Colistin Resistance Mechanisms in Human Salmonella enterica Strains Isolated by the National Surveillance Enter-Net Italia (2016–2018)
Abstract
1. Introduction
2. Results
2.1. Antimicrobial Susceptibility and Mcr-Gene Testing
2.2. Genomic Characterisation of Isolates
2.3. Chromosomal Mutations Study
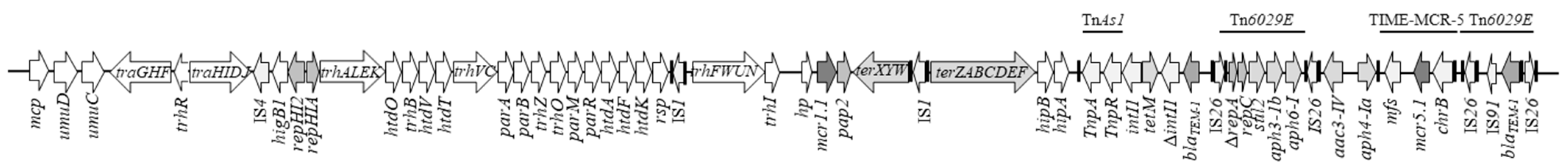
2.4. Characterisation of the Mcr-Positive Plasmids
3. Discussion
- The rhamnosyltransferase RfbN protein is one of the key factors of the O-antigen biosynthesis and showed the deleterious D107V mutation in the COL-R S. Enteritidis 45/7/18 (Table 2). The O-antigen oligosaccharide of S. Enteritidis and S. Typhimurium contains rhamnose [70]. The rhamnosyltransferases are identical in Salmonella groups A, B, D1 and D2 [71,72]. Proteins involved in the biosynthesis of the basic O-antigen, present in the same operon of RfbN, have been previously involved in conferring COL resistance [17].
- The R26L deleterious mutation found in the 45/7/18 strain involves the transcriptional regulatory protein ZraR activated by the sensor kinase ZraS in a zinc-dependent response regulation of a TCS (Table 2). It has been related to envelope environmental stress response, metabolism, protein synthesis, motility and biofilm formation. ZraR is also a bacterial enhancer-binding protein, controlling the multidrug export proteins AcrE, MdtE and MdtL. ZraR controls also LPS synthesis by RfaD and CpxP, the chaperone and modulator of CpxAR, respectively, that are directly involved in COL resistance [6,73]. CpxP-superfamily plays a role in resistance against polymyxin B in S. enterica [74]. Mutations in genes encoding regulators are essential in the adaptation process since mutations in regulatory elements can affect a broad range of targets [75].
- S. Enteritidis 58/10/16 showed a deleterious mutation R298C in MdsC, an outer membrane lipoprotein, part of the MdsAB-MdsC tripartite efflux pump, one of the efflux pump systems of the RND family (Table 2). The mutation found in this efflux pump component may result in overproduction of the pump or have other effects leading to COL resistance. In P. aeruginosa, the overexpression of the homolog RND efflux pump MexAB-OprM has been linked to COL resistance [76]. However, this mutation was not causing a significant increase in resistance to other antimicrobial compounds than COL.
- The deleterious mutation S91R, in S. Enteritidis 58/10/16 strain was detected in the outer membrane lipoprotein receptor LolB (Table 2). It is part of the system that transports lipoproteins, directly involved in outer membrane biogenesis and essential for cell viability [77]. A COL-R LPS-deficient Acinetobacter baumannii strain increased the exopolysaccharide production and the expression of lol genes to compensate for its deficit [78].
- The deleterious mutation R127L in the YdeI protein was observed in the 58/10/16 strain (Table 2). It encodes an oligosaccharide/oligonucleotide binding-fold (OB-fold) protein important for polymyxin B resistance. It is regulated by the RcsBCD, PhoP-PhoQ and PmrA-PmrB sensor-kinase systems that modify gene expression in the presence of cationic antimicrobial peptides (CAMPs) in S. enterica. Furthermore, it could interact with OmpD in the outer membrane to facilitate CAMP resistance [79].
- In the 58/10/16 strain, the Type IV pilus biogenesis protein PilN was also mutated (Table 2). It is part of the PilMNOP operon, promoting surface-associated twitching motility and virulence. In P. aeruginosa, it has been hypothesised that any mutation in PilN can destabilise the PilM-PilN interaction causing functionally significant structural changes in PilM [80,81]. In polymyxin B/COL-heteroresistant subpopulations colonies of Neisseria meningitidis, point mutations in pilM or pilQ were associated with the resistant phenotype [80]. Even if a direct relationship between these mutations and the COL resistance phenotype must be investigated, an association of different mutations in the same organism promoted a COL-R pattern. In P. aeruginosa, it has been asserted that the evolution of resistance is a complex, multistep process that requires a mutation in at least five independent loci that synergistically create the COL-R phenotype [75].
4. Materials and Methods
4.1. Settings and Bacterial Isolates
4.2. Antimicrobial Susceptibility Testing and Detection of Mcr Genes
4.3. Whole-Genome Sequencing (WGS)
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lima, T.; Domingues, S.; Da Silva, G.J. Plasmid-mediated colistin resistance in Salmonella enterica: A review. Microorganisms 2019, 7, 55. [Google Scholar] [CrossRef]
- Binsker, U.; Käsbohrer, A.; Hammerl, J.A. Global colistin use: A review of the emergence of resistant Enterobacterales and the impact on their genetic basis. FEMS Microbiol. Rev. 2021, fuab049, 1–37. [Google Scholar] [CrossRef]
- European Commission. A European One Health Action Plan against Antimicrobial Resistance (AMR) CONTENTS. 2021. Available online: https://ec.europa.eu/health/sites/default/files/antimicrobial_resistance/docs/amr_2017_action-plan.pdf (accessed on 9 December 2021).
- OIE. Oie List of Antimicrobials of Veterinary Importance. World Organ. Anim. Health 2007, 2007, 1–9. [Google Scholar]
- Zavascki, A.P.; Goldani, L.Z.; Li, J.; Nation, R.L. Polymyxin B for the treatment of multidrug-resistant pathogens: A critical review. J. Antimicrob. Chemother. 2007, 60, 1206–1215. [Google Scholar] [CrossRef] [PubMed]
- Aghapour, Z.; Gholizadeh, P.; Ganbarov, K.; Bialvaei, A.Z.; Mahmood, S.S.; Tanomand, A.; Yousefi, M.; Asgharzadeh, M.; Yousefi, B.; Kafil, H.S. Molecular mechanisms related to colistin resistance in Enterobacteriaceae. Infect. Drug Resist. 2019, 12, 965–975. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed Ahmed, M.A.E.G.; Zhong, L.L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.B. Colistin and its role in the Era of antibiotic resistance: An extended review (2000–2019). Emerg. Microbes Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef]
- Lin, Q.Y.; Tsai, Y.L.; Liu, M.C.; Lin, W.C.; Hsueh, P.R.; Liaw, S.J. Serratia marcescens arn, a PhoP-regulated locus necessary for polymyxin B resistance. Antimicrob. Agents Chemother. 2014, 58, 5181–5190. [Google Scholar] [CrossRef]
- Nang, S.C.; Azad, M.A.K.; Velkov, T.; Zhou, Q.T.; Li, J. Rescuing the last-line polymyxins: Achievements and challenges. Pharmacol. Rev. 2021, 73, 679–728. [Google Scholar] [CrossRef]
- Gunn, J.S.; Ryan, S.S.; Van Velkinburgh, J.C.; Ernst, R.K.; Miller, S.I. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 2000, 68, 6139–6146. [Google Scholar] [CrossRef] [PubMed]
- Olaitan, A.O.; Morand, S.; Rolain, J.M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef]
- Sekyere, J.O.; Govinden, U.; Bester, L.A.; Essack, S.Y. Colistin and tigecycline resistance in carbapenemase-producing Gram-negative bacteria: Emerging resistance mechanisms and detection methods. J. Appl. Microbiol. 2016, 121, 601–617. [Google Scholar] [CrossRef]
- Poirel, L.; Jayol, A.; Bontron, S.; Villegas, M.V.; Ozdamar, M.; Türkoglu, S.; Nordmann, P. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2015, 70, 75–80. [Google Scholar] [CrossRef]
- Moosavian, M.; Emam, N.; Pletzer, D.I.; Savari, M.I. Rough-type and loss of the LPS due to lpx genes deletions are associated with colistin resistance in multidrug-resistant clinical Escherichia coli isolates not harbouring mcr genes. PLoS ONE 2020, 15, e0233518. [Google Scholar] [CrossRef]
- Moffatt, J.H.; Harper, M.; Boyce, J.D. Mechanisms of Polymyxin Resistance. In Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2019; Volume 1145, pp. 55–71. [Google Scholar] [CrossRef]
- Barchiesi, J.; Espariz, M.; Checa, S.K.; Soncini, F.C. Downregulation of RpoN-controlled genes protects Salmonella cells from killing by the cationic antimicrobial peptide polymyxin B. FEMS Microbiol. Lett. 2009, 291, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Ricci, V.; Zhang, D.; Teale, C.; Piddock, L.J.V. The O-antigen epitope governs susceptibility to colistin in Salmonella enterica. MBio 2020, 11, e02831-19. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Jayol, A.; Nordmanna, P. Polymyxins: Antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Carroll, L.M.; Gaballa, A.; Guldimann, C.; Sullivan, G.; Henderson, L.O.; Wiedmann, M. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. MBio 2019, 10, e00853-19. [Google Scholar] [CrossRef]
- Carattoli, A.; Villa, L.; Feudi, C.; Curcio, L.; Orsini, S.; Luppi, A.; Pezzotti, G.; Magistrali, C.F. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Eurosurveillance 2017, 22, 30589. [Google Scholar] [CrossRef]
- Moreno, L.Z.; Gomes, V.T.M.; Moreira, J.; de Oliveira, C.H.; Peres, B.P.; Silva, A.P.S.; Thakur, S.; La Ragione, R.M.; Moreno, A.M. First report of mcr-1-harboring Salmonella enterica serovar Schwarzengrund isolated from poultry meat in Brazil. Diagn. Microbiol. Infect. Dis. 2019, 93, 376–379. [Google Scholar] [CrossRef]
- Quesada, A.; Ugarte-Ruiz, M.; Iglesias, M.R.; Porrero, M.C.; Martínez, R.; Florez-Cuadrado, D.; Campos, M.J.; García, M.; Píriz, S.; Sáez, J.L.; et al. Detection of plasmid mediated colistin resistance (MCR-1) in Escherichia coli and Salmonella enterica isolated from poultry and swine in Spain. Res. Vet. Sci. 2016, 105, 134–135. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhang, P.; Yang, X.; Wang, Z.; Fanning, S.; Wang, J.; Du, P.; Bai, L. Identification of a novel hybrid plasmid coproducing MCR-1 and MCR-3 variant from an Escherichia coli strain. J. Antimicrob. Chemother. 2019, 74, 1517–1520. [Google Scholar] [CrossRef] [PubMed]
- Borowiak, M.; Hammerl, J.A.; Deneke, C.; Fischer, J.; Szabo, I.; Malorny, B. Characterization of mcr-5-harboring Salmonella enterica subsp. enterica serovar Typhimurium isolates from animal and food origin in germany. Antimicrob. Agents Chemother. 2019, 63, 63. [Google Scholar] [CrossRef]
- Luo, Q.; Wang, Y.; Xiao, Y. Prevalence and transmission of mobilized colistin resistance (mcr) gene in bacteria common to animals and humans. Biosaf. Health 2020, 2, 71–78. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, J.; Liu, F.; Cheng, Y.; Su, J. Characterization of Salmonella enterica isolates from diseased poultry in northern China between 2014 and 2018. Pathogens 2020, 9, 95. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications [version 1; referees: 2 approved]. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Kawahara, R.; Hamamoto, K.; Hirai, I.; Khong, D.T.; Nguyen, T.N.; Tran, H.T.; Motooka, D.; Nakamura, S.; Yamamoto, Y. High prevalence of colistin-resistant Escherichia coli with chromosomally carried mcr-1 in healthy residents in vietnam. mSphere 2020, 5, e00117-20. [Google Scholar] [CrossRef]
- Sun, J.; Fang, L.X.; Wu, Z.; Deng, H.; Yang, R.S.; Li, X.P.; Li, S.M.; Liao, X.P.; Feng, Y.; Liu, Y.H. Genetic analysis of the IncX4 plasmids: Implications for a unique pattern in the mcr-1 acquisition. Sci. Rep. 2017, 7, 424. [Google Scholar] [CrossRef]
- Poirel, L.; Kieffer, N.; Brink, A.; Coetze, J.; Jayol, A.; Nordmann, P. Genetic features of MCR-1-producing colistin-resistant Escherichia coli isolates in South Africa. Antimicrob. Agents Chemother. 2016, 60, 4394–4397. [Google Scholar] [CrossRef]
- Zurfluh, K.; Nüesch-Inderbinen, M.; Klumpp, J.; Poirel, L.; Nordmann, P.; Stephan, R. Key features of mcr-1-bearing plasmids from Escherichia coli isolated from humans and food. Antimicrob. Resist. Infect. Control 2017, 6. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garciá-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In Silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Kieffer, N.; Nordmann, P.; Millemann, Y.; Poirel, L. Functional characterization of a miniature inverted transposable element at the origin of mcr-5 gene acquisition in Escherichia coli. Antimicrob. Agents Chemother. 2019, 63, e00559-19. [Google Scholar] [CrossRef]
- Chiou, C.S.; Alam, M.; Kuo, J.C.; Liu, Y.Y.; Wang, P.J. Chromosome-mediated multidrug resistance in Salmonella enterica serovar Typhi. Antimicrob. Agents Chemother. 2015, 59, 721–723. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lucarelli, C.; Dionisi, A.M.; Filetici, E.; Owczarek, S.; Luzzi, I.; Villa, L. Nucleotide sequence of the chromosomal region conferring multidrug resistance (R-type ASSuT) in Salmonella Typhimurium and monophasic Salmonella Typhimurium strains. J. Antimicrob. Chemother. 2012, 67, 111–114. [Google Scholar] [CrossRef]
- Pfeiffer, F.; Zamora-Lagos, M.A.; Blettinger, M.; Yeroslaviz, A.; Dahl, A.; Gruber, S.; Habermann, B.H. The complete and fully assembled genome sequence of Aeromonas salmonicida subsp. pectinolytica and its comparative analysis with other Aeromonas species: Investigation of the mobilome in environmental and pathogenic strains. BMC Genomics 2018, 19, 20. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority); ECDC (European Centre for Disease Prevention and Control). The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2018/2019. EFSA J. 2021, 19. [Google Scholar] [CrossRef]
- Kempf, I.; Jouy, E.; Chauvin, C. Colistin use and colistin resistance in bacteria from animals. Int. J. Antimicrob. Agents 2016, 48, 598–606. [Google Scholar] [CrossRef]
- Vidovic, N.; Vidovic, S. Antimicrobial resistance and food animals: Influence of livestock environment on the emergence and dissemination of antimicrobial resistance. Antibiotics 2020, 9, 52. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA). Technical specifications on arandomisation of sampling for the purpose of antimicrobial resistance monitoring from food-producinganimals and food as from 2021. EFSA J. 2020, 18, e06364. [Google Scholar] [CrossRef]
- Carnevali, C.; Morganti, M.; Scaltriti, E.; Bolzoni, L.; Pongolini, S.; Casadei, G. Occurrence of mcr-1 in colistin-resistant Salmonella enterica isolates recovered from humans and animals in Italy, 2012 to 2015. Antimicrob. Agents Chemother. 2016, 60, 7532–7534. [Google Scholar] [CrossRef] [PubMed]
- Elbediwi, M.; Li, Y.; Paudyal, N.; Pan, H.; Li, X.; Xie, S.; Rajkovic, A.; Feng, Y.; Fang, W.; Rankin, S.C.; et al. Global burden of colistin-resistant bacteria: Mobilized colistin resistance genes study (1980–2018). Microorganisms 2019, 7, 461. [Google Scholar] [CrossRef] [PubMed]
- Zingali, T.; Chapman, T.A.; Webster, J.; Chowdhury, P.R.; Djordjevic, S.P. Genomic characterisation of a multiple drug resistant IncHI2 ST4 plasmid in Escherichia coli ST744 in Australia. Microorganisms 2020, 8, 896. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Yu, W.; Zhou, K.; Guo, L.; Shen, P.; Lu, H.; Huang, C.; Xu, H.; Xu, S.; Xiao, Y.; et al. Molecular epidemiology and colistin resistant mechanism of mcr-positive and mcr-negative clinical isolated Escherichia coli. Front. Microbiol. 2017, 8, 2262. [Google Scholar] [CrossRef]
- Long, H.; Feng, Y.; Ma, K.; Liu, L.; McNally, A.; Zong, Z. The co-transfer of plasmid-borne colistin-resistant genes mcr-1 and mcr-3.5, the carbapenemase gene blaNDM-5 and the 16S methylase gene rmtB from Escherichia coli. Sci. Rep. 2019, 9, 696. [Google Scholar] [CrossRef]
- Campos, J.; Cristino, L.; Peixe, L.; Antunes, P. MCR-1 in multidrug-resistant and copper-tolerant clinically relevant Salmonella 1,4,[5],12:i:- and S. Rissen clones in Portugal, 2011 to 2015. Eurosurveillance 2016, 21, 30270. [Google Scholar] [CrossRef]
- Sia, C.M.; Greig, D.R.; Day, M.; Hartman, H.; Painset, A.; Doumith, M.; Meunier, D.; Jenkins, C.; Chattaway, M.A.; Hopkins, K.L.; et al. The characterization of mobile colistin resistance (mcr) genes among 33 000 Salmonella enterica genomes from routine public health surveillance in england. Microb. Genomics 2020, 6, e000331. [Google Scholar] [CrossRef]
- Harmer, C.J.; Hall, R.M. IS26-Mediated Formation of Transposons Carrying Antibiotic Resistance Genes. mSphere 2016, 1. [Google Scholar] [CrossRef]
- Reid, C.J.; Chowdhury, P.R.; Djordjevic, S.P. Tn6026 and Tn6029 are found in complex resistance regions mobilised by diverse plasmids and chromosomal islands in multiple antibiotic resistant Enterobacteriaceae. Plasmid 2015, 80, 127–137. [Google Scholar] [CrossRef]
- Shang, D.; Zhao, H.; Xu, X.; Arunachalam, K.; Chang, J.; Bai, L.; Shi, C. Conjugative IncHI2 plasmid harboring novel class 1 integron mediated dissemination of multidrug resistance genes in Salmonella Typhimurium. Food Control 2021, 122, 107810. [Google Scholar] [CrossRef]
- Hernández, M.; Iglesias, M.R.; Rodríguez-Lázaro, D.; Gallardo, A.; Quijada, N.M.; Miguela-Villoldo, P.; Campos, M.J.; Píriz, S.; López-Orozco, G.; de Frutos, C.; et al. Co-occurrence of colistin-resistance genes mcr-1 and mcr-3 among multidrug-resistant Escherichia coli isolated from cattle, Spain, September 2015. Eurosurveillance 2017, 22, 30586. [Google Scholar] [CrossRef]
- Xiang, R.; Liu, B.H.; Zhang, A.Y.; Lei, C.W.; Ye, X.L.; Yang, Y.X.; Chen, Y.P.; Wang, H.N. Colocation of the polymyxin resistance gene mcr-1 and a variant of mcr-3 on a plasmid in an Escherichia coli isolate from a chicken farm. Antimicrob. Agents Chemother. 2018, 62, e00501-18. [Google Scholar] [CrossRef]
- García, V.; García-Meniño, I.; Mora, A.; Flament-Simon, S.C.; Díaz-Jiménez, D.; Blanco, J.E.; Alonso, M.P.; Blanco, J. Co-occurrence of mcr-1, mcr-4 and mcr-5 genes in multidrug-resistant ST10 Enterotoxigenic and Shiga toxin-producing Escherichia coli in Spain (2006–2017). Int. J. Antimicrob. Agents 2018, 52, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Feng, Y.; Wang, G.; Zhang, Z.; Hu, H.; Yu, Y.; Liu, J.; Qiu, L.; Liu, H.; Guo, Z.; et al. Co-occurrence of the mcr-1.1 and mcr-3.7 genes in a multidrug-resistant Escherichia coli isolate from China. Infect. Drug Resist. 2020, 13, 3649–3655. [Google Scholar] [CrossRef] [PubMed]
- Doumith, M.; Godbole, G.; Ashton, P.; Larkin, L.; Dallman, T.; Day, M.; Day, M.; Muller-Pebody, B.; Ellington, M.J.; de Pinna, E.; et al. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J. Antimicrob. Chemother. 2016, 71, 2300–2305. [Google Scholar] [CrossRef]
- Simoni, S.; Morroni, G.; Brenciani, A.; Vincenzi, C.; Cirioni, O.; Castelletti, S.; Varaldo, P.E.; Giovanetti, E.; Mingoia, M. Spread of colistin resistance gene mcr-1 in Italy: Characterization of the mcr-1.2 allelic variant in a colistin-resistant blood isolate of Escherichia coli. Diagn. Microbiol. Infect. Dis. 2018, 91, 66–68. [Google Scholar] [CrossRef]
- Manageiro, V.; Clemente, L.; Romão, R.; Silva, C.; Vieira, L.; Ferreira, E.; Caniça, M. IncX4 plasmid carrying the new mcr-1.9 gene variant in a CTX-M-8 producing Escherichia coli isolate recovered from swine. Front. Microbiol. 2019, 10, 367. [Google Scholar] [CrossRef] [PubMed]
- Migura-Garcia, L.; González-López, J.J.; Martinez-Urtaza, J.; Aguirre Sánchez, J.R.; Moreno-Mingorance, A.; Perez de Rozas, A.; Höfle, U.; Ramiro, Y.; Gonzalez-Escalona, N. mcr-colistin resistance genes mobilized by IncX4, IncHI2, and IncI2 plasmids in Escherichia coli of pigs and white stork in Spain. Front. Microbiol. 2020, 10, 3072. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, Y.; Ling, Z.; Zhang, C.; Fu, M.; Wang, Y.; Wang, S.; Zhang, S.; Shen, Z. Peptide nucleic acid restores colistin susceptibility through modulation of MCR-1 expression in Escherichia coli. J. Antimicrob. Chemother. 2020, 75, 2059–2065. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Li, D.; Wang, L.; Bi, Y.; Wang, M.; Yang, F. Promoter variations associated with expression of mcr-1 gene and level of colistin resistance. Int. J. Antimicrob. Agents 2021, 58, 106371. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Wang, Y.; Fu, H.; Yu, X.; Zheng, B.; Chen, Y.; Berglund, B.; Xiao, Y. Serotype is associated with high rate of colistin resistance among clinical solates of Salmonella. Front. Microbiol. 2020, 11, 3295. [Google Scholar] [CrossRef]
- Agersø, Y.; Torpdahl, M.; Zachariasen, C.; Seyfarth, A.; Hammerum, A.M.; Nielsen, E.M. Tentative colistin epidemiological cut-off value for Salmonella spp. Foodborne Pathog. Dis. 2012, 9, 367–369. [Google Scholar] [CrossRef]
- Folkesson, A.; Haagensen, J.A.J.; Zampaloni, C.; Sternberg, C.; Molin, S. Biofilm induced tolerance towards antimicrobial peptides. PLoS ONE 2008, 3, 1891. [Google Scholar] [CrossRef]
- Bialvaei, A.Z.; Kafil, H.S. Colistin, mechanisms and prevalence of resistance. Curr. Med. Res. Opin. 2015, 31, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Trimble, M.J.; Mlynárčik, P.; Kolář, M.; Hancock, R.E.W. Polymyxin: Alternative mechanisms of action and resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025288. [Google Scholar] [CrossRef]
- Rule, R.; Mbelle, N.; Sekyere, J.O.; Kock, M.; Hoosen, A.; Said, M. A rare case of colistin-resistant Salmonella Enteritidis meningitis in an HIV-seropositive patient. BMC Infect. Dis. 2019, 19, 806. [Google Scholar] [CrossRef]
- Pitt, M.E.; Elliott, A.G.; Cao, M.D.; Ganesamoorthy, D.; Karaiskos, I.; Giamarellou, H.; Abboud, C.S.; Blaskovich, M.A.T.T.; Cooper, M.A.; Coin, L.J.M.M.; et al. Multifactorial chromosomal variants regulate polymyxin resistance in extensively drug-resistant Klebsiella pneumoniae. Microb. Genomics 2018, 4, e000158. [Google Scholar] [CrossRef] [PubMed]
- Samuel, G.; Reeves, P. Biosynthesis of O-antigens: Genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 2003, 338, 2503–2519. [Google Scholar] [CrossRef]
- Liu, D.; Lindqvist, L.; Reeves, P.R. Transferases of O-antigen biosynthesis in Salmonella enterica: Dideoxyhexosyltransferases of groups B and C2 and acetyltransferase of group C2. J. Bacteriol. 1995, 177, 4084–4088. [Google Scholar] [CrossRef]
- Xiang, S.H.; Haase, A.M.; Reeves, P.R. Variation of the rfb gene clusters in Salmonella enterica. J. Bacteriol. 1993, 175, 4877–4884. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rome, K.; Borde, C.; Taher, R.; Cayron, J.; Lesterlin, C.; Gueguen, E.; De Rosny, E.; Rodrigue, A. The two-component system ZraPSR is a novel ESR that contributes to intrinsic antibiotic tolerance in Escherichia coli. J. Mol. Biol. 2018, 430, 4971–4985. [Google Scholar] [CrossRef]
- Mlynarcik, P.; Kolar, M. Molecular mechanisms of polymyxin resistance and detection of mcr genes. Biomed. Pap. 2019, 163, 28–38. [Google Scholar] [CrossRef]
- Jochumsen, N.; Marvig, R.L.; Damkiær, S.; Jensen, R.L.; Paulander, W.; Molin, S.; Jelsbak, L.; Folkesson, A. The evolution of antimicrobial peptide resistance in Pseudomonas aeruginosa is shaped by strong epistatic interactions. Nat. Commun. 2016, 7, 1–10. [Google Scholar] [CrossRef]
- Pamp, S.J.; Gjermansen, M.; Johansen, H.K.; Tolker-Nielsen, T. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol. Microbiol. 2008, 68, 223–240. [Google Scholar] [CrossRef]
- Choi, U.; Lee, C.R. Antimicrobial agents that inhibit the outer membrane assembly machines of Gram-negative bacteria. J. Microbiol. Biotechnol. 2019, 29, 1–10. [Google Scholar] [CrossRef]
- Henry, R.; Vithanage, N.; Harrison, P.; Seemann, T.; Coutts, S.; Moffatt, J.H.; Nation, R.L.; Li, J.; Harper, M.; Adler, B.; et al. Colistin-resistant, lipopolysaccharide-deficient Acinetobacter baumannii responds to lipopolysaccharide loss through increased expression of genes involved in the synthesis and transport of lipoproteins, phospholipids, and poly-β-1,6-N-acetylglucosamine. Antimicrob. Agents Chemother. 2012, 56, 59–69. [Google Scholar] [CrossRef]
- Pilonieta, M.C.; Erickson, K.D.; Ernst, R.K.; Detweiler, C.S. A protein important for antimicrobial peptide resistance, YdeI/OmdA, is in the periplasm and interacts with OmpD/NmpC. J. Bacteriol. 2009, 191, 7243–7252. [Google Scholar] [CrossRef] [PubMed]
- Tzeng, Y.L.; Berman, Z.; Toh, E.; Bazan, J.A.; Turner, A.N.; Retchless, A.C.; Wang, X.; Nelson, D.E.; Stephens, D.S. Heteroresistance to the model antimicrobial peptide polymyxin B in the emerging Neisseria meningitidis lineage 11.2 urethritis clade: Mutations in the pilMNOPQ operon. Mol. Microbiol. 2019, 111, 254–268. [Google Scholar] [CrossRef] [PubMed]
- McCallum, M.; Tammam, S.; Little, D.J.; Robinson, H.; Koo, J.; Shah, M.; Calmettes, C.; Moraes, T.F.; Burrows, L.L.; Howell, P.L. PilN binding modulates the structure and binding partners of the Pseudomonas aeruginosa type IVa pilus protein PilM. J. Biol. Chem. 2016, 291, 11003–11015. [Google Scholar] [CrossRef] [PubMed]
- Song, T.; Park, Y.; Shamputa, I.C.; Seo, S.; Lee, S.Y.; Jeon, H.S.; Choi, H.; Lee, M.; Glynne, R.J.; Barnes, S.W.; et al. Fitness costs of rifampicin resistance in Mycobacterium tuberculosis are amplified under conditions of nutrient starvation and compensated by mutation in the β’ subunit of RNA polymerase. Mol. Microbiol. 2014, 91, 1106–1119. [Google Scholar] [CrossRef]
- Beceiro, A.; Moreno, A.; Fernández, N.; Vallejo, J.A.; Aranda, J.; Adler, B.; Harper, M.; Boyce, J.D.; Bou, G. Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2014, 58, 518–526. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Wang, N.; Li, X.; Shi, K.; Zhou, Z.; Yu, Y.; Hua, X. The effect of colistin resistance-associated mutations on the fitness of Acinetobacter baumannii. Front. Microbiol. 2016, 7, 1715. [Google Scholar] [CrossRef]
- Choi, Y.; Lee, J.Y.; Lee, H.; Park, M.; Kang, K.; Lim, S.K.; Shin, D.; Ko, K. Comparison of fitness cost and virulence in chromosome- and plasmid-mediated colistin-resistant Escherichia coli. Front. Microbiol. 2020, 11, 798. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I. The biological cost of mutational antibiotic resistance: Any practical conclusions? Curr. Opin. Microbiol. 2006, 9, 461–465. [Google Scholar] [CrossRef]
- Li, W.; Liu, Z.; Yin, W.; Yang, L.; Qiao, L.; Song, S.; Ling, Z.; Zheng, R.; Wu, C.; Wang, Y.; et al. MCR expression conferring varied fitness costs on host bacteria and affecting bacteria virulence. Antibiotics 2021, 10, 872. [Google Scholar] [CrossRef]
- Yang, Q.E.; Craig, M.C.; Papkou, A.; Pritchard, M.; Powell, L.; Thomas, D.; Andrey, D.O.; Li, M.; Spiller, B.; Yang, W. Compensatory mutations modulate the competitiveness and dynamics of plasmid-mediated colistin resistance in Escherichia coli clones. ISME J. 2020, 14, 861–865. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Yi, L.; Yu, L.; Wang, J.; Liu, Y.; Chen, X.; Lv, L.; Yang, J.; Liu, J.-H. Fitness Advantage of mcr-1–bearing IncI2 and IncX4 plasmids in vitro. Front. Microbiol. 2018, 9, 331. [Google Scholar] [CrossRef]
- Peyclit, L.; Sophie, A.B.; Rolain, J.M. Drug repurposing to fight colistin and carbapenem-resistant bacteria. Front. Cell. Infect. Microbiol. 2019, 9, 193. [Google Scholar] [CrossRef]
- Callaway, T.R.; Lillehoj, H.; Chuanchuen, R.; Gay, C.G. Conference report alternatives to antibiotics: A symposium on the challenges and solutions for animal health and production. Antibiotics. 2021, 10, 471. [Google Scholar] [CrossRef]
- EMA 2016. Updated Advice on the Use of Colistin Products in Animals within the European Union: Development of Resistance and Possible Impact on Human and Animal Health EMA/CVMP/CHMP/231573/2016. 2016. Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/updated-advice-use-colistin-products-animals-within-european-union-development-resistance-possible_en-0.pdf (accessed on 10 December 2021).
- EUCAST. EUCAST Breakpoints. Available online: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_8.1_Breakpoint_Tables.pdf (accessed on 4 June 2018).
- Rebelo, A.R.; Bortolaia, V.; Kjeldgaard, J.S.; Pedersen, S.K.; Leekitcharoenphon, P.; Hansen, I.M.; Guerra, B.; Malorny, B.; Borowiak, M.; Hammerl, J.A.; et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Eurosurveillance 2018, 23, 17-00672. [Google Scholar] [CrossRef]
- Lei, C.W.; Zhang, Y.; Wang, Y.T.; Wang, H.N. Detection of mobile colistin resistance gene mcr-10.1 in a conjugative plasmid from Enterobacter roggenkampii of chicken origin in China. Antimicrob. Agents Chemother. 2020, 64, 64. [Google Scholar] [CrossRef] [PubMed]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.H.; McDermott, P.F.; et al. Using the NCBI AMRFinder tool to determine antimicrobial resistance genotype-phenotype correlations within a collection of NARMS isolates. bioRxiv 2019, 550707. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Zhou, Z.; Sergeant, M.J.; Achtman, M. A genomic overview of the population structure of Salmonella. PLoS Genet. 2018, 14, e1007261. [Google Scholar] [CrossRef]
- Kaas, R.S.; Leekitcharoenphon, P.; Aarestrup, F.M.; Lund, O. Solving the Problem of Comparing Whole Bacterial Genomes across Different Sequencing Platforms. PLoS ONE 2014, 9, e104984. [Google Scholar] [CrossRef]
- Jeannot, K.; Bolard, A.; Plésiat, P. Resistance to polymyxins in Gram-negative organisms. Int. J. Antimicrob. Agents 2017, 49, 526–535. [Google Scholar] [CrossRef]
- Overbeek, R.; Begley, T.; Butler, R.M.; Choudhuri, J.V.; Chuang, H.Y.; Cohoon, M.; de Crécy-Lagard, V.; Diaz, N.; Disz, T.; Edwards, R.; et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005, 33, 5691–5702. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef]
| Strain | Serogroup | Isolation Year | MIC (mg/L) | MLST (ST) | Resfinder Detected Acquired Antimicrobial-Resistant Genes | Mutations in the Chromosome of COL-R Strains a | ResFinder Detected Mutations in COL-R Strains b | Metal Resistant Genes | Plasmid Finder [pMLST/pDLST/ FAB] c | Genbank acc. Number (mcr Plasmid acc. Number) c |
|---|---|---|---|---|---|---|---|---|---|---|
| 58/10/16 | S. Enteritidis | 2016 | 4 | ST3645 | neg | MdsC R298C PilN P107L YdeI R127L LolB S91R | neg | golTS | IncFII(S), IncFIB(S) [S1:A-:B22] | SAMN13046498 |
| 83/46/17 | S. Enteritidis | 2017 | 4 | ST3233 | neg | wt | neg | golTS | IncFII(S), IncFIB(S) [S1:A-:B22] | SAMN13046552 |
| 45/7/18 | S. Enteritidis | 2018 | 4 | ST11 | neg | ZraR R26L RfbN D107V | neg | golTS | IncFII(S) IncFIB(S) [S1:A-:B22] | SAMN13047690 |
| 77/84/18 | S. Typhimurium monophasic var. | 2016 | 4 | ST34 | mcr-1.1, mcr-5.1, sul2, aph(6)-Id, aph(4)-Ia, aac(3)-I v, aph(3’’)-Ib, tet(M)-like, blaTEM-1 | nd | neg | golTS, pcoSRDCA, silPABFCRSE, arsCBADRST, terWZD | IncHI2, IncHI2A, IncQ1 [ST-4] ColRNAI | SAMN13046551 (MZ666126) |
| 56/1/16 | S. Enteritidis | 2016 | 2 | ST11 | neg | wt | neg | golTS | IncFII(S), IncFIB(S) [S1:A-:B22] | SAMN13046483 |
| 4/23/16 | S. Enteritidis | 2016 | 1 | ST11 | neg | wt | neg | golTS | IncFII(S), IncFIB(S) [S1:A-:B22] | SAMN13039343 |
| 61/4/09 | S. Enteritidis | 2009 | 4 | ST11 | mcr-1.1, sul2, dfrA14, aph(3′′)-Ib,aph(6)-Id, tet(A), blaTEM-1 | nd | GyrA_D87Y | golTS | IncFII(S), IncFIB(S) [S1:A-:B22] IncX4 IncN [ST-3] | SAMN13046518 (OK605084) |
| Protein Name | PROVEAN | 58/10/16 | 83/46/17 | 45/7/18 |
|---|---|---|---|---|
| Multidrug efflux system, outer membrane factor lipoprotein of OprM/OprM family, MdsC | −7.963 | R298C | no mutation | no mutation |
| Type IV pilus biogenesis protein, PilN | −9.667 | P107L | no mutation | no mutation |
| Yde family stress tolerance OB-fold protein, YdeI | −2.914 | R127L | no mutation | no mutation |
| Outer membrane lipoprotein component of the lipoprotein transport system, LolB | −3.279 | S91R | no mutation | no mutation |
| Response regulator of zinc sigma-54-dependent two-component system, ZraR | −2.709 | no mutation | no mutation | R26L |
| O antigen biosynthesis rhamnosyltransferase, RfbN | −7.898 | no mutation | no mutation | D107V |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fortini, D.; Owczarek, S.; Dionisi, A.M.; Lucarelli, C.; Arena, S.; Carattoli, A.; Enter-Net Italia Colistin Resistance Study Group; Villa, L.; García-Fernández, A. Colistin Resistance Mechanisms in Human Salmonella enterica Strains Isolated by the National Surveillance Enter-Net Italia (2016–2018). Antibiotics 2022, 11, 102. https://doi.org/10.3390/antibiotics11010102
Fortini D, Owczarek S, Dionisi AM, Lucarelli C, Arena S, Carattoli A, Enter-Net Italia Colistin Resistance Study Group, Villa L, García-Fernández A. Colistin Resistance Mechanisms in Human Salmonella enterica Strains Isolated by the National Surveillance Enter-Net Italia (2016–2018). Antibiotics. 2022; 11(1):102. https://doi.org/10.3390/antibiotics11010102
Chicago/Turabian StyleFortini, Daniela, Slawomir Owczarek, Anna Maria Dionisi, Claudia Lucarelli, Sergio Arena, Alessandra Carattoli, Enter-Net Italia Colistin Resistance Study Group, Laura Villa, and Aurora García-Fernández. 2022. "Colistin Resistance Mechanisms in Human Salmonella enterica Strains Isolated by the National Surveillance Enter-Net Italia (2016–2018)" Antibiotics 11, no. 1: 102. https://doi.org/10.3390/antibiotics11010102
APA StyleFortini, D., Owczarek, S., Dionisi, A. M., Lucarelli, C., Arena, S., Carattoli, A., Enter-Net Italia Colistin Resistance Study Group, Villa, L., & García-Fernández, A. (2022). Colistin Resistance Mechanisms in Human Salmonella enterica Strains Isolated by the National Surveillance Enter-Net Italia (2016–2018). Antibiotics, 11(1), 102. https://doi.org/10.3390/antibiotics11010102







