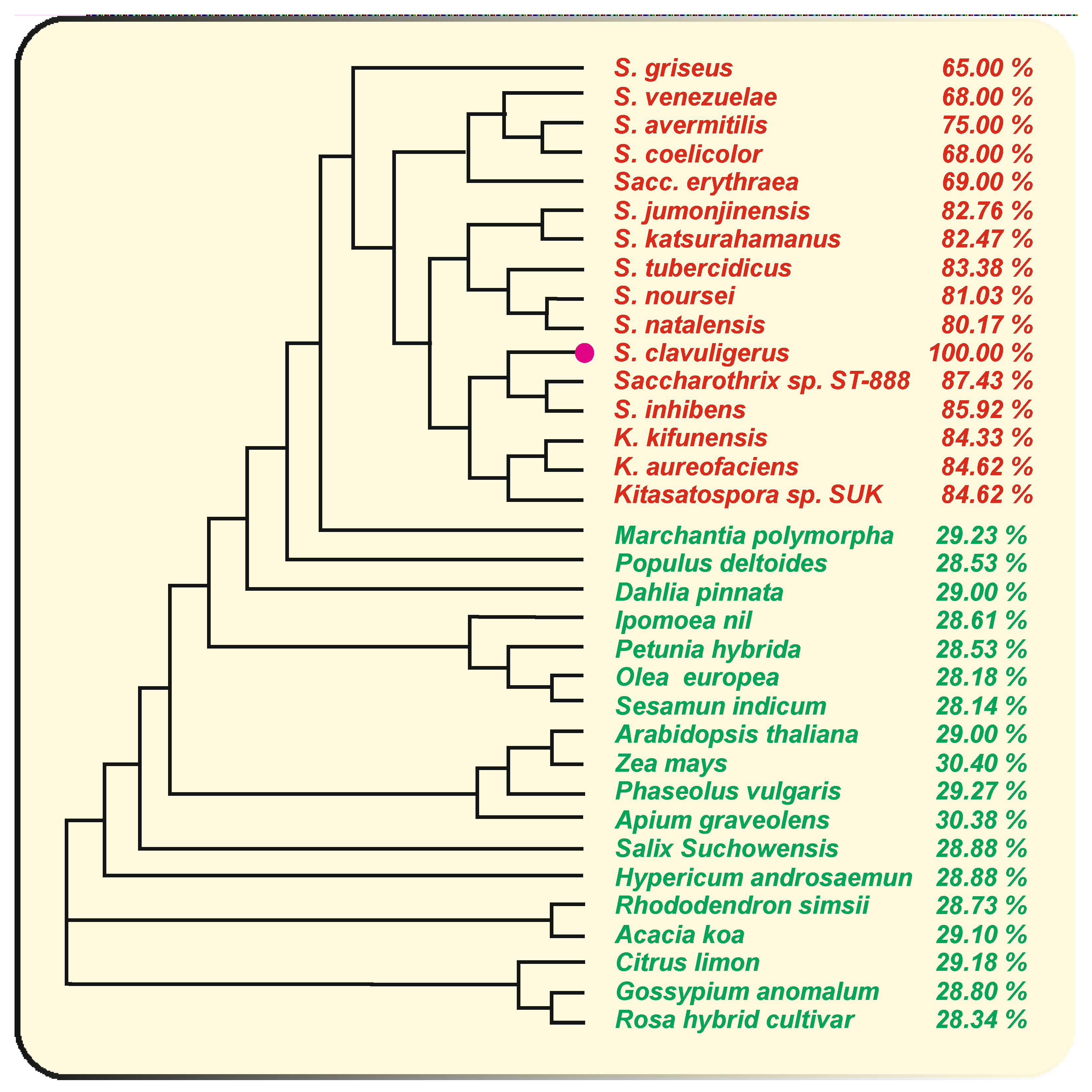
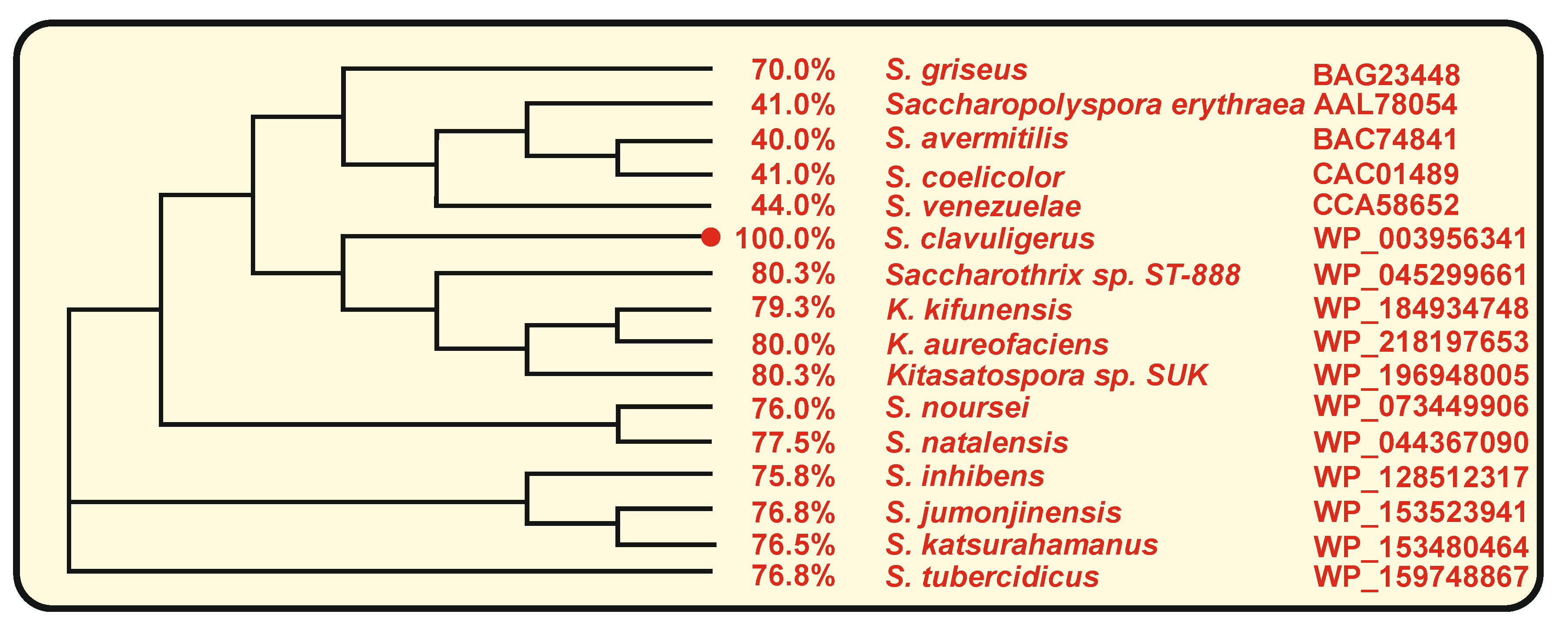
3. The Naringenin Chalcone Synthase of S. clavuligerus: Comparison with Other Bacterial Chalcone Synthases
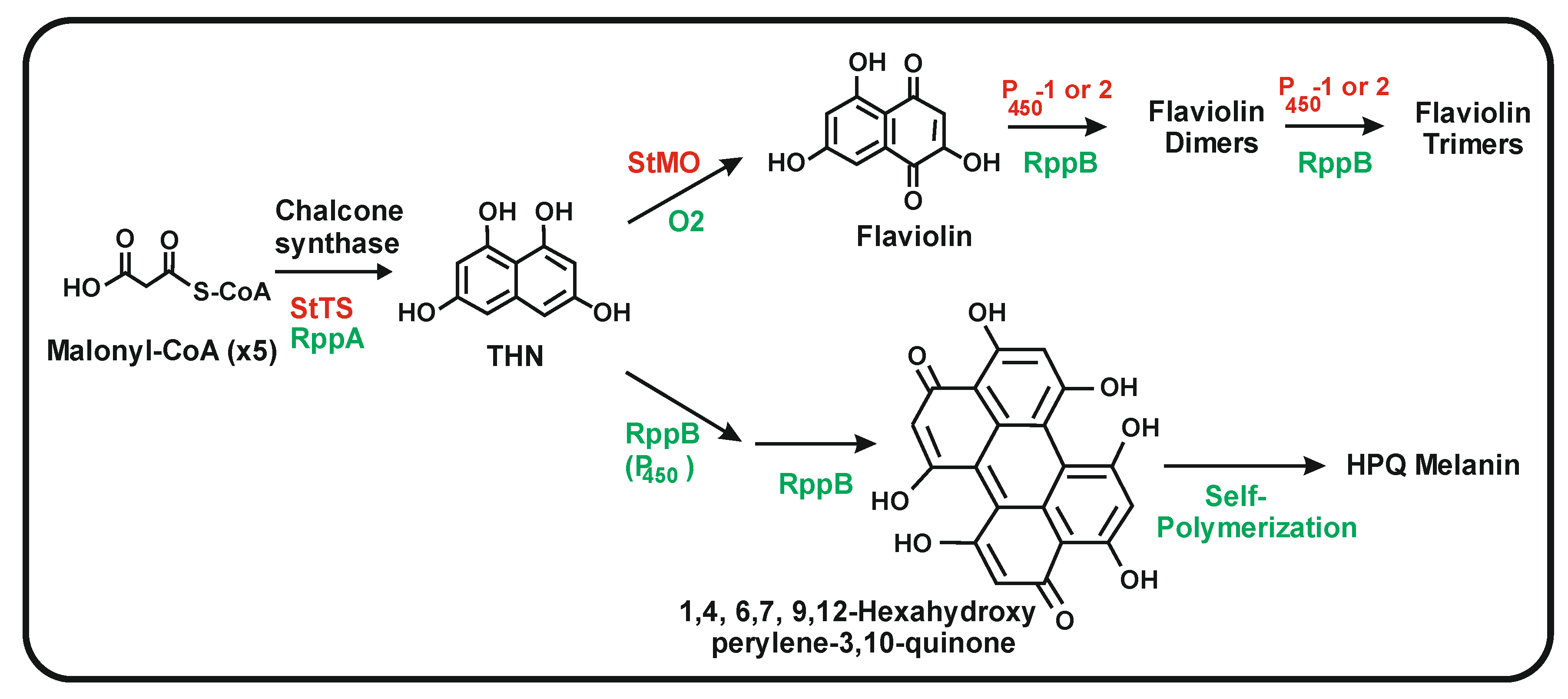
3.1. Bacterial Chalcone Synthases That Use Aromatic or Aliphatic Starter Units
3.2. Role of the P450 Monooxygenase (NcyP) Encoded by a Gene Linked to ncs
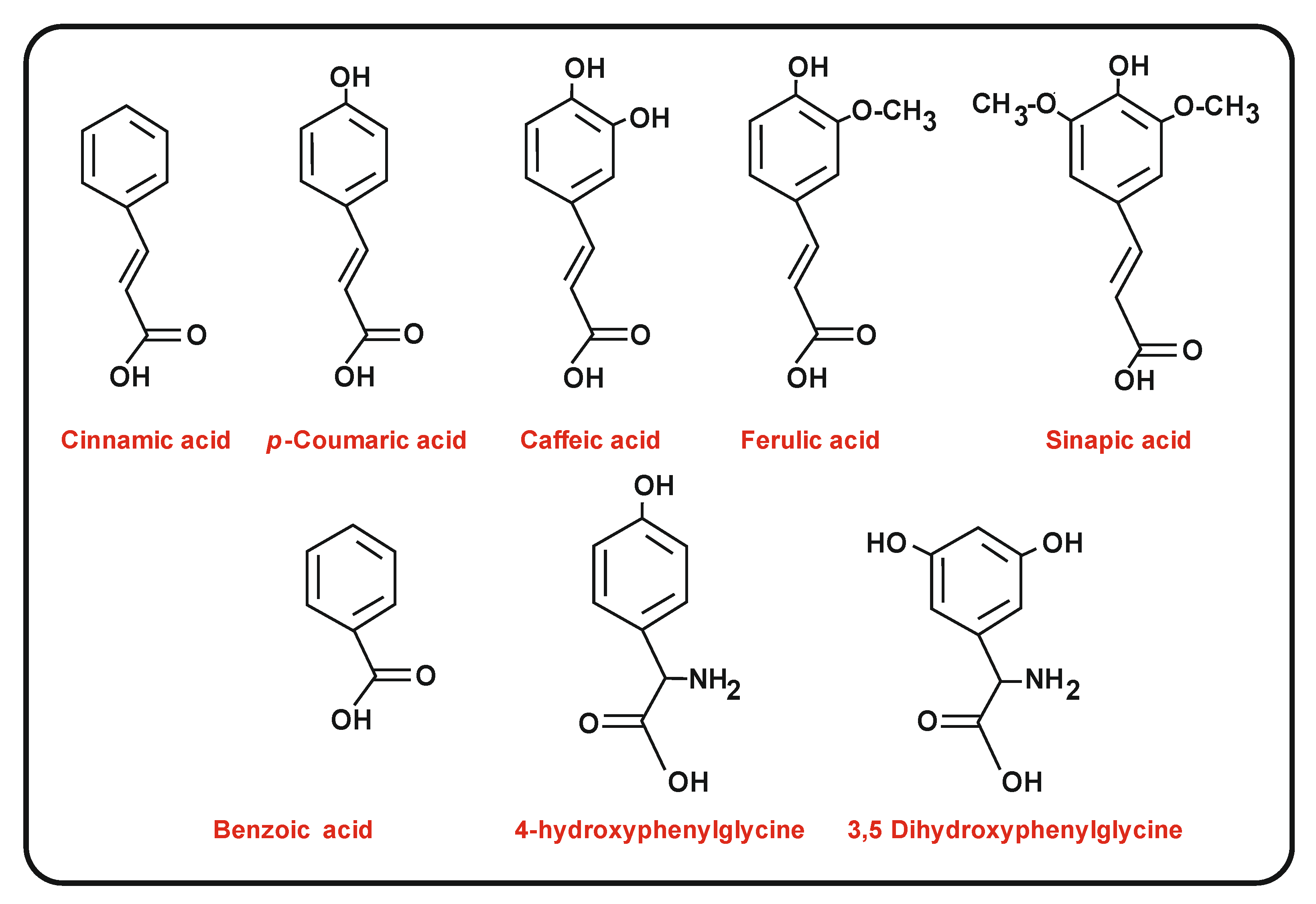
4. Biosynthesis of p-Coumaric Acid and Other Related Starter Molecules: Role of Ammonia Lyases
4.1. Biosynthesis of p-Coumaric Acid in Plants and Yeasts
4.2. Bacterial Amino Acid Ammonia Lyases
4.3. The Aromatic Ammonia Lyases and the Aminomutases Contain a Methylidene Imidazol-5-one (MIO) Prostetic Group
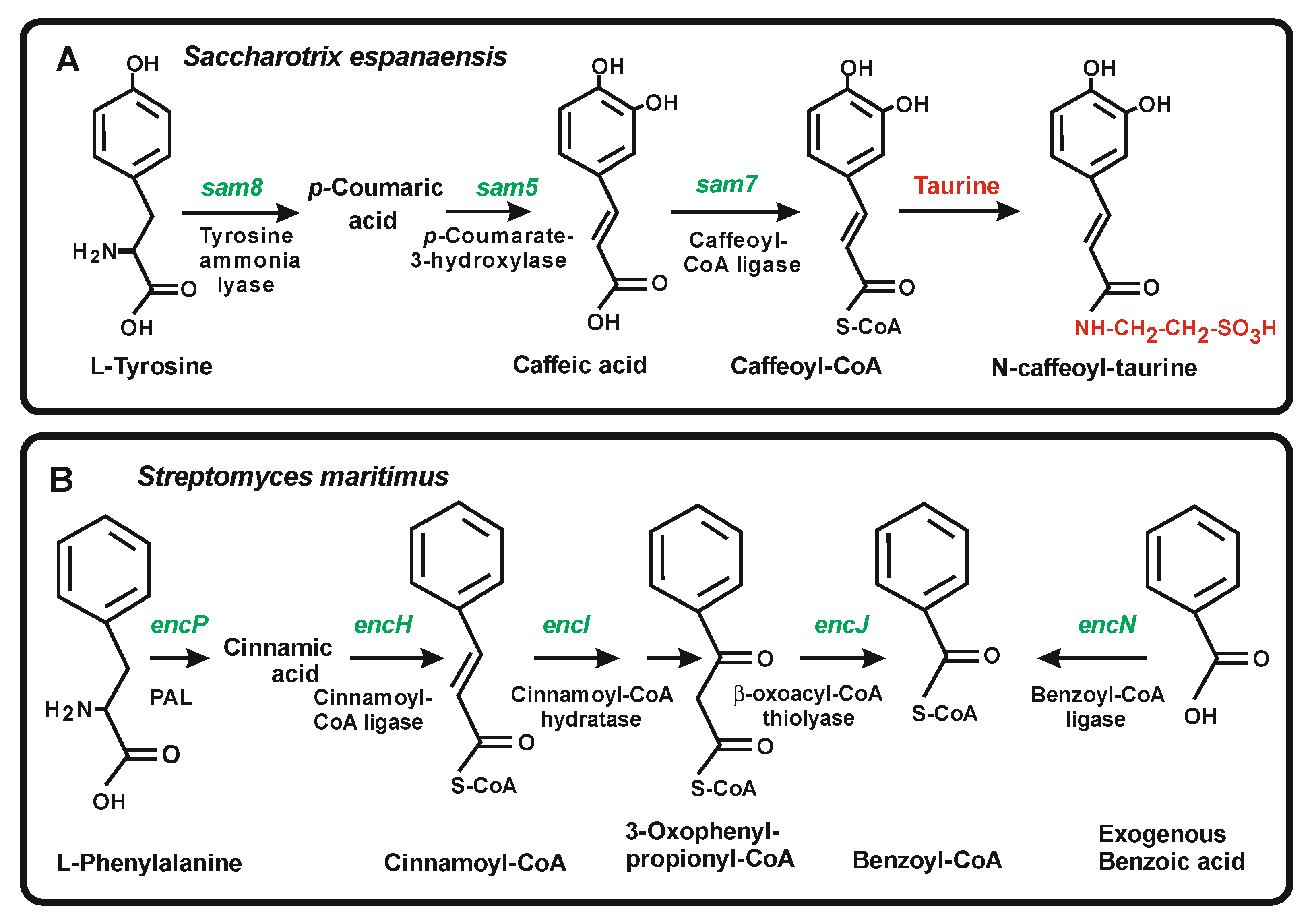
4.4. Enzymes for the Conversion of Trans-Cinnamic Acid and p-Coumaric Acid into Caffeic Acid and Ferulic Acid
5. Activation of p-Coumaric Acid and Related Chalcone Precursors by aryl-CoA Ligases
5.1. Substrate Specificity of p-Coumaroyl-CoA Ligases in Plants and Filamentous Fungi
5.2. Activation of Benzoic Acid, a Rare Aromatic Precursor in the Biosynthesis of the Polyketide Enterocins in Streptomyces Maritimus
6. Biosynthesis of 4-Hydroxyphenylglycine and (S) 3,5 Dihydroxyphenylglycine, Related to Aromatic Starter Units of Chalcones
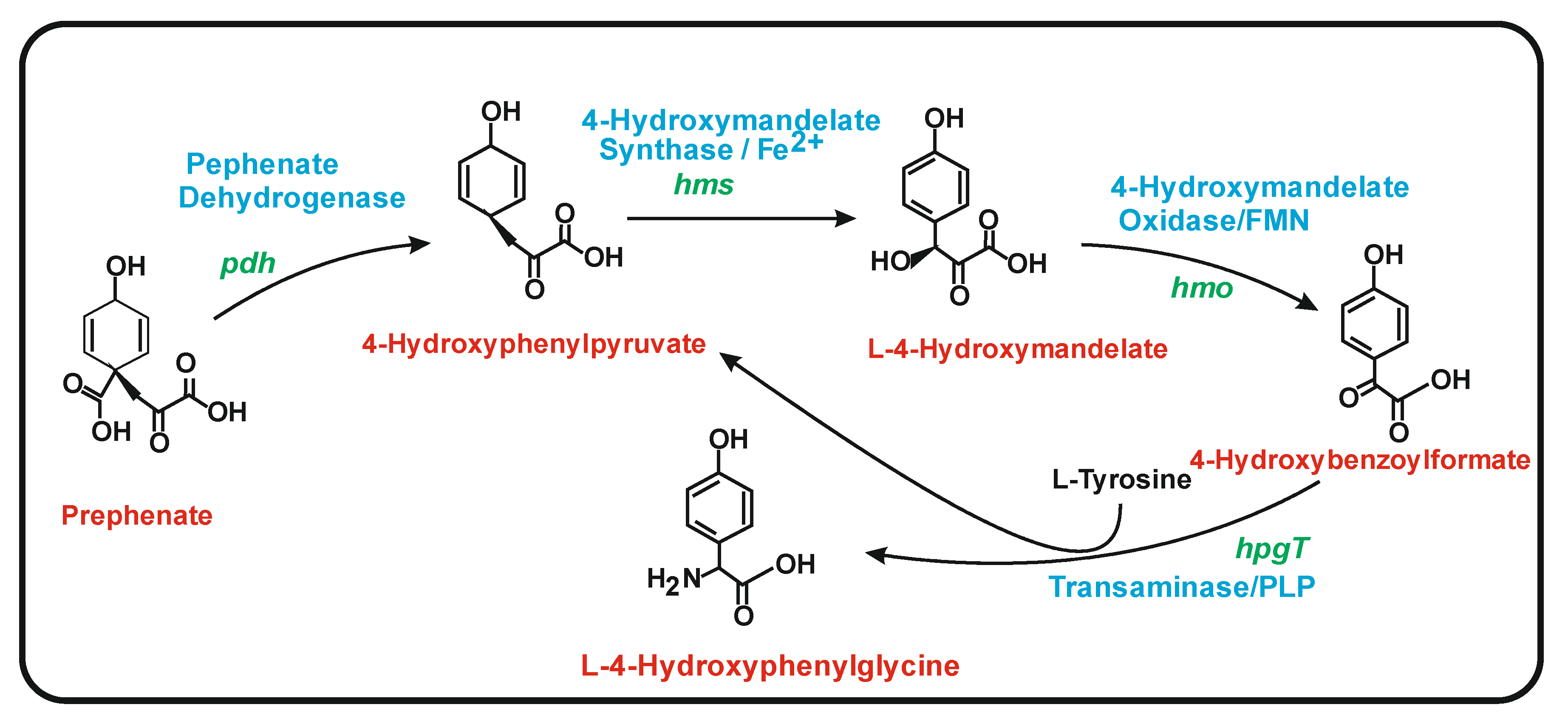
6.1. Biosynthesis of 4-Hydroxyphenylglycine (HPG)
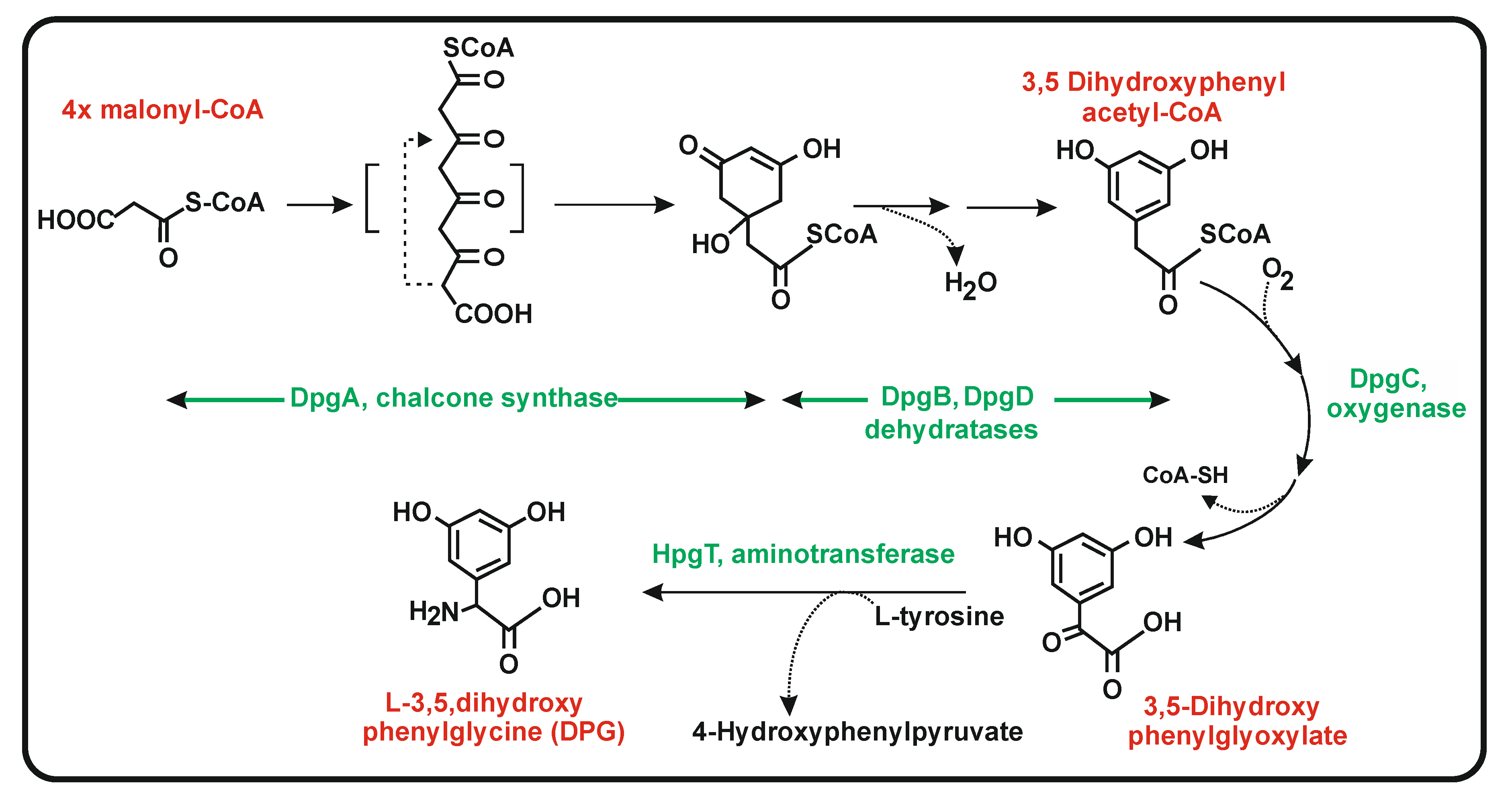
6.2. Biosynthesis of 3,5-Dihydroxyphenylglycine (DPG)
7. Heterologous Production of Naringenin in Yeasts: Biotechnological Applications
8. Conclusions and Future Outlook
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CoA | coenzyme A |
| 4CL | p-coumaroyl-CoA ligases |
| ACP | acyl carrier protein |
| CHS | chalcone synthase |
| DAHP | 3-deoxy-D-arabinoheptulosonate 7-phosphate synthase |
| DPG | 3,5 dihydroxyphenylglycine |
| HPG | 4-hydroxyphenylglycine |
| HPQ | 1,4,6,7,9,12-hexahydroxyperylene-3,10-quinone |
| O-MT | O-methyltransferase |
| PAL | phenylalanine ammonia lyase |
| PKS | polyketide synthase |
| TAL | tyrosine ammonia lyase |
| THN | 1,3,6,8-tetrahydroxynaphtalene |
| Sac | genus Saccharothrix |
| Sac | genus Saccharopolyspora |
References
- Forkmann, G.; Heller, W. Biosynthesis of flavonoids. In Comprehensive Natural Products Chemistry; Barton, D., Nakanishi, K., Eds.; Elsevier Sci. Ltd.: Oxford, UK, 1999; pp. 713–748. [Google Scholar]
- Dixon, R.A. Phytoestrogens. Annu. Rev. Plant Biol. 2004, 55, 225–261. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef]
- Tong, Y.; Lyu, Y.; Xu, S.; Zhang, L.; Zhou, J. Optimum chalcone synthase for flavonoid biosynthesis in microorganisms. Crit. Rev. Biotechnol. 2021, 41, 1194–1208. [Google Scholar] [CrossRef] [PubMed]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as Antioxidants in Plants: Location and Functional Significance. Plant Sci. Int. J. Exp. Plant Biol. 2012, 196, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.A.; Grayer, R.J. Anthocyanins and other flavonoids. Nat. Prod. Rep. 2004, 21, 539–573. [Google Scholar] [CrossRef] [PubMed]
- Fowler, Z.L.; Koffas, M.A.G. Biosynthesis and biotechnological production of flavanones: Current state and perspectives. Appl. Microbiol. Biotechnol. 2009, 83, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Cavia-Saiz, M.; Busto, M.D.; Pilar-Izquierdo, M.C.; Ortega, N.; Pérez-Mateos, M. Muñiz properties, radical scavenging activity and biomolecule protection capacity of flavonoid naringenin and its glycoside naringin: A comparative study. J. Sci. Food Agric. 2010, 90, 1238–1244. [Google Scholar] [CrossRef] [PubMed]
- Jagetia, A.; Jagetia, G.C.; Jha, S. Naringenin, a grapefruit flavanone protects V79 cells agains the bleomycin-induced genotoxicity and decline in survival. J. Appl. Toxicol. 2007, 27, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhou, T.; Du, G.; Zhou, J.; Chen, J. Modular optimization of heterologous pathways for the novo synthesis of (2S)-naringenin in Escherichia coli. PLoS ONE 2014, 9, e101492. [Google Scholar] [CrossRef]
- D’Amore, A.; Gradogna, A.; Palombi, F.; Minicozzi, V.; Ceccarelli, M.; Carpaneto, A.; Filippini, A. The Discovery of Naringenin as Endolysosomal Two-Pore Channel Inhibitor and Its Emerging Role in SARS-CoV-2 Infection. Cells 2021, 10, 1130. [Google Scholar] [CrossRef]
- Salehi, B.; Fokou, P.V.T.; Sharifi-Rad, M.; Zucca, P.; Pezzani, R.; Martins, N.; Sharifi-Rad, J. The Therapeutic Potential of Naringenin: A Review of Clinical Trials. Pharmaceuticals 2019, 12, 11. [Google Scholar] [CrossRef] [PubMed]
- Heller, W.; Hahlbrock, K. Highly purified “flavanone synthase” from parsley catalyzes the formation of naringenin chalcone. Arch. Biochem. Biophys. 1980, 200, 617–619. [Google Scholar] [CrossRef]
- Burbulis, I.E.; Winkel-Shirley, B. Interactions among enzymes of the Arabidopsis flavonoid biosynthetic pathway. Proc. Natl. Acad. Sci. USA 1999, 96, 12929–12934. [Google Scholar] [CrossRef] [PubMed]
- Yun, C.S.; Yamamoto, T.; Nozawa, A.; Tozawa, Y. Expression of parsley flavone synthase I establishes the flavone biosynthetic pathway in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2008, 72, 968–973. [Google Scholar] [CrossRef] [PubMed]
- Austin, M.B.; Noel, J.P. The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 2003, 20, 79–110. [Google Scholar] [CrossRef]
- Imaizumi, R.; Mameda, R.; Takeshita, K.; Kubo, H.; Sakai, N.; Nakata, S.; Takahashi, S.; Kataoka, K.; Yamamoto, M.; Nakayama, T.; et al. Crystal structure of chalcone synthase, a key enzyme for isoflavonoid biosynthesis in soybean. Proteins 2020, 89, 126–131. [Google Scholar] [CrossRef]
- Abe, I.; Morita, H. Structure and function of the chalcone synthase superfamily of plant type III polyketide synthases. Nat. Prod. Rep. 2010, 27, 809–838. [Google Scholar] [CrossRef]
- Schröder, J. Probing plant polyketide biosynthesis. J. Nat. Struct. Biol. 1999, 6, 714–716. [Google Scholar] [CrossRef]
- Moore, B.S.; Hertweck, C. Biosynthesis and attachment of novel bacterial polyketide synthase starter units. Nat. Prod. Rep. 2002, 19, 70–99. [Google Scholar]
- Álvarez-Álvarez, R.; Botas, A.; Albillos, S.M.; Rumbero, A.; Martín, J.F.; Liras, P. Molecular genetics of naringenin biosynthesis, a typical plant secondary metabolite produced by Streptomyces clavuligerus. Microb. Cell Factories 2015, 14, 178. [Google Scholar] [CrossRef]
- Liras, P.; Martín, J.F. Streptomyces clavuligerus: The Omics Era. J. Ind. Microbiol. Biotechnol. 2021, in press. [Google Scholar] [CrossRef]
- Shaikh, A.A.; Nothias, L.-F.; Srivastava, S.K.; Dorrestein, P.C.; Tahlan, K. Specialized Metabolites from Ribosome Engineered Strains of Streptomyces clavuligerus. Metabolites 2021, 11, 239. [Google Scholar] [CrossRef]
- Wang, Y.C.; Chen, S.M.; Yu, O. Metabolic engineering of flavonoid in plants and microorganisms. Appl. Microbiol. Biotechnol. 2011, 91, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Pandith, S.A.; Ramazan, S.; Khan, M.I.; Reshi, Z.A.; Shah, M.A. Chalcone synthases (CHSs): The symbolic type III polyketide synthases. Planta 2019, 251, 15. [Google Scholar] [CrossRef] [PubMed]
- Waki, T.; Mameda, R.; Nakano, T.; Yamada, S.; Terashita, M.; Ito, K.; Tenma, N.; Li, Y.; Fujino, N.; Uno, K.; et al. A conserved strategy of chalcone isomerase-like protein to rectify promiscuous chalcone synthase specificity. Nat. Commun. 2020, 11, 870. [Google Scholar] [CrossRef]
- Jez, J.M.; Bowman, M.E.; Noel, J.P. Role of hydrogen bonds in the reaction mechanism of chalcone isomerase. Biochemistry 2002, 41, 5168–5176. [Google Scholar] [CrossRef] [PubMed]
- Ni, R.; Zhu, T.T.; Zhang, X.S.; Wang, P.Y.; Sun, C.J.; Qiao, Y.N.; Lou, H.X.; Cheng, A.X. Identification and evolutionary analysis of chalcone isomerase-fold proteins in ferns. J. Exp. Bot. 2020, 71, 290–304. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Shen, H.; Xu, H.; Tang, X.; Tang, M.; Ju, Z.; Yi, Y. Chalcone isomerase a key enzyme for anthocyanin biosynthesis in Ophiorrhiza japonica. Front. Plant Sci. 2019, 10, 865. [Google Scholar] [CrossRef]
- Muir, S.R.; Collins, G.J.; Robinson, S.; Hughes, S.; Bovy, A.; de Vos, C.H.R.; van Tunen, A.J.; Verhoeyen, M.E. Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels offlavonols. Nat. Biotechnol. 2001, 19, 470–474. [Google Scholar] [CrossRef]
- Moore, B.S.; Hertweck, C.; Hopke, J.N.; Izumikawa, M.; Kalaizis, J.A.; Nilsen, G.; O’Hare, T.; Piel, J.; Shipley, P.R.; Xiang, L.; et al. Plant-like biosynthetic pathways in bacteria: From benzoic acid to chalcone. J. Nat. Prod. 2002, 65, 1956–1962. [Google Scholar] [CrossRef]
- Xiang, L.; Moore, B.S. Characterization of benzoyl coenzyme A biosynthesis genes in the enterocin-producing bacterium Streptomyces maritimus. J. Bacteriol. 2003, 185, 399–404. [Google Scholar] [CrossRef] [PubMed]
- Berner, M.; Krug, D.; Bihlmaier, C.; Vente, A.; Müller, R.; Bechthold, A. Genes and enzymes involved in caffeic acid biosynthesis in the actinomycete Saccharothrix espanaensis. J. Bacteriol. 2006, 188, 2666–2673. [Google Scholar] [CrossRef] [PubMed]
- Ueda, K.; Kim, K.M.; Beppu, T.M.; Horinoushi, S. Overexpression of a gene cluster encoding a chalcone synthase-like protein confers red-brown pigment production in Streptomyces griseus. J. Antibiot. 1995, 48, 638–646. [Google Scholar] [CrossRef][Green Version]
- Funa, N.; Ohnishi, Y.; Ebizuka, Y.; Horinouchi, S. Properties and substrate specificity of RppA, a chalcone synthase-related polyketide synthase in Streptomyces griseus. J. Biol. Chem. 2002, 277, 4628–4635. [Google Scholar] [CrossRef] [PubMed]
- Cortés, J.; Velasco, J.; Foster, G.; Blackaby, A.P.; Rudd, B.A.M.; Wilkinson, B. Identification and cloning of a type III polyketide synthase required for diffusible pigment biosynthesis in Saccharopolyspora erythraea. Mol. Microbiol. 2002, 44, 1213–1224. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Grüschov, S.; Dordik, J.; Sherman, D.H. Molecular analysis of the role of tyrosine 224 in the active site of Streptomyces coelicolor RppA, a bacterial type III polyketide synthase. J. Biol. Chem. 2007, 282, 12765–12772. [Google Scholar] [CrossRef]
- Funa, N.; Funabashi, M.; Ohnishi, Y.; Horinouchi, S. Biosynthesis of hexahydroxyperylene quinone melanin via oxidative aryl coupling by cytochrome P450 in Streptomyces griseus. J. Bacteriol. 2005, 187, 8149–8155. [Google Scholar] [CrossRef]
- Zeng, J.; Decker, R.; Zhan, J. Biochemical characterization of a type III polyketide biosynthetic gene cluster from Streptomyces toxytricini. Appl. Biochem. Biotechnol. 2012, 166, 1020–1033. [Google Scholar] [CrossRef]
- AbuSara, N.F.; Piercey, B.M.; Moore, M.A.; Shaikh, A.A.; Nothias, L.F.; Srivastava, S.K.; Cruz-Morales, P.; Dorrestein, P.C.; Barona-Gómez, F.; Tahlan, K. Comparative Genomics and Metabolomics Analyses of Clavulanic Acid-Producing Streptomyces Species Provides Insight into Specialized Metabolism. Front. Microbiol. 2019, 10, 2550. [Google Scholar] [CrossRef]
- Funa, N.; Ohnishi, Y.; Ebizuka, Y.; Horinouchi, S. Alteration of reaction and substrate specificity of a bacterial type III polyketide synthase by site-directed mutagenesis. Biochem. J. 2002, 367, 781–789. [Google Scholar] [CrossRef]
- Ferrer, J.-L.; Jez, J.M.; Bowman, M.E.; Dixon, R.A.; Noel, J.P. Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat. Struct. Biol. 1999, 6, 775–784. [Google Scholar] [PubMed]
- Jez, J.M.; Noel, J.P. Mechanism of chalcone synthase: pKa of the catalytic cysteine and the role of the conserved histidine in a plant polyketide synthase. J. Biol. Chem. 2000, 275, 39640–39646. [Google Scholar] [CrossRef] [PubMed]
- Louie, G.V.; Bowman, M.E.; Moffitt, M.C.; Baiga, T.J.; Moore, B.S.; Noel, J.P. Structural determinants and modulation of substrate specificity in phenylalanine-tyrosine ammonia-lyases. Chem. Biol. 2006, 13, 1327–1338. [Google Scholar] [CrossRef]
- Rosler, J.; Krekel, F.; Amrhein, N.; Schmid, J. Maize phenylalanine ammonia-lyases has tyrosine ammonia lyase activity. Plant Physiol. 1997, 113, 175–179. [Google Scholar] [CrossRef]
- Calabrese, J.C.; Jordan, D.B.; Boodhoo, A.; Sariaslani, S.; Vannelli, T. Crystal structure of phenylalanine ammonia lyase: Multiple helix dipoles implicated in catalysis. Biochemistry 2004, 43, 11403–11416. [Google Scholar] [CrossRef] [PubMed]
- Hyun, M.W.; Yun, Y.H.; Kim, J.Y.; Kim, S.H. Fungal and plant phenylalanine ammonia-lyase. Microbiology 2011, 39, 257–265. [Google Scholar] [CrossRef]
- Cochrane, F.C.; Davin, L.B.; Lewis, N.G. The Arabidopsis phenylalanine ammonia lyase gene family; kinetics characterization of the four PAL isoforms. Phytochemistry 2004, 65, 1557–1564. [Google Scholar] [CrossRef]
- Hsieh, L.S.; Hsieh, Y.L.; Yeh, C.S.; Cheng, C.Y.; Yang, C.C.; Lee, P.D. Molecular characterization of a phenylalanine ammonia lyase gene (BoPAL1) from Bambuse oldhamii. Mol. Biol. Rep. 2011, 38, 283–290. [Google Scholar] [CrossRef]
- Li, C.L.; Bai, Y.C.; Chen, H.; Zhao, H.X.; Shao, J.R.; Wu, Q. Cloning, characterization and functional analysis of a phenylalanine ammonia-lyase gene (FtPAL) from Fagopyrum tataricum Gaertn. Plant Mol. Biol. Report. 2012, 30, 1172–1182. [Google Scholar] [CrossRef]
- Zang, Y.; Jiang, T.; Cong, Y.; Zheng, Z.; Ouyang, J. Molecular characterization of a recombinant Zea mays phenylalanine ammonia-lyase (ZmPAL2) and its application in trans-cinnamic acid production from L-phenylalanine. Appl. Biochem. Biotechnol. 2015, 176, 923–937. [Google Scholar] [CrossRef]
- Jun, S.-Y.; Sattler, S.A.; Cortez, G.S.; Vermerris, W.; Sattler, S.E.; Kang, C. Biochemical and structural analysis of substrate specificity of a phenylalanine ammonia-lyase. Plant Physiol. 2018, 176, 1452–1468. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Woods, K.V.; Morgan, J.A. Metabolic engineering of the phenylpopanoid pathway in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2005, 71, 2962–2969. [Google Scholar] [CrossRef] [PubMed]
- Poppe, L.; Rétey, J. Friedel-Crafts-type mechanism for the enzymatic elimination of ammonia from histidine and phenylalanine. Angew. Chem. Int. Ed. 2005, 44, 3668–3688. [Google Scholar] [CrossRef] [PubMed]
- Röther, D.; Poppe, L.; Viergutz, S.; Langer, B.; Rétey, J. Characterization of the active site of histidine ammonia-lyase from Pseudomonas putida. Eur. J. Biochem. 2001, 268, 6011–6019. [Google Scholar] [CrossRef]
- Xiang, L.; Moore, B.S. Biochemical characterization of a prokaryotic phenylalanine ammonia lyase. J. Bacteriol. 2005, 187, 4286–4289. [Google Scholar] [CrossRef]
- Williams, J.S.; Tomas, M.; Clarke, D.J. The gene stlA encodes a phenylalanine ammonia-lyase that is involved in the production of a stilbene antibioti in Photorhabdus luminiscens TT01. Microbiology 2005, 151, 2543–2550. [Google Scholar] [CrossRef]
- Hill, A.M.; Thompson, B.L.; Harris, J.O.; Segret, R. Investigation of the early stages in soraphen A biosynthesis. ChemBioChem 2003, 4, 1358–1359. [Google Scholar] [CrossRef]
- Emes, A.V.; Vining, L.C. Partial purification and properties of 1-phenylalanine ammonia lyase from Streptomyces verticillatus. Can. J. Biochem. 1970, 48, 613–6122. [Google Scholar] [CrossRef]
- Kyndt, J.A.; Meyer, T.E.; Cusanovich, M.A.; van Beeumen, J.J. Characterization of a bacterial tyrosine ammonia lyase, a biosynthetic enzyme for the photoactive yellow protein. FEBS Lett. 2002, 512, 240–244. [Google Scholar] [CrossRef]
- Watts, K.T.; Mijts, B.N.; Lee, P.C.; Manning, A.J.; Schmidt-Dannert, C. Discovery of a substrate selectivity switch in tyrosine ammonia-lyase, a member of the aromatic amino acid lyase family. Chem. Biol. 2006, 13, 1317–1326. [Google Scholar] [CrossRef]
- Nishiyama, Y.; Yun, C.-S.; Matsuda, F.; Sasaki, T.; Saito, K.; Tozawa, Y. Expression of bacterial tyrosine ammonia-lyase creates a novel p-coumaric acid pathway in the biosynthesis of phenylpropanoids in Arabidopsis. Planta 2010, 232, 20–218. [Google Scholar] [CrossRef]
- Christenson, S.D.; Liu, W.; Toney, M.D.; Shen, B. A novel 4-methylideneimidazole-5-one-containing tyrosine aminomutase in enediyne antitumor antibiotic C-1027 biosynthesis. J. Am. Chem. Soc. 2003, 125, 6062–6063. [Google Scholar] [CrossRef] [PubMed]
- Baedeker, M.; Schulz, G.E. Autocatalytic peptide cyclization during chain folding of histidine ammonia-lyase. Structure 2002, 10, 61–67. [Google Scholar] [CrossRef][Green Version]
- Retey, J. Discovery and role of methylidene imidazolone, a highly electrophilic prostetic group. Biochim. Biophys. Acta 2003, 1647, 179–184. [Google Scholar] [CrossRef]
- Walker, K.D.; Klettke, K.; Akiyama, T.; Croteau, R. Cloning, heterologous expression and characterization of a phenylalanine aminomutase involved in taxol biosynthesis. J. Biol. Chem. 2004, 279, 56854–63947. [Google Scholar] [CrossRef] [PubMed]
- Steele, C.L.; Chen, Y.; Dougherty, B.A.; Li, W.; Hofstead, S.; Lam, K.S.; Xing, Z.; Chiang, S.J. Purification, cloning and functional expression of phenylalanine aminomutase: The first committed step in Taxol side-chain biosynthesis. Arch. Biochem. Biophys. 2005, 438, 1–10. [Google Scholar] [CrossRef]
- Boniwell, J.M.; Butt, V.S. Flavin Nucleotide-dependent 3-hydroxylation of 4-hydroxyphenylpropanoid carboxylic acids by particulate preparations from potato-tubers. Z. Nat. C 1986, 41, 56–60. [Google Scholar] [CrossRef]
- Kneusel, R.E.; Matern, U.; Nicolay, K. Formation of trans-caffeoyl-CoA from trans-4-coumaroyl-CoA by Zn2+-dependent enzymes in cultured plant cells and its activation by an elicitor induced pH shift. Arch. Biochem. Biophys. 1989, 269, 455–462. [Google Scholar] [CrossRef]
- Kojima, M.; Takeuchi, W. Detection and characterization of p-coumaric acid hydroxylase in mung bean, Vigna mungo, seedlings. J. Biochem. 1989, 105, 265–270. [Google Scholar] [CrossRef]
- Heller, W.; Kühnl, T. Elicitor induction of a microsomal 5-O-(4-coumaroyl) shikimate 3-hydroxylase in parsley cell suspension cultures. Arch. Biochem. Biophys. 1985, 241, 453–460. [Google Scholar] [CrossRef]
- Humphreys, J.M.; Chapple, C. Rewriting the lignin roadmap. Curr. Opin. Plant Biol. 2002, 5, 224–229. [Google Scholar] [CrossRef]
- Kühnl, T.; Koch, U.; Heller, W.; Wellmann, E. Chlorogenic acid biosynthesis- characterization of a light induced microsomal 5-O-(4-coumaroyl-D-quinate/shikimate) 3´hydroxylase from carrot (Daucus carota L.) cell suspension cultures. Arch. Biochem. Biophys. 1987, 258, 226–232. [Google Scholar] [CrossRef]
- Franke, R.; Humphreys, J.M.; Hemm, M.R.; Denult, J.W.; Ruegger, M.O.; Cusumano, J.C.; Chapple, C. The Arabidopsis ref8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J. 2002, 30, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Schoch, G.; Goepfert, S.; Morant, M.; Hehn, A.; Meyer, D.; Ullmann, P.; Werck-Reichhart, D. CYP98A3 from Arabidosis thaliana is a 3´hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J. Biol. Chem. 2001, 276, 36566–36574. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Byeon, Y.; Lee, H.J.; Back, K. Cloning of Arabidopsis serotonin N acetyl transferase and its role with caffeic acid O-methyltransferase in the biosynthesis of melatonin in vitro despite their different subcellular localizations. J. Pineal Res. 2014, 57, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Pruthi, V. Potential application of ferulic acid from natural sources. Biotechnol. Rep. 2014, 4, 86–93. [Google Scholar] [CrossRef]
- Li, J.J.; Zhang, G.; Yu, J.-H.; Li, Y.-Y.; Huang, X.-H.; Wang, W.-J.; Tan, R.; Zhou, J.-Y.; Liao, H. Molecular cloning and characterization of caffeic acid 3-O-methyltransferase from the rhizome of Ligusticum chuanxiong. Biotechnol. Lett. 2015, 37, 2295–2302. [Google Scholar] [CrossRef]
- Narnoliya, L.K.; Sangwan, N.; Jadaum, J.S.; Bansai, S.; Sangwan, R.S. Defining the role of a caffeic acid 3-O-methyltransferase from Azadirachta indica fruits in the biosynthesis of ferulic acid through heterologous over-expression in Ocimum species and Withania somnifera. Planta 2021, 253, 20. [Google Scholar] [CrossRef]
- Youngdae, Y.; Park, Y.; Yi, Y.S.; Lee, Y.; Jo, G.; Park, J.C.; Ahn, J.-H.; Lim, Y. Characterization of an O-Methyltransferase from Streptomyces avermitilis MA-4680. J. Microbiol. Biotechnol. 2010, 20, 1359–1366. [Google Scholar]
- Zhang, Y.; Han, M.-Z.; Zhu, S.-L.; Li, M.; Luo, Z.-C.; Kong, Z.; Lu, Y.-X.; Wang, S.-Y.; Tong, W.-Y. Studies on the function and catalytic mechanism of O-methyltransferases SviOMT02, SviOMT03 and SviOMT06 from Streptomyces virginiae IBL14. Enzym. Microb. Technol. 2015, 73–74, 72–79. [Google Scholar] [CrossRef]
- Pietrowska-Borek, M.; Stuible, H.-P.; Kombrink, E.; Guranowski, A. 4-coumarate: Coenzyme A ligase has the catalytic capacity to synthesize and reuse various (di)adenosine polyphosphates. Plant Physiol. 2003, 131, 1401–1410. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K.; Hövel, K.; Witzel, K.; Hamberger, B.; Schomburg, D.; Kombrink, E.; Stuible, H.-P. The substrate specificity-determining amino acid code of 4-coumarate: CoA ligase. Proc. Nat. Acad. Sci. USA 2003, 100, 8601–8606. [Google Scholar] [CrossRef] [PubMed]
- Ehlting, J.; Büttner, D.; Wang, Q.; Douglas, C.J.; Somssich, I.E.; Kombrink, E. Three 4-coumarate: Coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 1999, 19, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Lindermayr, C.; Fliegmann, J.; Ebel, J. Deletion of a single amino acid residue from different 4-coumarate:CoA ligases from soybean results in the generation of new substrate specificities. J. Biol. Chem. 2003, 278, 2781–2786. [Google Scholar] [CrossRef] [PubMed]
- Lamas-Maceiras, M.; Vaca, I.; Rodríguez, E.; Casqueiro, J.; Martín, J.F. Amplification and disruption of the phenylacetyl-CoA ligase gene of Penicillium chrysogenum encoding an aryl-capping enzyme that supplies phenylacetic acid to the isopenicillin N acyltransferase. Biochem. J. 2006, 395, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.Q.; Liu, J.; Dai, M.; Ren, Z.H.; Su, C.-Y.; He, J.G. Molecular cloning and functional identification of a novel phenylacetyl.CoA ligase gene from Penicillium chrysogenum. Biochem. Biophys. Res. Commun. 2007, 360, 453–458. [Google Scholar] [CrossRef]
- Koetsier, M.J.; Gombert, A.K.; Fekken, S.; Bovenberg, R.A.; Van den Berg, M.A.; Kiel, J.A.; Jekel, P.A.; Janssen, D.B.; Pronk, J.T.; Van der Klei, I.J.; et al. The Penicillium chrysogenum aclA gene encodes a broad-substrate-specificity acyl-coenzyme A ligase involved in activation of adipic acid, a side-chain precursor for cephem antibiotics. Fungal Genet. Biol. 2010, 47, 33–42. [Google Scholar] [CrossRef]
- Koetsier, M.J.; Jekel, P.A.; van den Berg, M.A.; Bovenberg, R.A.; Jansen, D.B. Characterization of a phenylacetate-CoA ligase from Penicillium chrysogenum. Biochem. J. 2009, 417, 467–476. [Google Scholar] [CrossRef]
- Martín, J.F.; Ullán, R.V.; García-Estrada, C. Role of peroxisomes in the biosynthesis and secretion of b-lactams and other secondary metabolites. J. Ind. Microbiol. Biotechnol. 2012, 39, 367–382. [Google Scholar] [CrossRef]
- Piel, J.; Hertweck, C.; Shipley, P.R.; Hunt, D.M.; Newman, M.S.; Moore, B.S. Cloning, sequencing and analysis of the enterocin biosynthesis gene cluster from the marine isolate Streptomyces maritimus: Evidence for the derailment of an aromatic polyketide synthase. Chem. Biol. 2000, 7, 943–955. [Google Scholar] [CrossRef]
- Xiang, L.; Moore, B.S. Inactivation, complementation and heterologous expression of encP, a novel bacterial phenylalanine ammonia-lyase gene. J. Biol. Chem. 2002, 277, 32505–32509. [Google Scholar] [CrossRef]
- Hubbard, B.K.; Thomas, M.G.; Walsh, C.T. Biosynthesis of L-p-hydroxyphenylglycine, a non-proteinogenic amino acid constituent of peptide antibiotics. Chem. Biol. 2000, 7, 931–942. [Google Scholar] [CrossRef]
- Choroba, O.W.; Williams, D.H.; Spencer, J.B. Biosynthesis of the vancomycin group of antibiotics: Involvement of an unusual dioxygenase in the pathway to (S)-4-hydroxyphenylglycine. J. Am. Chem. Soc. 2000, 122, 5389–5390. [Google Scholar] [CrossRef]
- Sandercock, A.M.; Charles, E.H.; Scaife, W.; Kirkpatrick, P.N.; O’Brien, S.W.; Papageorgiou, E.A.; Spencer, J.B.; Williams, D.H. Characterisation of a hydroxymandelate oxidase involved in the biosynthesis of two unusual amino acids occurring in the vancomycin group of antibiotics. Chem. Commun. 2001, 18, 1252–1253. [Google Scholar] [CrossRef]
- Chung, S.K.; Taylor, P.; Oh, Y.K.; DeBrosse, C.; Jeffs, P.W. Biosynthetic studies of aridicin antibiotics. I. Labeling patterns and overall pathways. J. Antibiot. 1986, 39, 642–651. [Google Scholar] [CrossRef] [PubMed]
- Nicas, T.I.; Cooper, R.D.G. Vancomycin and other glycopeptides. In Biotechnology of Antibiotics; Strohl, W.R., Ed.; Marcel Dekker: New York, NY, USA, 1997; pp. 363–392. [Google Scholar]
- Toma, A.R.S.; Brieke, C.; Cryle, M.J.; Süssmuth, R.D. Structural aspects of phenylglycines, their biosynthesis and occurrence in peptide natural products. Nat. Prod. Rep. 2015, 32, 1207–1235. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, V.; Nicholson, G.J.; Ries, J.; Recktenwald, J.; Schefer, A.B.; Shawky, R.M.; Schöder, J.; Wohlleben, W.; Pelzer, S. A polyketide synthase in glycopeptide biosynthesis: The biosynthesis of the non-proteinogenic amino acid (S)-3,5-dihydroxyphenylglycine. J. Biol. Chem. 2001, 276, 38370–38377. [Google Scholar] [CrossRef]
- Chen, H.; Tseng, C.C.; Hubbard, B.K.; Walsh, C.T. Glycopeptide antibiotic biosynthesis: Enzymatic assembly of the dedicated amino acid monomer (S9-3,5-dihydroxyphenylglicine. Proc. Nat. Acad. Sci. USA 2001, 98, 14901–14906. [Google Scholar] [CrossRef]
- Tseng, C.C.; McLoughlin, S.M.; Kelleher, N.L.; Walsh, C.T. Role of the active site cysteine of DpgA, a bacterial type III polyketide synthase. Biochemistry 2004, 43, 970–980. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, B.; Jiang, R. Improving acetyl-CoA biosynthesis in Saccharomyces cerevisiae via the overexpression of pantothenate kinase and PDH bypass. Biotechnol. Biofuels 2017, 10, 41–49. [Google Scholar] [CrossRef]
- Palmer, C.M.; Miller, K.K.; Nguyen, A.; Alper, H.S. Engineering 4-coumaroyl-CoA derived polyketide production in Yarrowia lipolytica through a β-oxidation mediated strategy. Metab. Eng. 2020, 57, 174–181. [Google Scholar] [CrossRef]
- Koopman, F.; Beekwilder, J.; Crimi, B.; van Houwelingen, A.; Hall, R.D.; Bosch, D.; van Maris, A.J.; Pronk, J.T.; Daran, J.M. De novo production of the flavonoid naringenin in engineered Saccharomyces cerevisiae. Microb. Cell Factories 2012, 11, 155–169. [Google Scholar] [CrossRef]
- Lyu, X.; Ng, K.R.; Lee, J.L.; Mark, R.; Chen, W.N. Enhancement of Naringenin Biosynthesis from Tyrosine by Metabolic Engineering of Saccharomyces cerevisiae. J. Agric. Food Chem. 2017, 65, 6638–6646. [Google Scholar] [CrossRef]
- Wei, W.; Zhang, P.; Shang, Y.; Zhou, Y.; Ye, B.C. Metabolically engineering of Yarrowia lipolytica for the biosynthesis of naringenin from a mixture of glucose and xylose. Bioresour. Technol. 2020, 314, 123726. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, H.; Li, X.; Liu, D.; Dong, X.T.; Li, F.F.; Wang, E.X.; Li, B.Z.; Yuan, Y.J. Production of naringenin from D-xylose with co-culture of E. coli and Saccharomyces cerevisiae. Eng. Life Sci. 2017, 17, 1021–1029. [Google Scholar] [CrossRef]
- Li, H.; Gao, S.; Zhang, S.; Zeng, W.; Zhou, J. Effects of metabolic pathway gene copy numbers on the biosynthesis of naringenin in Saccharomyces cerevisiae. J. Biotechnol. 2021, 325, 119–127. [Google Scholar] [CrossRef]
- Gao, S.; Lyu, Y.; Zeng, W.; Du, G.; Zhou, J.; Chen, J. Efficient Biosynthesis of (2S)-Naringenin from p-Coumaric Acid in Saccharomyces cerevisiae. J. Agric. Food Chem. 2020, 68, 1015–1021. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Marsafari, M.; Koffas, M.; Zhou, J.; Xu, P. Optimizing Oleaginous Yeast Cell Factories for Flavonoids and Hydroxylated Flavonoids Biosynthesis. ACS Synth. Biol. 2019, 8, 2514–2523. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Tang, H.; Ni, J.; Wu, Q.; Hua, D.; Tao, F.; Xu, P. Characterization of two Streptomyces enzymes that convert ferulic acid to vanillin. PLoS ONE 2013, 8, e67339. [Google Scholar]
- Schmidt, S.; Rainieri, S.; Witte, S.; Matern, U.; Martens, S. Identification of a Saccharomyces cerevisiae glucosidase that hydrolyzes flavonoid glucosides. Appl. Environ. Microbiol. 2011, 77, 1751–1757. [Google Scholar] [CrossRef]
- Ohashi, T.; Hasegawa, Y.; Misaki, R.; Fujiyama, K. Substrate preference of citrus naringenin rhamnosyltransferases and their application to flavonoid glycoside production in fission yeast. Appl. Microbiol. Biotechnol. 2016, 100, 687–696. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martín, J.F.; Liras, P. Comparative Molecular Mechanisms of Biosynthesis of Naringenin and Related Chalcones in Actinobacteria and Plants: Relevance for the Obtention of Potent Bioactive Metabolites. Antibiotics 2022, 11, 82. https://doi.org/10.3390/antibiotics11010082
Martín JF, Liras P. Comparative Molecular Mechanisms of Biosynthesis of Naringenin and Related Chalcones in Actinobacteria and Plants: Relevance for the Obtention of Potent Bioactive Metabolites. Antibiotics. 2022; 11(1):82. https://doi.org/10.3390/antibiotics11010082
Chicago/Turabian StyleMartín, Juan F., and Paloma Liras. 2022. "Comparative Molecular Mechanisms of Biosynthesis of Naringenin and Related Chalcones in Actinobacteria and Plants: Relevance for the Obtention of Potent Bioactive Metabolites" Antibiotics 11, no. 1: 82. https://doi.org/10.3390/antibiotics11010082
APA StyleMartín, J. F., & Liras, P. (2022). Comparative Molecular Mechanisms of Biosynthesis of Naringenin and Related Chalcones in Actinobacteria and Plants: Relevance for the Obtention of Potent Bioactive Metabolites. Antibiotics, 11(1), 82. https://doi.org/10.3390/antibiotics11010082





