Abstract
In a report by WHO (2014), it was stated that antimicrobial resistance is an arising challenge that needs to be resolved. This resistance is a critical issue in terms of disease or infection treatment and is usually caused due to mutation, gene transfer, long-term usage or inadequate use of antimicrobials, survival of microbes after consumption of antimicrobials, and the presence of antimicrobials in agricultural feeds. One of the solutions to this problem is antimicrobial peptides (AMPs), which are ubiquitously present in the environment. These peptides are of concern due to their special mode of action against a wide spectrum of infections and health-related problems. The biomedical field has the highest need of AMPs as it possesses prominent desirable activity against HIV-1, skin cancer, breast cancer, in Behcet’s disease treatment, as well as in reducing the release of inflammatory cells such as TNFα, IL-8, and IL-1β, enhancing the production of anti-inflammatory cytokines such as IL-10 and GM-CSF, and in wound healing properties. This review has highlighted all the major functions and applications of AMPs in the biomedical field and concludes the future potential of AMPs.
1. Introduction
Microorganisms can be useful or hazardous, and some microbes show unrecognizable effects. Microbes with beneficial effects are important and are utilized for CO2 fixations, the degradation of complex organic compounds into basic molecules, fermentation, and many more applications. Furthermore, microorganisms can have a harmful effect, causing infections or diseases such as anthrax, conjunctivitis, ringworm, influenza, and many more [1]. Treatment of such health issues caused by microbes can be performed by appropriate diagnosis followed by adequate medication or therapy [2]. Antimicrobial compounds are considered a convenient solution for such health issues and can be of low molecular weight (LMW) or high molecule weight (HMW). Diverse relationships have been observed between the molecular weight and antimicrobial activity of these compounds. However, the majority of researchers report an increase in antimicrobial activity with the decrease in the degree of polymerization (i.e., molecular weight) [3]. It is expected that the adhesion of HMW antimicrobial polymers to a negatively charged bacterial cell membrane should be highly effective in comparison to the adhesion of LMW antimicrobial polymers. Instead, contradicting results are found, according to which LMW antimicrobial polymers have greater biocidial activity [4]. Usually for bacterial infections, antibiotics are the most commonly used drug for treatment, which show adverse effects such as reducing immunity, increasing susceptibility to infections, and developing resistance against antimicrobial agents [5].
Excess use of antibiotics results in antimicrobial resistance, which has devastating effects. This has led to the increase in demand for low molecular weight antimicrobial peptides, which are effective against antimicrobial-resistant bacteria [6].
Antimicrobial peptides (AMPs) are short-chain (5 to 100) amino acids that possess the ability to counter microbial attacks or any infective agent in all living organisms [7]. AMPs act as the first line of defense to disrupt bacterial, fungal, yeast, viral, and even cancer cells [8]. Most AMPs are found to be cationic, and are considered promising agents due to their distinctive mode of action and the inability of microbes to develop resistivity against them [9,10]. However, some of the AMPs are also anionic in nature due to the acidic polar residues, and these consist of short Asp-rich sequences [11]. More than 2500 AMPs are present in nature, which are broadly grouped into four categories: α helical, β sheet, extended, and αβ mixed antimicrobial peptides [12,13]. α helical AMPs possess a distance of about 0.15 nm between two adjacent bond angles (for example, LL-37) [14]. The β Sheet peptides are rigid structures that are more organized in an aqueous solution and do not show conformational change on membrane linkage (for example, Gomesin) [15]. Extended peptides comprise a high amount of arginine, tryptophan, proline, and/or histidine residues (for example, indolicidin and α1-purothionin) [16,17,18]. On the basis of activity, AMPs can also be classified as antimicrobial, antiprotozoal, anticancer, insecticidal, and antiparasitic [19]. AMPs can be harvested from invertebrates, vertebrates, and plants [20]. The first AMPs derived from certain sources were as follows: Cecropin from Hyalophora cecropia [21], defensis from rabbit [22], purothionin from wheat flour [23], and Gramicidin D from Bacillus brevis [24]. Among all the sources, some of the most valuable sources of AMPs with future prospects are marine animals [25], wild plants and weeds [26], and the black soldier fly [27]. A comparison between the antibiotics and AMPs is mentioned in Table 1.

Table 1.
Comparative study between antibiotics and AMPs.
Similar to AMPs, a group of polymers exists, known as synthetic antimicrobial oligomers, which are cationic and amphiphilic in nature [32]. These antimicrobial agents are fabricated to imitate AMPs [33]. However, these oligomers have a drawback of heterogeneity and innate toxicity [34]. Thus, antimicrobial peptides have the advantage of low toxicity over the synthetic antimicrobial oligomers and are highly effective in microbial disruption [35]. Recent studies have shown the impactful results of using AMPs in the biomedical sector. This article is different from various other papers on AMPs because it not only includes information about the factors affecting the functioning of AMPs and their important role, but along with it, focuses on the implementation techniques of AMPs in the biomedical sector. In this review, smart and intelligent delivery methods are also considered. Thus, this review could be helpful for readers that require compact information about the role of AMPs in biomedical applications. The schematic representation of this review is shown in Figure 1.
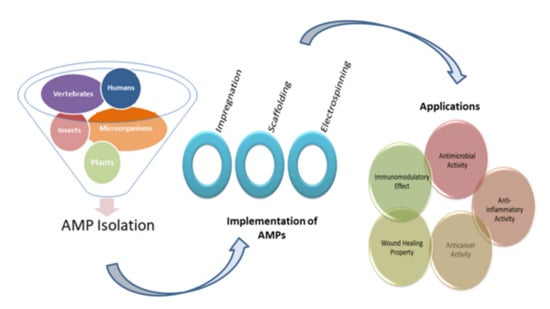
Figure 1.
Diagrammatic representation of the five main sources for the isolation of AMPs, three widely used implementation techniques, and their applications.
2. Factors Affecting the Functioning of AMPs
An upgraded database of the Collection of Antimicrobial Peptide comprises 18.7% helical structures of AMPs, 18.9% β-strand, and 60.1% mixed or coil [36]. The assortment of AMP structures suggests the diverse mechanism of action in the inhibition of microbial cells beyond pore formation. Characteristics such as the secondary structure, charge, hydrophobicity, hydrophobic moment, amphipathicity, polar angle, and peptide length are responsible for modulating the inhibitory mechanism of AMPs against microbes [37,38].
2.1. Secondary Structure
Secondary structure plays a significant role in improving the antimicrobial activity of AMPs. It is observed that among all the structures, the α helical structure promotes the insertion of antimicrobial peptides into the cellular membrane due to its facially amphiphilic structure [39]. In a previous investigation, two AMPs were compared, C5A (single helix) and AH (structure consisting of a hinge in between two short helixes), among which C5A had a more uniform helical structure. Analysis showed that a 10 nm concentration of C5A was capable of lysing lipid vesicles, whereas AH showed comparatively less potency [40].
2.2. Charge
Most of the AMPs are cationic in nature ranging between +2 and +9 [41]. In addition, Porto reported a range between +3 and +9 [42]. Charge plays a critical role in AMPs’ activity, which is reduced beyond the optimum level of charge [43]. Removal of the cationic terminus from melittin reduced the hemolytic activity and the ability to bind the membrane and can be used to modulate membrane selectivity [44]. Cationic AMPs bind with membranes consisting of negatively charged lipids due to an electrostatic interaction [45]. Anionic AMPs have a net positive charge but also consist of a few cationic residues, which interact with the membrane in a similar manner to cationic AMPs [46,47].
2.3. Hydrophobicity
Microbial membranes are protected from exogenous matter such as polysaccharides, proteins, and peptides due to its hydrophobic characteristics, but AMP has the ability to interact with the microbial membrane [48]. For instance, magainin has the ability to inhibit the growth of Gram-negative bacteria, but the analogs with improved hydrophobicity have shown effectiveness against Gram-positive bacteria as well [49].
2.4. Amphipathicity
Amphipathic peptides are ubiquitously present in nature with variation in their attributes [50,51]. The amphipathicity of AMPs can be defined as the ability to survive in solution under both hydrophobic and hydrophilic conditions [52]. Amphipathicity helps in improving inhibitory activity against microorganisms [16]. Narayana designed a trypsin-rich AMP (WW291) and studied the antimicrobial activity of eight permutated sequences (WW291 to WW298). From the results, he concluded that WW295 has increased antimicrobial activity four-fold against Klebsiella pneumonia. The appreciable activity of WW295 was supposed to be due to the presence of hydrophobic residues at the bottom and two hydrophilic amino acids at the top [53].
2.5. Hydrophobic Moment
The hydrophobic moment is similar to the electric dipole moment, and it can be defined as the lack of equality of hydrophobicity in the AMP’s structure [54]. The hydrophobic moment shows changes in rearranging the sequence [55]. With an increasing hydrophobic moment, antimicrobial peptides’ activity is found to be enhanced. Myxinidin was harvested from hagfish, then its analogues were developed, which are proved to serve as templates for the therapeutic agent. In this research, four AMPs were studied—myxinidin, myxinidin1, myxinidin2, and myxinidin3. Among all the antimicrobial peptides, myxinidin2 and myxinidin3 had the highest hydrophobic moments, which were 0.410 and 0.414, respectively. This factor has shown direct proportionality with the minimum inhibitory concentration (MIC) to kill microorganisms [56].
2.6. Polar Angle
The polar angle of AMPs can be defined as the relative proportion of polar to non-polar residues of the helix. The polar angle will be 180° if the number of polar residues is equal to non-polar residues. The polar angle is higher, if the number of hydrophobic residues is higher than hydrophilic residues, and visa-versa [57]. A lower polar angle signifies the better ability of the peptide to permeate the membrane [58]. This is also proved by another study, in which two staphylococcal peptides, warnericin RK and PSMα, were isolated and their anti-legionella activity was analyzed. Results suggested that the higher polar angle (around 140°) of both the peptides led to the need for aggregates of a bigger size to disrupt the microbial membrane [59].
2.7. Peptide Length
Peptide length is also among the critical attributes that affect the activity efficiency of AMPs. Juba investigated the effect of peptide length on membrane disruption using full-length cationic AMPs (NA- CATH) and truncated isomers (L- ATRA-1A and D- ATRA-1A). The activity of these peptides was tested against Escherichia coli and Bacillus cereus. It was observed that NA-CATH has higher antimicrobial potency than truncated peptides. It was also noted the peptide length affected the mechanism of action of peptides, and the full-length peptide disrupted the membrane via liposome lysing, whereas the truncated peptide caused liposomes’ exudation followed by fusion and aggregation [60].
3. Functions of AMPs
3.1. Disruption of Bacteria
AMPs have antimicrobial activity against a vast range of microorganisms and follow membrane disruption or non-membranolytic destruction of microorganisms [61,62]. In a previous study, stomoxynZH1 was extracted from Hermetia illucens via the RNA extraction method, which showed the inhibitory activity against Staphylococcus aureus, E. coli, Rhi-zoctonia solani, and Sclero-tinia sclerotiorum [63]. AMPs also play an important role in fighting against antibiotic-resistant bacteria. Polymyxin B has the capability of inhibiting the growth of multidrug-resistant Pseudomonas aeruginosa, Klebsiella pneumoniae, and Acinetobacter baumannii [64,65,66]. LS-sarcotoxin and LS-stomoxyn isolated from Lucilia sericata were found to inhibit 90% of Salmonella enterica, E. coli, Acinetobacter baumannii, and Enterobacter cloacae [67]. Some of the research proving the antimicrobial activity of AMPs are mentioned in Table 2.

Table 2.
List of some of the AMPs with putative antibacterial activity.
3.2. Antifungal Activity
AMPs possess inhibitory activity against fungi [78]. In previous research, Roscetto isolated VLL-28 from an archaeal protein and examined the antimicrobial efficacy against 10 variants of Candida species. Among all the species, VLL-28 showed the lowest minimum inhibition concentration of 44.25 μg/mL against C.tropicalis 54 and C.tropicalis 2 and the highest MIC of 177 μg/mL against C. glabrata 28 and C. glabrata 34. Hence, it was concluded that VLL-28 has antifungal activity against planktonic cells and mature biofilms of Candida species [79]. AMPs with an antifungal property are mentioned in Table 3.

Table 3.
List of some of the AMPs with a putative antifungal activity.
3.3. Antiviral Activity
The competency of the virus to replicate overwhelmingly is one of the causes of concern for life-threatening effects [92]. AMPs are effective and have minor side effects, which make them a potential alternative for the treatment of viral infection [93]. Zhang examined the antiviral potency of DP7 (VQWRIRVAVIRK) against SARS coronavirus. He claimed that DP7 has the ability to inhibit SARS-CoV and SARS-CoV-2 infection by examining its activity via cell receptor ACE2. He also reported that 104 μg/mL and 73.625 μg/mL (50% inhibitory concentration) of DP7 is required for inhibiting the SARS-CoV and SARS-CoV-2 pseudovirus [94]. Some of the research proving the antiviral activity of AMPs is mentioned in Table 4.

Table 4.
List of some of the AMPs with putative antiviral activity.
3.4. Inhibition of Cancer Cell Growth
Treatment of cancer has unbounded usage due to the easy development of resistance and the toxicity issue [104]. This has led the focus towards AMPs, which have the ability to resist cancer growth. Zhao reported the anticancer activity of the HPRP-A1 peptide isolated from Helicobacter pylori [105]. Further, the combined effect of iRGD (homing peptide) and HPRP-A1 were examined for enhancement in anticancer activity. Furthermore, the results suggested that iRGD helped in improving the penetration of HPRP-A1 on A549 MCS [106]. L-K6 is reported to be capable of killing MCF-7 breast cancer cells via nuclear disruption without cell surface disruption [107]. AMPs with the ability to inhibit cancer cell growth are mentioned in Table 5.

Table 5.
List of some of the AMPs that can be used in cancer therapeutic studies.
3.5. Immunomodulatory Effect
Several cationic AMPs are observed to have immunomodulatory characteristics due to two main reasons: Stimulation capability to induce chemokines and decrease in the release of undesirable pro-inflammatory cytokines [119,120,121,122]. In an investigation, clavanin A was modified to clavanin MO by inserting hydrophobic residues into the conserved oligopeptide FLPII to obtain both antimicrobial and immunomodulatory properties in the same peptide. The immunomodulatory effect of AMPs clavanin-MO isolated from marine tunicate was studied on the murine macrophage. It was observed from the results that clavanin-A and clavanin MO-treated samples showed an increase in the production of IL-10 (anti-inflammatory cytokine) and a decrease in IL-12 (a pro-inflammatory cytokine). Hence, it is concluded that clavanin-MO modulates the innate immune system due to its ability to stimulate leukocyte recruitment at the infection area and produce GM-CSF, IFN-γ, and MCP-1 [123]. A short synthetic peptide of 12 amino acids (RR) was designed with improved selectivity, antibacterial activity, and lesser toxicity. The immunomodulatory effect was analyzed by the results of acidity on antibacterial activity via measuring MIC, and the mechanism of bacterial inhibition was also explored in this study. Among all the analogues of RR, D-RR4 showed the lowest MIC50 and MIC90, which were 2 and 4 µM against P. aeruginosa and A. baumannii [124]. Some of the studies proving the immunomodulatory activity of AMPs are mentioned in Table 6.

Table 6.
List of some of the antimicrobial peptides suggesting an immunomodulatory effect.
3.6. Anti-Inflammatory Effect
AMPs are good candidates for suppressing inflammatory activity. The anti-inflammatory property of melectin was harvested from Melecta albifrons (bee venom), and cationic AMPs was studied via qRT-PCR. In this assessment, four groups were considered: Control (untreated human fibroblast), melectin-treated fiberoblast, S. aureus-infected fibroblast, and fibroblast treated with melecitin in the presence of S. aureus infection. In the melictin-treated group, no effect was observed in the reduction of pro-inflammatory cytokines. Meanwhile, the S. aureus-infected human fibroblast showed the maximum reduction in the release of TNFα, IL-8, IL-6, and IL-1β in melectin (5 µM) treated samples [134]. AMPs possessing anti- inflammatory activity are mentioned in Table 7.

Table 7.
List of some of the AMPs suggesting an anti-inflammatory effect.
3.7. Wound Healing
Several reports have suggested a wound healing property of AMPs [141]. Wound healing activity of recombinant P-LL37, which is derived from the human cathelicidin antimicrobial peptide LL37, was studied on dexamethasone-treated mice. Mice were cleaned and a 5 mm-diameter wound was created on the dorsal surface under aseptic conditions. Two times a day, wounds were treated with LL37 and sterile water. A re-epithelialization study suggests that the keranocytes layer is complete in AMPs-treated samples, whereas in untreated samples, the keranocytes layer is incomplete. A higher number of blood vessels was observed in treated samples compared to untreated samples [142]. Several research studies on the ability to effectively heal wounds are mentioned in Table 8.

Table 8.
List of some of the AMPs that suggest wound-healing properties.
4. Implementation Techniques
4.1. Impregnation of AMPs
The incorporation of AMPs into multilayers is the most commonly studied technique because of its ability to reduce the loss of antimicrobial activity and the fact that it requires a low quantity of AMPs [150,151]. The layer-by-layer technique was used to incorporate AMP (nisin Z) into polyelectrolyte multilayers (PEM) made up of chitosan and carrageenan (CAR). This study involved three groups: PEM without AMP, PEM with AMP, and PEM with an AMP/CAR outer layer. Among all the groups, PEM consisting of nisin showed the highest antimicrobial activity against S. aureus and MRSA. During adsorption, some of the nisin Z was lost, and 0.89 ± 0.064 µg cm−2 was retained [152]. Antimicrobial peptides-embedded cotton gauze (18 mm × 18 mm), functionalized with chitosan and alginic acid sodium salt, was examined as a novel method of antimicrobial wound-dressing. This method involved the soaking of cotton gauze in chitosan and alginic acid sodium salt for a 5-min duration. It was then immersed in AMP, followed by washing with deonized water and coating with chitosan. Four antimicrobial peptides were studied: hBD-1, β-Defensin-1, human dermaseptin, Cys-LC-LL-37, and Magainin 1. The lowest MIC was observed for Magainin 1 against S. aureus and K. pneumoniae. The release study recorded the release after 6 h of immersion and also noted that Dermaseptin had the fastest release and Magainin had the slowest among all the samples. It was found that after 24 h, only 75% of AMPS was utilized from the cotton gauzes [150]. Cubosome is a nanoparticle representing a lipid biomimteric environment that has been studied for the incorporation of AMPs [153]. Gramicidin A and alamethicin were incorporated into cubosome (composed of monoolein, monopalmitolein, monovaccenin, and 1-(7Ztetradecenoyl)-rac-glycerol). The range of AMP concentration was considered to be from 0 to 10% and it is observed that with the increase in concentration, cubic symmetry was decreased. The loss of structure can be prevented by using the lipid bilayer, which is approximately equal to the peptide length [154]. Methods of incorporating AMPs into a matrix are shown in Figure 2.
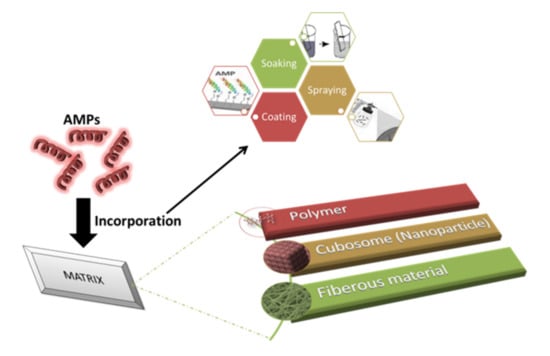
Figure 2.
Impregnation of AMPs into compatible matrices. Incorporation of AMPs in a matrix is usually performed via soaking, coating, or spraying. The three common matrices are polymer, cubosome, and fibrous material.
4.2. Scaffolding
Scaffolding is a three-dimensional porous solid biomaterial that acts as a matrix to hold bioactive compounds [155]. This method can be used to incorporate AMP to obtain the desired features. In an investigation, LL37, melittin, buforin, MM1, AR23, and RV23 were scaffolded in protofibril to study the immunomodulatory potency. All of the AMPs formed square columnar lattices. These scaffolds were incubated with human pDCs and the production of IFN-α was recorded. The results suggest that LL37-dsDNA and buforin-dsDNA stimulate significantly higher production of IFN-α compared to the control. Scaffolding of AMPs in ordered nanocrystalline complexes helps in amplifying the production of cytokines to modulate immunity [156]. Ye prepared a scaffold from intrafibrillar mineralized collagen via the biomimetic technique incorporated with GL13K peptides to treat bone defects. This research involves the study of collagen mineralized for four different durations (1, 2, 4, and 8 days). The morphology suggested that 4 days and 8 days of mineralization led to homogeneity and a high degree of mineralization. Mineralization was more useful in the proper loading of AMP in comparison to non-mineralized scaffolds. The release of GL13K was not observed for up to 14 days, but a drastic release was reported from 21 to 28 days, which may be due to collagen hydrolysis. The hydrophobicity of the scaffold was improved by coating it with GL13K, which suggests stability in the presence of water. GL13K scaffolds, which were mineralized for two days or more, reduced S. gordonii viability in a better way compared to others [157]. Rabanal and a few others developed a scaffold from lipopeptide, which is found to possess improved antimicrobial activity against P. aeruginosa, E. coli, S. aureus, and E. faecalis [158].
4.3. Electrospinning
Electrospinning is a novel technology that consists of a high-voltage power supply, a syringe, which is the producer of nanofiber from the polymeric solution (includes pump and needle), and a collector surface on which the polymeric solution is coated during the process [159]. Electrospinning is an effective and inexpensive technique used in biological and medical applications, which involves the incorporation of a strong electric field to develop nanofibers [160]. Electrospinning technology is used for biomedical purposes such as electrospun prosthetic heart valves [161]. Shortcomings in such innovations concern microbial deterioration, which can be avoided with the use of antimicrobial agents. Various studies have proved the use of AMPs instead of antimicrobial agents to prevent the risk of infection of electrospun materials [162]. For instance, Cm-p1 is derived from the Caribbean Sea mollusk, which was added in PVA and used in electrospinning a solubilized solution of Cm-p1 with 2.5%, 5%, and 10%, w/v concentrations in 0.5 mL of deionized water. Further, a polymeric solution was prepared by adding 50 g of PVA [163]. This solution is pumped at a flow rate of 0.2 mL.h−1 via a syringe at 15 kV of voltage with a distance of 10 cm between the needle tip and collector. The releasing property of the nanofiber was studied using RP-HPLC and the sustainable release of Cm-p1 was observed for up to 48 h. Only Cm-p1 10%-PVA inhibited C. albinas growth, resulting in a 3.94 mm diameter of the zone. The immune response was studied via the release of cytokines IL-6 and TNF-α by RAW 264.7 macrophages cells. These cytokines had the ability of low induction by Cm-p1 10%-PVA [164]. Similar electrospinning conditions were considered in another study. Pleurocidin in four different concentrations (0.03 wt.%, 0.06 wt.%, 0.12 wt.%, and 0.25 wt.%) was mixed with 12% PVA. PVA with 0.25% pleurocidin inhibited E. coli growth within 5 h of storage. The antimicrobial property in apple cider was also assessed, where results suggest that within 14 days, PVA with 0.25% pleurocidin inhibited E. coli growth to an undetectable level. They concluded that electrospinning is useful in controlling the release of antimicrobial peptides [165]. Figure 3 represents the schematic diagram for the electrospinning process.
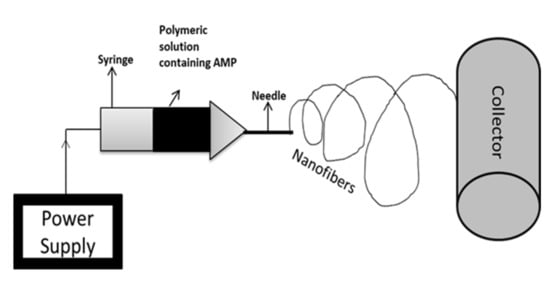
Figure 3.
Schematic representation of electrospinning technique. Electrospinning is a nanofabrication method that involves the controlled flow of a polymeric solution placed in a syringe via a needle. The collector is the material on which nanofibers are coated.
5. Applications in Biomedical Sector
5.1. Vertebrate-Derived Antimicrobial Peptides
Vertebrates are mammals with spinal cords, such as humans, marine animals, birds, and other animals. AMPs isolated from vertebrates are studied due to their potency against a wide range of microorganism. The most commonly found and highly investigated antimicrobial peptides are defensins and cathelicidins [166]. Defensins are positively charged antimicrobial peptides composed of 29–34 amino acids in a β-sheet structure [167]. They are categorized into three groups—α-, β, and θ-defensins—and possess antimicrobial activity in the range of 0.5–5 μm [166,168]. A-Defensins are found in inflamed tissues [169], β-defensins are present in bovine neutrophils [170], and θ-defensins can be harvested from neutrophils and bone marrow [171]. Cathelicidin is a multifunctional peptide that has conserved pro-peptide sequences and is identified as an N-terminal signal peptide [172,173]. They are harvested from porcine intestines and bovine neutrophils [174,175]. These peptides have shown wound healing activity, immunomodulatory effect and apoptosis [176].
Shaat investigated serum α-defensins 1–3 and salivary α-defensins 1–3 for Behcet’s disease treatment. The receiver operating characteristic was noted as 0.743 and 0.936 for serum and salivary α-defensins, respectively, suggesting potential in the pathogenesis of Behcet’s disease [177]. Human defensins-5 has shown beneficial effects in treating dysbiosis [178]. Tracheal AMP (β-defensins) has shown resistance to Mycoplasma bovis, which is a respiratory pathogen [179]. Crohn’s disease can be eliminated by inducing β- defensins [180]. In patients suffering from periodontitis, β-defensins play a critical role in bone repairment [181]. Cole studied a peptide named retrocyclin (θ-defensins), which prevents infection by T-and M-tropic strains of HIV-1 [182]. θ-defensins has the capability to suppress inflammatory cytokines and is also used in therapy for the herpes simplex virus [183,184]. Retrocylin-2 has potency in treating Influenza A Virus [185].
The potency of cathelicidins LL-37 was examined on human MDMs and THP-1 cells, which were infected with Mycobacterium tuberculosis. These cells were infected with the M. tuberculosis H37Rv for four hours, followed by treatment with 1 µg/m of LL-37 for 24 h. It was concluded that LL-37 mediates the activation of autophagy via the P2RX7 receptor and inhibits the growth of Mycobacterium tuberculosis [186]. In another study, the Cathelicidins-inspired peptides have shown antifungal activity against Fusarium, Aspergillus, Cryptococcus, Malassezia, Candida, and Talaromyces [187]. Applications of vertebrate-derived AMPs are not limited to the few above-discussed AMPs; instead, there are many AMPs with great potential to treat infections and health issues. For instance, Scapularisin-3 and Scapularisin-6 isolated from Ixodes scapularis are reported to have a strong inhibitory property against Fusarium culmorum and F. graminearum [188].
5.2. Insect Derived AMPs
Among all sources of AMPs, insects are resistant to a wide range of microorganisms due to their tolerance to harsh living conditions [189]. Insect AMPs are found to have great potential in treating skin cancer [190]. CopA3 isolated from Copris tripartitus has proved the ability to treat gastric cancer and leukemia as well [117,191]. An evaluation of antibacterial and inflammatory properties of DLP2 and DLP4 isolated from Hermetia illucens and expressed in P. pastoris was performed. The time-dependent relation was recorded for antimicrobial activity against MRSA. It was observed that Log10 (CFU/mL) of S. aureus was reduced to 1.68‒1.89 and 1.06‒1.34 for DLP2 and DLP4, respectively. DLP4 destroyed 99% of MRSA within 2 h, while DLP2 took 6 h. It was also concluded that DLP4 has two- to four-fold higher activity than DLP2 against MRSA. In experimentation on mice, it was observed that DLP4 (7.5 mg/kg) has better efficiency against S. aureus inhibition than DLP2 (7.5 mg/kg). A smaller antibiotic effect was reported in treatment with DLP2 compareed to DLP4 [128].
5.3. Plant Derived AMPs
The smallest AMPs were derived from jatropha curcas with seven amino acids [192]. Commonly found AMPs in plants are the cyclotide family, thionins, the α-Hairpinin family, Hevein-like peptides, lipid transfer protein, knottin-type peptides, snakins, and plant defensins [193]. Thionin is a useful AMP for the development of a glucose detection biosensor. Salimi and coworkers induced thionin in multiwall carbon nanotubes to develop a sensor that selectively detects glucose depending on the cathodic peak current [194]. Another probe was fabricated as a lung cancer biomarker using the nanohybrid of graphene oxide-thionin-hemin-Au with a low detection limit of 0.026 pg mL−1. In this probe, graphene oxide acted as a supporting material in which thionin and hemin were immobilized followed by a reduction of silver particles by thionin [195]. Hu and coworkers harvested three novel cyclotides from roots and leaves of Hedyotis diffusa. Experimental data suggest the inhibition of invasion of LNCap cells, which confirms the anti-cancer effect [196]. An analogue of Cp-thionin II (KT43C) was isolated from cowpea and the study showed that the growth of F. culmorum, P. expansum, and A. niger is inhibited [197]. Neutrophil elastase-associated diseases can be prevented by using roseltide rT1, which is derived from Hibiscus sabdariffa [198]. Plant defensins MtDef5 extracted from Medicago truncatula have shown the ability to permeate the membrane of F. graminearum and Neurospora crassa [199]. Hence, AMPs harvested from plant sources have a wide spectrum of applications in the biomedical field with future prospects.
5.4. Microorganism Derived AMPs
AMPs derived from bacteria are usually termed bacteriocin, such as nisin [200]. Nisin is isolated from Gram-positive bacteria [201,202]. The anticancer effect of nisin has been reported [203,204]. Begde reported the immunomodulatory effect of nisin due to its ability to activate neutrophils [205]. Other AMPs from microorganisms such as enterocins DD28 and DD93, derived from Enterococcus faecalis, have shown inhibitory activity against MRSA [206]. Table 9 represents the biomedical applications of nisin.

Table 9.
Biomedical applications of nisin (AMPs derived from bacteria).
6. Smart and Intelligent Delivery of AMPs
AMPs, though capable enough to prevent various biological and medical issues, still have limited commercial applications. The release of antimicrobial molecules in the presence or absence of infections (i.e., uncontrolled release) is one of the major concerns [210]. Novel drug delivery systems include smart and intelligent mechanisms that are capable of adjusting the release rate of drugs according to the physiological conditions of the patients [211]. Novel delivery systems are classified into four groups: Systems in which the release rate depends on a specific targeting moiety, activation-modulated delivery systems, systems in which the release rate depends on triggering agents, and systems with the programmed release of the drug [212].
Other than the uncontrolled release rate, factors such as low biocompatibility, low metabolic stability, low solubility, high susceptibility to degradation, and toxicity act as a hurdle in applications of AMPs (Figure 4) [213].
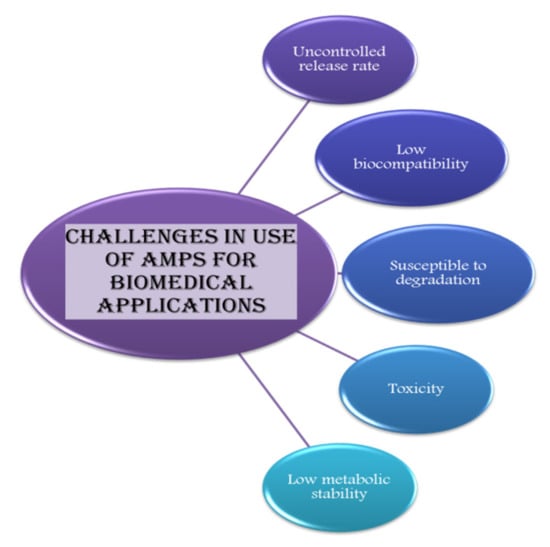
Figure 4.
Amps can be used for various applications in the biomedical sector but its use is limited due to certain challenges, which are mentioned in this diagrammatic illustration.
The incorporation of antimicrobial peptides into a nanostructure can work as a solution to these issues. Some of the nanosystems researched are metal nanoparticles [214], carbon nanotubes [215], liposomes [216], liquid crystalline particles [217], dendritic systems [218], aptamers [219], hydrogels [220], polymers [221], and cyclodextrins [222].
Among all, to combat barriers in the delivery of AMPs, nanofibers are considered an excellent method that can be produced by electrospinning or self-assembly [223]. Electrospinning is a process that involves the deposition of a polymeric solution on the substrate surface via an electric field [224]. Various applications of electrospinning involved in AMPs’ incorporation are mentioned in Section 4.3. The self-assembled system has a precise structure that involves nanocarriers for target delivery [225]. AMPs L-5 was incorporated into poly (ethylene glycol)-co-acrylic acid microgel, and the results suggest that this self-assembly significantly limited the release of AMPs, improving the anti-adherent property and antimicrobial activity [226]. These novel techniques have the potential to overcome challenges in applications of AMPs in the biomedical field.
7. Conclusions
AMPs act as bioactive compounds with huge potential in the field of the biomedical sector. AMPs are harvested from various sources, consisting of varying structures and characteristics. These AMPs are studied in depth to obtain a better understanding of the structure, mechanism of action, and applications. Various studies have proved the significance of AMP over other drugs for the treatment of infections and health issues. It has been claimed to be a better option for treatment due to its eminent anti-inflammatory effect, immunomodulatory activity, antimicrobial activity, anticancer activity, and wound-healing properties. On the contrary, AMPs have several disadvantages that limit the application of AMPs in the biomedical field, such as the uncontrolled release rate, which results in loss of the antimicrobial peptide and reduces its efficiency. The isolation of antimicrobial peptides is another challenge, which is a tedious and costly process. Preservation of activity of AMPs throughout its expected use is also a point of concern. AMPs are further under investigation to overcome these challenges by designing a better system with a controlled release rate, efficient functioning at desired conditions, and minimum or no sensitivity, to deliver expected results. The incorporation of AMPs into a matrix can be performed in several ways by altering the preparation conditions, which affects the end result and activity of AMPs. This article has discussed most of the recent studies on the properties and impregnation methods of AMPs, along with their scope in the biomedical sector.
Author Contributions
A.S. carried out the literature search, analyzed, and wrote the manuscript. H.L. commented on and edited the manuscript. S.R. supervised the project and provided scope and directions. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gupta, A.; Gupta, R.; Singh, R.L. Microbes and environment. In Principles and Applications of Environmental Biotechnology for a Sustainable Future; Springer: Singapore, 2017; pp. 43–84. [Google Scholar]
- Leekha, S.; Terrell, C.L.; Edson, R.S. General principles of antimicrobial therapy. In Mayo Clinic Proceedings; Elsevier: Amsterdam, The Netherlands, 2011; Volume 86, pp. 156–167. [Google Scholar]
- Grace, J.L.; Huang, J.X.; Cheah, S.E.; Truong, N.P.; Cooper, M.A.; Li, J.; Davis, T.P.; Quinn, J.F.; Velkov, T.; Whittaker, M.R. Antibacterial low molecular weight cationic polymers: Dissecting the contribution of hydrophobicity, chain length and charge to activity. RSC Adv. 2016, 6, 15469–15477. [Google Scholar] [CrossRef]
- Ahmed, M.S. Synthesis of Antimicrobial Polymers to Overcome Antimicrobial Resistance. Ph.D. Thesis, Florida International University, Miami, FL, USA, 12 June 2017. [Google Scholar]
- Willing, B.P.; Russell, S.L.; Finlay, B.B. Shifting the balance: Antibiotic effects on host–microbiota mutualism. Nat. Rev. Microbiol. 2011, 9, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.B.; Seo, J. Antimicrobial peptides under clinical investigation. Pept. Sci. 2019, 111, e24122. [Google Scholar] [CrossRef]
- Jenssen, H.; Hamill, P.; Hancock, R.E. Peptide antimicrobial agents. Clin. Microbiol. Rev. 2006, 19, 491–511. [Google Scholar] [CrossRef] [PubMed]
- Mondal, N.R.; Jagdale, D.M. AntimicrobialPeptides A Review on its Types, Mechanism of Action, Synthesis and Therapeutic Applications. Int. J. For. Pharm. Res. Sch. 2016, 5, 72–81. [Google Scholar]
- Hultmark, D. Drosophila immunity: Paths and patterns. Curr. Opin. Immunol. 2003, 15, 12–19. [Google Scholar] [CrossRef]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.; Schneider, G. Designing antimicrobial peptides: Form follows function. Nat. Rev. Drug Discov. 2012, 11, 37–51. [Google Scholar] [CrossRef]
- Almarwani, B.; Phambu, N.; Hamada, Y.Z.; Sunda-Meya, A. Interactions of an Anionic Antimicrobial Peptide with Zinc (II): Application to Bacterial Mimetic Membranes. Langmuir 2020, 36, 14554–14562. [Google Scholar] [CrossRef]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar]
- Bahar, A.A.; Ren, D. Antimicrobial peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef]
- Takahashi, D.; Shukla, S.K.; Prakash, O.; Zhang, G. Structural determinants of host defense peptides for antimicrobial activity and target cell selectivity. Biochimie 2010, 92, 1236–1241. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Haney, E.F.; Vogel, H.J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011, 29, 464–472. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef]
- Kang, X.; Dong, F.; Shi, C.; Liu, S.; Sun, J.; Chen, J.; Li, H.; Xu, H.; Lao, X.; Zheng, H. DRAMP 2.0, an updated data repository of antimicrobial peptides. Sci. Data 2019, 6, 1–10. [Google Scholar] [CrossRef]
- Reddy, K.; Yedery, R.; Aranha, C. Antimicrobial peptides: Premises and promises. Int. J. Antimicrob. Agents 2004, 24, 536–547. [Google Scholar] [CrossRef]
- Hultmark, D.; Steiner, H.; Rasmuson, T.; Boman, H.G. Insect Immunity. Purification and Properties of Three Inducible Bactericidal Proteins from Hemolymph of Immunized Pupae of Hyalophora cecropia. Eur. J. Biochem. 2005, 106, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, J.G. Phagocytin: A bactericidal substance from polymorphonuclear leucocytes. J. Exp. Med. 1956, 103, 589–611. [Google Scholar] [CrossRef] [PubMed]
- De Caleya, R.F.; Gonzalez-Pascual, B.; Garcia-Olmedo, F.; Carbonero, P. Susceptibility of phytopathogenic bacteria to wheat purothionins in vitro. Appl. Microbiol. 1972, 23, 998–1000. [Google Scholar] [CrossRef] [PubMed]
- Hotchkiss, R.D.; Dubos, R.J. The Isolation of Bactericidal Substances from Cultures of Bacillus Brevis. J. Biol. Chem. 1941, 141, 155–162. [Google Scholar] [CrossRef]
- Ohtsuka, Y.; Inagaki, H. In silico identification and functional validation of linear cationic α-helical antimicrobial peptides in the ascidian Ciona intestinalis. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Odintsova, T.; Egorov, T. Plant antimicrobial peptides. Plant. Signal. Pept. 2012, 16, 107–133. [Google Scholar] [CrossRef]
- Thomas, S.; Karnik, S.; Barai, R.; Jayaraman, V.K.; Idicula-Thomas, S. CAMP: A useful resource for research on antimicrobial peptides. Nucleic Acids Res. 2009, 38, D774–D780. [Google Scholar] [CrossRef] [PubMed]
- McKenna, M. Antibiotics set to flood Florida’s troubled orange orchards. Nature 2019, 567, 302–303. [Google Scholar] [CrossRef]
- Zhang, L.J.; Gallo, R.L. Antimicrobial peptides. Curr. Biol. 2016, 26, R14–R19. [Google Scholar] [CrossRef]
- Meng, S.; Xu, H.; Wang, F. Research advances of antimicrobial peptides and applications in food industry and agriculture. Curr. Protein Pept. Sci. 2010, 11, 264–273. [Google Scholar] [CrossRef]
- Peschel, A.; Sahl, H.G. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 2006, 4, 529–536. [Google Scholar] [CrossRef]
- Yang, L.; Gordon, V.; Mishra, A.; Som, A.; Purdy, K.R.; Davis, M.A.; Tew, G.N.; Wong, G.C.L. Synthetic Antimicrobial Oligomers Induce a Composition-Dependent Topological Transition in Membranes. J. Am. Chem. Soc. 2007, 129, 12141–12147. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zhou, F.; Su, H.; Zhang, Y. Structural design of microbicidal cationic oligomers and their synergistic interaction with azoles against Candida albicans. Sci. Rep. 2019, 9, 1–11. [Google Scholar]
- Palermo, E.F.; Kuroda, K. Structural determinants of antimicrobial activity in polymers which mimic host defense peptides. Appl. Microbiol. Biotechnol. 2010, 87, 1605–1615. [Google Scholar] [CrossRef]
- Li, P.; Li, X.; Saravanan, R.; Li, C.M.; Leong, S.S.J. Antimicrobial macromolecules: Synthesis methods and future applications. RSC Adv. 2012, 2, 4031–4044. [Google Scholar] [CrossRef]
- Waghu, F.H.; Gopi, L.; Barai, R.S.; Ramteke, P.; Nizami, B.; Idicula-Thomas, S. CAMP: Collection of sequences and structures of antimicrobial peptides. Nucleic Acids Res. 2013, 42, D1154–D1158. [Google Scholar] [CrossRef]
- Lee, T.-H.; Hall, K.N.; Aguilar, M.-I. Antimicrobial Peptide Structure and Mechanism of Action: A Focus on the Role of Membrane Structure. Curr. Top. Med. Chem. 2015, 16, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Schmidtchen, A.; Pasupuleti, M.; Malmsten, M. Effect of hydrophobic modifications in antimicrobial peptides. Adv. Colloid Interface Sci. 2014, 205, 265–274. [Google Scholar] [CrossRef]
- McKay, M.J.; Afrose, F.; Koeppe II, R.E.; Greathouse, D.V. Helix formation and stability in membranes. Biochim. Biophys. Acta (BBA) Biomembr. 2018, 1860, 2108–2117. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Jackman, J.A.; Cho, N.J. Comparing the membrane-interaction profiles of two antiviral peptides: Insights into structure–function relationship. Langmuir 2019, 35, 9934–9943. [Google Scholar] [CrossRef]
- Sánchez-Vásquez, L.; Silva-Sanchez, J.; Jiménez-Vargas, J.M.; Rodriguez-Romero, A.; Muñoz-Garay, C.; Rodriguez, M.D.C.; Gurrola, G.B.; Possani, L.D. Enhanced antimicrobial activity of novel synthetic peptides derived from vejovine and hadrurin. Biochim. Biophys. Acta (BBA) Gen. Subj. 2013, 1830, 3427–3436. [Google Scholar] [CrossRef] [PubMed]
- Porto, W.F.; Pires, Á.S.; Franco, O.L. CS-AMPPred: An updated SVM model for antimicrobial activity prediction in cysteine-stabilized peptides. PLoS ONE 2012, 7, e51444. [Google Scholar] [CrossRef]
- Dathe, M.; Nikolenko, H.; Meyer, J.; Beyermann, M.; Bienert, M. Optimization of the antimicrobial activity of magainin peptides by modification of charge. FEBS Lett. 2001, 501, 146–150. [Google Scholar] [CrossRef]
- Hall, K.; Lee, T.H.; Aguilar, M.I. The role of electrostatic interactions in the membrane binding of melittin. J. Mol. Recognit. 2011, 24, 108–118. [Google Scholar] [CrossRef]
- Sengupta, D.; Leontiadou, H.; Mark, A.; Marrink, S.-J. Toroidal pores formed by antimicrobial peptides show significant disorder. Biochim. et Biophys. Acta (BBA) Biomembr. 2008, 1778, 2308–2317. [Google Scholar] [CrossRef] [PubMed]
- Laverty, G.; Gorman, S.P.; Gilmore, B.F. The Potential of Antimicrobial Peptides as Biocides. Int. J. Mol. Sci. 2011, 12, 6566–6596. [Google Scholar] [CrossRef] [PubMed]
- Sowa-Jasiłek, A.; Zdybicka-Barabas, A.; Stączek, S.; Pawlikowska-Pawlęga, B.; Grygorczuk-Płaneta, K.; Skrzypiec, K.; Gruszecki, W.I.; Mak, P.; Cytryńska, M. Antifungal Activity of Anionic Defense Peptides: Insight into the Action of Galleria mellonella Anionic Peptide 2. Int. J. Mol. Sci. 2020, 21, 1912. [Google Scholar] [CrossRef] [PubMed]
- Copolovici, D.; Langel, K.; Eriste, E.; Langel, U. Cell-Penetrating Peptides: Design, Synthesis, and Applications. ACS Nano 2014, 8, 1972–1994. [Google Scholar] [CrossRef]
- Futaki, S.; Ohashi, W.; Suzuki, T.; Niwa, M.; Tanaka, S.; Ueda, K.; Harashima, H.; Sugiura, Y. Stearylated Arginine-Rich Peptides: A New Class of Transfection Systems. Bioconjugate Chem. 2001, 12, 1005–1011. [Google Scholar] [CrossRef]
- Dathe, M.; Wieprecht, T.; Nikolenko, H.; Handel, L.; Maloy, W.; Macdonald, D.L.; Beyermann, M.; Bienert, M. Hydrophobicity, hydrophobic moment and angle subtended by charged residues modulate antibacterial and haemolytic activity of amphipathic helical peptides. FEBS Lett. 1997, 403, 208–212. [Google Scholar] [CrossRef]
- Brogden, N.K.; Brogden, K.A. Will new generations of modified antimicrobial peptides improve their potential as pharmaceuticals? Int. J. Antimicrob. Agents 2011, 38, 217–225. [Google Scholar] [CrossRef]
- Rathinakumar, R.; Walkenhorst, W.F.; Wimley, W.C. Broad-Spectrum Antimicrobial Peptides by Rational Combinatorial Design and High-Throughput Screening: The Importance of Interfacial Activity. J. Am. Chem. Soc. 2009, 131, 7609–7617. [Google Scholar] [CrossRef]
- Narayana, J.L.; Mishra, B.; Lushnikova, T.; Wu, Q.; Chhonker, Y.S.; Zhang, Y.; Zarena, D.; Salnikov, E.S.; Dang, X.; Wang, F.; et al. Two distinct amphipathic peptide antibiotics with systemic efficacy. Proc. Natl. Acad. Sci. USA 2020, 117, 19446–19454. [Google Scholar] [CrossRef]
- Eisenberg, D.; Weiss, R.M.; Terwilliger, T.; Wilcox, W. Hydrophobic moments and protein structure. Faraday Symp. Chem. Soc. 1982, 17, 109–120. [Google Scholar] [CrossRef]
- Porto, W.F.; do Vale Ferreira, K.C.; Ribeiro, S.M.; Franco, O.L. Sense the Moment: A highly sensitive antimicrobial activity predictor based on hydrophobic moment. bioRxiv 2020. [Google Scholar] [CrossRef]
- Han, H.M.; Gopal, R.; Park, Y. Design and membrane-disruption mechanism of charge-enriched AMPs exhibiting cell selectivity, high-salt resistance, and anti-biofilm properties. Amino Acids 2015, 48, 505–522. [Google Scholar] [CrossRef] [PubMed]
- Ebenhan, T.; Gheysens, O.; Kruger, H.G.; Zeevaart, J.R.; Sathekge, M.M. Antimicrobial Peptides: Their Role as Infection-Selective Tracers for Molecular Imaging. BioMed Res. Int. 2014, 2014, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Uematsu, N.; Matsuzaki, K. Polar Angle as a Determinant of Amphipathic α-Helix-Lipid Interactions: A Model Peptide Study. Biophys. J. 2000, 79, 2075–2083. [Google Scholar] [CrossRef]
- Marchand, A.; Augenstreich, J.; Loiseau, C.; Verdon, J.; Lecomte, S.; Berjeaud, J.M. Effect of amino acid substitution in the staphylococcal peptides warnericin RK and PSMα on their anti-Legionella and hemolytic activities. Mol. Cell. Biochem. 2015, 405, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Juba, M.L.; Porter, D.K.; Williams, E.H.; Rodriguez, C.A.; Barksdale, S.M.; Bishop, B.M. Helical cationic antimicrobial peptide length and its impact on membrane disruption. Biochim. et Biophys. Acta (BBA) Biomembr. 2015, 1848, 1081–1091. [Google Scholar] [CrossRef]
- Epand, R.; Vogel, H.J. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta (BBA) Biomembr. 1999, 1462, 11–28. [Google Scholar] [CrossRef]
- Podda, E.; Benincasa, M.; Pacor, S.; Micali, F.; Mattiuzzo, M.; Gennaro, R.; Scocchi, M. Dual mode of action of Bac7, a proline-rich antibacterial peptide. Biochim. Biophys. Acta (BBA) Gen. Subj. 2006, 1760, 1732–1740. [Google Scholar] [CrossRef]
- Elhag, O.; Zhou, D.; Song, Q.; Soomro, A.A.; Cai, M.; Zheng, L.; Yu, Z.; Zhang, J. Screening, Expression, Purification and Functional Characterization of Novel Antimicrobial Peptide Genes from Hermetia illucens (L.). PLoS ONE 2017, 12, e0169582. [Google Scholar] [CrossRef]
- Zavascki, A.P.; Goldani, L.Z.; Li, J.; Nation, R.L. Polymyxin B for the treatment of multidrug-resistant pathogens: A critical review. J. Antimicrob. Chemother. 2007, 60, 1206–1215. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Nation, R.L.; Turnidge, J.D.; Milne, R.W.; Coulthard, K.; Rayner, C.R.; Paterson, D.L. Colistin: The re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet. Infect. Dis. 2006, 6, 589–601. [Google Scholar] [CrossRef]
- Nation, R.L.; Li, J.; Cars, O.; Couet, W.; Dudley, M.N.; Kaye, K.S.; Mouton, J.W.; Paterson, D.; Tam, V.H.; Theuretzbacher, U.; et al. Framework for optimisation of the clinical use of colistin and polymyxin B: The Prato polymyxin consensus. Lancet Infect. Dis. 2015, 15, 225–234. [Google Scholar] [CrossRef]
- Hirsch, R.; Wiesner, J.; Marker, A.; Pfeifer, Y.; Bauer, A.; Hammann, E.P.; Vilcinskas, A. Profiling antimicrobial peptides from the medical maggot Lucilia sericata as potential antibiotics for MDR Gram-negative bacteria. J. Antimicrob. Chemother. 2018, 74, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Kerenga, B.K.; McKenna, J.; Harvey, P.J.; Quimbar, P.; Garcia-Ceron, D.; Lay, F.; Phan, T.K.; Veneer, P.K.; Vasa, S.; Parisi, K.; et al. Salt-Tolerant Antifungal and Antibacterial Activities of the Corn Defensin ZmD32. Front. Microbiol. 2019, 10, 795. [Google Scholar] [CrossRef]
- Shurko, J.F.; Galega, R.S.; Li, C.; Lee, G.C. Evaluation of LL-37 antimicrobial peptide derivatives alone and in combination with vancomycin against S. aureus. J. Antibiot. 2018, 71, 971–974. [Google Scholar] [CrossRef]
- Yasir, M.; Dutta, D.; Willcox, M.D. Comparative mode of action of the antimicrobial peptide melimine and its derivative Mel4 against Pseudomonas aeruginosa. Sci. Rep. 2019, 9, 1–2. [Google Scholar]
- Fenner, A. Antimicrobial peptide derived from moths can eradicate UPEC biofilms and could offer a novel therapeutic option. Nat. Rev. Urol. 2020, 17, 191. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.; Kwok, S.H.; Humble, J.L.; Liang, Y.; Tang, S.W.; Tang, K.H.; Tse, M.K.; Lei, J.H.; Ramalingam, R.; Koohi-Moghadam, M.; et al. BING, a novel antimicrobial peptide isolated from Japanese medaka plasma, targets bacterial envelope stress response by suppressing cpxR expression. Sci. Rep. 2021, 11, 1–17. [Google Scholar] [CrossRef]
- Zaet, A.; Dartevelle, P.; Daouad, F.; Ehlinger, C.; Quilès, F.; Francius, G.; Boehler, C.; Bergthold, C.; Frisch, B.; Prévost, G.; et al. D-Cateslytin, a new antimicrobial peptide with therapeutic potential. Sci. Rep. 2017, 7, 15199. [Google Scholar] [CrossRef]
- Porto, W.F.; Irazazabal, L.; Alves, E.S.; Ribeiro, S.M.; Matos, C.O.; Pires, Á.S.; Fensterseifer, I.C.; Miranda, V.J.; Haney, E.F.; Humblot, V.; et al. In silico optimization of a guava antimicrobial peptide enables combinatorial exploration for peptide design. Nat. Commun. 2018, 9, 1–2. [Google Scholar] [CrossRef]
- Ma, B.; Fang, C.; Lu, L.; Wang, M.; Xue, X.; Zhou, Y.; Li, M.; Hu, Y.; Luo, X.; Hou, Z. The antimicrobial peptide thanatin disrupts the bacterial outer membrane and inactivates the NDM-1 metallo-β-lactamase. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Piras, A.M.; Maisetta, G.; Sandreschi, S.; Gazzarri, M.; Bartoli, C.; Grassi, L.; Esin, S.; Chiellini, F.; Batoni, G. Chitosan nanoparticles loaded with the antimicrobial peptide temporin B exert a long-term antibacterial activity in vitro against clinical isolates of Staphylococcus epidermidis. Front. Microbiol. 2015, 6, 372. [Google Scholar] [CrossRef]
- Knappe, D.; Piantavigna, S.; Hansen, A.; Mechler, A.; Binas, A.; Nolte, O.; Martin, L.L.; Hoffmann, R. Oncocin (VDKPPYLPRPRPPRRIYNR-NH2): A novel antibacterial peptide optimized against gram-negative human pathogens. J. Med. Chem. 2010, 53, 5240–5247. [Google Scholar] [CrossRef]
- De Lucca, A.J.; Walsh, T.J. Antifungal peptides: Novel therapeutic compounds against emerging pathogens. Antimicrob. Agents Chemother. 1999, 43, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Roscetto, E.; Contursi, P.; Vollaro, A.; Fusco, S.; Notomista, E.; Catania, M.R. Antifungal and anti-biofilm activity of the first cryptic antimicrobial peptide from an archaeal protein against Candida spp. clinical isolates. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Utkina, L.L.; Andreev, Y.A.; Rogozhin, E.A.; Korostyleva, T.V.; Slavokhotova, A.A.; Oparin, P.B.; Vassilevski, A.A.; Grishin, E.V.; Egorov, T.A.; Odintsova, T.I. Genes encoding 4-Cys antimicrobial peptides in wheat T riticum kiharae Dorof. et Migush.: Multimodular structural organization, instraspecific variability, distribution and role in defence. FEBS J. 2013, 280, 3594–3608. [Google Scholar] [CrossRef]
- Ochiai, A.; Ogawa, K.; Fukuda, M.; Ohori, M.; Kanaoka, T.; Tanaka, T.; Taniguchi, M.; Sagehashi, Y. Rice Defensin OsAFP1 is a New Drug Candidate against Human Pathogenic Fungi. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Luo, X.-L.; Li, J.-X.; Huang, H.-R.; Duan, J.-L.; Dai, R.-X.; Tao, R.-J.; Yang, L.; Hou, J.-Y.; Jia, X.-M.; Xu, J.-F. LL37 Inhibits Aspergillus fumigatus Infection via Directly Binding to the Fungus and Preventing Excessive Inflammation. Front. Immunol. 2019, 10, 283. [Google Scholar] [CrossRef] [PubMed]
- Do Nascimento Dias, J.; de Souza Silva, C.; de Araújo, A.R.; Souza, J.M.; Júnior, P.H.; Cabral, W.F.; da Silva, M.D.; Eaton, P.; de Almeida, J.R.; Nicola, A.M.; et al. Mechanisms of action of antimicrobial peptides ToAP2 and NDBP-5.7 against Candida albicans planktonic and biofilm cells. Sci. Rep. 2020, 10, 1–4. [Google Scholar]
- Velivelli, S.L.S.; Czymmek, K.J.; Li, H.; Shaw, J.B.; Buchko, G.W.; Shah, D.M. Antifungal symbiotic peptide NCR044 exhibits unique structure and multifaceted mechanisms of action that confer plant protection. Proc. Natl. Acad. Sci. USA 2020, 117, 16043–16054. [Google Scholar] [CrossRef]
- Nakamura, I.; Yoshimura, S.; Masaki, T.; Takase, S.; Ohsumi, K.; Hashimoto, M.; Furukawa, S.; Fujie, A. ASP2397: A novel antifungal agent produced by Acremonium persicinum MF-347833. J. Antibiot. 2016, 70, 45–51. [Google Scholar] [CrossRef]
- Colombo, M.; Masiero, S.; Rosa, S.; Caporali, E.; Toffolatti, S.L.; Mizzotti, C.; Tadini, L.; Rossi, F.; Pellegrino, S.; Musetti, R.; et al. NoPv1: A synthetic antimicrobial peptide aptamer targeting the causal agents of grapevine downy mildew and potato late blight. Sci. Rep. 2020, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Moghaddam, M.R.; Gross, T.; Becker, A.; Vilcinskas, A.; Rahnamaeian, M. The selective antifungal activity of Drosophila melanogaster metchnikowin reflects the species-dependent inhibition of succinate–coenzyme Q reductase. Sci. Rep. 2017, 7, 1–9. [Google Scholar]
- De Azevedo, P.O.; Mendonça, C.M.; Moreno, A.C.; Bueno, A.V.; de Almeida, S.R.; Seibert, L.; Converti, A.; Watanabe, I.S.; Gierus, M.; de Souza Oliveira, R.P. Antibacterial and antifungal activity of crude and freeze-dried bacteriocin-like inhibitory substance produced by Pediococcus pentosaceus. Sci. Rep. 2020, 10, 1–4. [Google Scholar]
- Gong, Z.; Karlsson, A.J. Translocation of cell-penetrating peptides into Candida fungal pathogens. Protein Sci. 2017, 26, 1714–1725. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Sun, J.; Xia, S.; Tian, X.; Cheserek, M.J.; Le, G. Mechanism of antifungal activity of antimicrobial peptide APP, a cell-penetrating peptide derivative, against Candida albicans: Intracellular DNA binding and cell cycle arrest. Appl. Microbiol. Biotechnol. 2016, 100, 3245–3253. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Jia, F.; Dang, W.; Zhao, Y.; Zhu, R.; Sun, M.; Qiu, S.; An, X.; Ma, Z.; Zhu, Y.; et al. Antifungal effect and action mechanism of antimicrobial peptide polybia-CP. J. Pept. Sci. 2015, 22, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Domingo, E. Mechanisms of viral emergence. Vet. Res. 2010, 41, 38. [Google Scholar] [CrossRef]
- Castel, G.; Chtéoui, M.; Heyd, B.; Tordo, N. Phage Display of Combinatorial Peptide Libraries: Application to Antiviral Research. Molecules 2011, 16, 3499–3518. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Jiang, X.; Qiao, J.; Wang, Z.; Tong, A.; Yang, J.; Yang, S.; Yang, L. Antimicrobial peptide DP7 with potential activity against SARS coronavirus infections. Signal Transduct. Target. Ther. 2021, 6, 1–3. [Google Scholar]
- Zhang, Z.; Zhang, H.; Peng, T.; Li, D.; Xu, J. Melittin suppresses cathepsin S-induced invasion and angiogenesis via blocking of the VEGF-A/VEGFR-2/MEK1/ERK1/2 pathway in human hepatocellular carcinoma. Oncol. Lett. 2016, 11, 610–618. [Google Scholar] [CrossRef][Green Version]
- Elnagdy, S.; AlKhazindar, M. The Potential of Antimicrobial Peptides as an Antiviral Therapy against COVID-19. ACS Pharmacol. Transl. Sci. 2020, 3, 780–782. [Google Scholar] [CrossRef]
- Wiens, M.E.; Smith, J.G. α-Defensin HD5 Inhibits Human Papillomavirus 16 Infection via Capsid Stabilization and Redirection to the Lysosome. mBio 2017, 8, e02304-16. [Google Scholar] [CrossRef]
- Mohan, K.V.; Rao, S.S.; Atreya, C.D. Antiviral activity of selected antimicrobial peptides against vaccinia virus. Antivir. Res. 2010, 86, 306–311. [Google Scholar] [CrossRef]
- Nigro, E.; Colavita, I.; Sarnataro, D.; Scudiero, O.; Zambrano, G.; Granata, V.; Daniele, A.; Carotenuto, A.; Galdiero, S.; Folliero, V.; et al. An ancestral host defence peptide within human β-defensin 3 recapitulates the antibacterial and antiviral activity of the full-length molecule. Sci. Rep. 2015, 5, 18450. [Google Scholar] [CrossRef]
- Methatham, T.; Boonchuen, P.; Jaree, P.; Tassanakajon, A.; Somboonwiwat, K. Antiviral action of the antimicrobial peptide ALFPm3 from Penaeus monodon against white spot syndrome virus. Dev. Comp. Immunol. 2017, 69, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, J.M.; Oliveira, M.D.; Dias, R.S.; Nacif-Marçal, L.; Feio, R.N.; Ferreira, S.O.; Oliveira, L.L.; Silva, C.C.; Paula, S.O. The antimicrobial peptide HS-1 inhibits dengue virus infection. Virology 2018, 514, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Novoa, B.; Romero, A.; Álvarez, Á.L.; Moreira, R.; Pereiro, P.; Costa, M.M.; Dios, S.; Estepa, A.; Parra, F.; Figueras, A. Antiviral activity of myticin C peptide from mussel: An ancient defense against herpesviruses. J. Virol. 2016, 90, 7692–7702. [Google Scholar] [CrossRef]
- Zhao, H.; Zhou, J.; Zhang, K.; Chu, H.; Liu, D.; Poon, V.K.-M.; Chan, C.C.-S.; Leung, H.-C.; Fai, N.; Lin, Y.-P.; et al. A novel peptide with potent and broad-spectrum antiviral activities against multiple respiratory viruses. Sci. Rep. 2016, 6, 22008. [Google Scholar] [CrossRef] [PubMed]
- Vanneman, M.; Dranoff, G. Combining immunotherapy and targeted therapies in cancer treatment. Nat. Rev. Cancer 2012, 12, 237–251. [Google Scholar] [CrossRef]
- Zhao, L.; Huang, Y.; Gao, S.; Cui, Y.; He, D.; Wang, L.; Chen, Y. Comparison on effect of hydrophobicity on the antibacterial and antifungal activities of α-helical antimicrobial peptides. Sci. China Chem. 2013, 56, 1307–1314. [Google Scholar] [CrossRef]
- Hu, C.; Chen, X.; Huang, Y.; Chen, Y. Co-administration of iRGD with peptide HPRP-A1 to improve anticancer activity and membrane penetrability. Sci. Rep. 2018, 8, 2274. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.; Haney, E.F.; Gill, E.E. The immunology of host defence peptides: Beyond antimicrobial activity. Nat. Rev. Immunol. 2016, 16, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Pinto, M.E.F.; Najas, J.Z.G.; Magalhães, L.G.; Bobey, A.F.; Mendonça, J.N.; Lopes, N.P.; Leme, F.; Teixeira, S.P.; Trovó, M.; Andricopulo, A.D.; et al. Inhibition of Breast Cancer Cell Migration by Cyclotides Isolated from Pombalia calceolaria. J. Nat. Prod. 2018, 81, 1203–1208. [Google Scholar] [CrossRef]
- Rai, D.; Qian, S. Interaction of the Antimicrobial Peptide Aurein 1.2 and Charged Lipid Bilayer. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, X.; Li, Y.; Lan, X.; Leung, P.; Li, J.; Li, G.; Xie, M.; Han, Y.; Lin, X. Composite Membranes of Recombinant Silkworm Antimicrobial Peptide and Poly (L-lactic Acid) (PLLA) for biomedical application. Sci. Rep. 2016, 6, 31149. [Google Scholar] [CrossRef]
- Baindara, P.; Gautam, A.; Raghava, G.; Korpole, S. Anticancer properties of a defensin like class IId bacteriocin Laterosporulin10. Sci. Rep. 2017, 7, srep46541. [Google Scholar] [CrossRef]
- Shi, D.; Hou, X.; Wang, L.; Gao, Y.; Wu, D.; Xi, X.; Zhou, M.; Kwok, H.F.; Duan, J.; Chen, T.; et al. Two Novel Dermaseptin-Like Antimicrobial Peptides with Anticancer Activities from the Skin Secretion of Pachymedusa dacnicolor. Toxins 2016, 8, 144. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, I.-W.; Kim, S.-H.; Kim, M.-A.; Yun, E.-Y.; Nam, S.-H.; Ahn, M.-Y.; Kang, D.; Hwang, J.S. Anticancer Activity of the Antimicrobial Peptide Scolopendrasin VII Derived from the Centipede, Scolopendra subspinipes mutilans. J. Microbiol. Biotechnol. 2015, 25, 1275–1280. [Google Scholar] [CrossRef]
- Li, C.; Liu, H.; Yang, Y.; Xu, X.; Lv, T.; Zhang, H.; Liu, K.; Zhang, S.; Chen, Y. N-myristoylation of Antimicrobial Peptide CM4 Enhances Its Anticancer Activity by Interacting with Cell Membrane and Targeting Mitochondria in Breast Cancer Cells. Front. Pharmacol. 2018, 9, 1297. [Google Scholar] [CrossRef]
- Chu, H.-L.; Yip, B.-S.; Chen, K.-H.; Yu, H.-Y.; Chih, Y.-H.; Cheng, H.-T.; Chou, Y.-T.; Cheng, J.-W. Novel Antimicrobial Peptides with High Anticancer Activity and Selectivity. PLoS ONE 2015, 10, e0126390. [Google Scholar] [CrossRef]
- Gaglione, R.; Pirone, L.; Farina, B.; Fusco, S.; Smaldone, G.; Aulitto, M.; Dell’Olmo, E.; Roscetto, E.; Del Gatto, A.; Fattorusso, R.; et al. Insights into the anticancer properties of the first antimicrobial peptide from Archaea. Biochim. Biophys. Acta (BBA) Gen. Subj. 2017, 1861, 2155–2164. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, I.-W.; Kim, S.-H.; Yun, E.-Y.; Nam, S.-H.; Ahn, M.-Y.; Kang, D.-C.; Hwang, J.S. Anticancer activity of CopA3 dimer peptide in human gastric cancer cells. BMB Rep. 2015, 48, 324–329. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Cui, Z.; Li, Y.-H.; Hsu, W.-H.; Lee, B.-H. In Vitro and in Vivo Anticancer Activity of Pardaxin against Proliferation and Growth of Oral Squamous Cell Carcinoma. Mar. Drugs 2015, 14, 2. [Google Scholar] [CrossRef] [PubMed]
- Reffuveille, F.; De La Fuente-Núñez, C.; Mansour, S.; Hancock, R.E.W. A Broad-Spectrum Antibiofilm Peptide Enhances Antibiotic Action against Bacterial Biofilms. Antimicrob. Agents Chemother. 2014, 58, 5363–5371. [Google Scholar] [CrossRef]
- Haney, E.F.; Mansour, S.C.; Hilchie, A.L.; de la Fuente-Núñez, C.; Hancock, R.E. High throughput screening methods for assessing antibiofilm and immunomodulatory activities of synthetic peptides. Peptides 2015, 71, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Mansour, S.C.; De La Fuente-Núñez, C.; Hancock, R. Peptide IDR-1018: Modulating the immune system and targeting bacterial biofilms to treat antibiotic-resistant bacterial infections. J. Pept. Sci. 2014, 21, 323–329. [Google Scholar] [CrossRef]
- Nijnik, A.; Hancock, R.E.W. The roles of cathelicidin LL-37 in immune defences and novel clinical applications. Curr. Opin. Hematol. 2009, 16, 41–47. [Google Scholar] [CrossRef]
- Silva, O.; De La Fuente-Núñez, C.; Haney, E.F.; Fensterseifer, I.C.M.; Ribeiro, S.; Porto, W.; Brown, P.; Faria-Junior, C.; Rezende, T.; Moreno, S.E.; et al. An anti-infective synthetic peptide with dual antimicrobial and immunomodulatory activities. Sci. Rep. 2016, 6, 35465. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, M.; Brezden, A.; Mohammad, H.; Chmielewski, J.; Seleem, M. A short D-enantiomeric antimicrobial peptide with potent immunomodulatory and antibiofilm activity against multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii. Sci. Rep. 2017, 7, 6953. [Google Scholar] [CrossRef] [PubMed]
- Kindrachuk, J.; Jenssen, H.; Elliott, M.; Nijnik, A.; Magrangeas-Janot, L.; Pasupuleti, M.; Thorson, L.; Ma, S.; Easton, D.M.; Bains, M.; et al. Manipulation of innate immunity by a bacterial secreted peptide: Lantibiotic nisin Z is selectively immunomodulatory. Innate Immun. 2012, 19, 315–327. [Google Scholar] [CrossRef]
- Mookherjee, N.; Brown, K.; Bowdish, D.; Doria, S.; Falsafi, R.; Hokamp, K.; Roche, F.M.; Mu, R.; Doho, G.H.; Pistolic, J.; et al. Modulation of the TLR-Mediated Inflammatory Response by the Endogenous Human Host Defense Peptide LL-37. J. Immunol. 2006, 176, 2455–2464. [Google Scholar] [CrossRef] [PubMed]
- Veldhuizen, J.A.E.; Scheenstra, M.R.; Tjeerdsma-van Bokhoven, L.M.J.; Coorens, M.; Schneider, A.F.V.; Bikker, F.J.; van Dijk, A.; Haagsman, H.P. Antimicrobial and immunomodulatory activity of PMAP-23 derived peptides. Protein Pept. Lett. 2017, 24, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Mao, R.; Teng, D.; Hao, Y.; Chen, H.; Wang, X.; Wang, X.; Yang, N.; Wang, J. Antibacterial and immunomodulatory activities of insect defensins-DLP2 and DLP4 against multidrug-resistant Staphylococcus aureus. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Neshani, A.; Zare, H.; Eidgahi, M.R.A.; Khaledi, A.; Ghazvini, K. Epinecidin-1, a highly potent marine antimicrobial peptide with anticancer and immunomodulatory activities. BMC Pharmacol. Toxicol. 2019, 20, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.H.; Lillehoj, H.S.; Min, W. Evaluation of the Immunomodulatory Activity of the Chicken NK-Lysin-Derived Peptide cNK-2. Sci. Rep. 2017, 7, srep45099. [Google Scholar] [CrossRef]
- Ting, C.-H.; Pan, C.-Y.; Chen, Y.-C.; Lin, Y.-C.; Chen, T.-Y.; Rajanbabu, V.; Chen, J.-Y. Impact of Tilapia hepcidin 2-3 dietary supplementation on the gut microbiota profile and immunomodulation in the grouper (Epinephelus lanceolatus). Sci. Rep. 2019, 9, 1–17. [Google Scholar] [CrossRef]
- Wang, X.; Chen, L.; Zhao, X.; Xiao, L.; Yi, S.; Kong, Y.; Jiang, Y.; Zhang, J. A cathelicidin-related antimicrobial peptide suppresses cardiac hypertrophy induced by pressure overload by regulating IGFR1/PI3K/AKT and TLR9/AMPKα. Cell Death Dis. 2020, 11, 96-15. [Google Scholar] [CrossRef]
- Daneshmand, A.; Kermanshahi, H.; Sekhavati, M.H.; Javadmanesh, A.; Ahmadian, M. Antimicrobial peptide, cLF36, affects performance and intestinal morphology, microflora, junctional proteins, and immune cells in broilers challenged with E. coli. Sci. Rep. 2019, 9, 1–9. [Google Scholar]
- Ko, S.J.; Park, E.; Asandei, A.; Choi, J.-Y.; Lee, S.-C.; Seo, C.H.; Luchian, T.; Park, Y. Bee venom-derived antimicrobial peptide melectin has broad-spectrum potency, cell selectivity, and salt-resistant properties. Sci. Rep. 2020, 10, 1–12. [Google Scholar]
- Wei, L.; Huang, C.; Yang, H.; Li, M.; Yang, J.; Qiao, X.; Mu, L.; Xiong, F.; Wu, J.; Xu, W. A potent anti-inflammatory peptide from the salivary glands of horsefly. Parasites Vectors 2015, 8, 556. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brunetti, J.; Carnicelli, V.; Ponzi, A.; Di Giulio, A.; Lizzi, A.R.; Cristiano, L.; Cresti, L.; Cappello, G.; Pollini, S.; Mosconi, L.; et al. Antibacterial and Anti-Inflammatory Activity of an Antimicrobial Peptide Synthesized with D Amino Acids. Antibiotics 2020, 9, 840. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Jacob, B.; Jang, M.; Kwak, C.; Lee, Y.; Son, K.; Lee, S.; Jung, I.D.; Jeong, M.S.; Kwon, S.-H.; et al. Development of a novel short 12-meric papiliocin-derived peptide that is effective against Gram-negative sepsis. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Leake, I. Cathelicidin can reverse intestinal fibrosis in models of colitis. Nat. Rev. Gastroenterol. Hepatol. 2014, 12, 3. [Google Scholar] [CrossRef]
- Qiu, X.; Macchietto, M.G.; Liu, X.; Lu, Y.; Ma, Y.; Guo, H.; Saqui-Salces, M.; Bernlohr, D.A.; Chen, C.; Shen, S.; et al. Identification of gut microbiota and microbial metabolites regulated by an antimicrobial peptide lipocalin 2 in high fat diet-induced obesity. Int. J. Obes. 2020, 45, 143–154. [Google Scholar] [CrossRef]
- Carlile, S.R.; Shiels, J.; Kerrigan, L.; Delaney, R.; Megaw, J.; Gilmore, B.F.; Weldon, S.; Dalton, J.P.; Taggart, C.C. Sea snake cathelicidin (Hc-cath) exerts a protective effect in mouse models of lung inflammation and infection. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef]
- Silva, J.P.; Dhall, S.; Garcia, M.; Chan, A.; Costa, C.; Gama, M.; Martins-Green, M. Improved burn wound healing by the antimicrobial peptide LLKKK18 released from conjugates with dextrin embedded in a carbopol gel. Acta Biomater. 2015, 26, 249–262. [Google Scholar] [CrossRef]
- Ramos, R.; Silva, J.P.; Rodrigues, A.C.; Costa, R.; Guardão, L.; Schmitt, F.; Soares, R.; Vilanova, M.; Domingues, L.; Gama, M. Wound healing activity of the human antimicrobial peptide LL37. Peptides 2011, 32, 1469–1476. [Google Scholar] [CrossRef]
- Song, D.W.; Kim, S.H.; Kim, H.H.; Lee, K.H.; Ki, C.S.; Park, Y.H. Multi-biofunction of antimicrobial peptide-immobilized silk fibroin nanofiber membrane: Implications for wound healing. Acta Biomater. 2016, 39, 146–155. [Google Scholar] [CrossRef]
- Jiao, J.; Peng, C.; Li, C.; Qi, Z.; Zhan, J.; Pan, S. Dual bio-active factors with adhesion function modified electrospun fibrous scaffold for skin wound and infections therapeutics. Sci. Rep. 2021, 11, 1–13. [Google Scholar] [CrossRef]
- Di Grazia, A.; Cappiello, F.; Imanishi, A.; Mastrofrancesco, A.; Picardo, M.; Paus, R.; Mangoni, M.L. The frog skin-derived antimicrobial peptide esculentin-1a (1-21) NH2 promotes the migration of human HaCaT keratinocytes in an EGF receptor-dependent manner: A novel promoter of human skin wound healing? PLoS ONE 2015, 10, e0128663. [Google Scholar]
- Bucekova, M.; Sojka, M.; Valachova, I.; Martinotti, S.; Ranzato, E.; Szep, Z.; Majtan, V.; Klaudiny, J.; Majtan, J. Bee-derived antibacterial peptide, defensin-1, promotes wound reepithelialisation in vitro and in vivo. Wound Heal. S. Afr. 2017, 10, 25–35. [Google Scholar]
- Chung, E.M.C.; Dean, S.N.; Propst, C.N.; Bishop, B.M.; Van Hoek, M.L. Komodo dragon-inspired synthetic peptide DRGN-1 promotes wound-healing of a mixed-biofilm infected wound. NPJ Biofilms Microbiomes 2017, 3, 1–13. [Google Scholar] [CrossRef]
- Tang, J.; Liu, H.; Gao, C.; Mu, L.; Yang, S.; Rong, M.; Zhang, Z.; Liu, J.; Ding, Q.; Lai, R. A Small Peptide with Potential Ability to Promote Wound Healing. PLoS ONE 2014, 9, e92082. [Google Scholar]
- Liu, S.; Long, Q.; Xu, Y.; Wang, J.; Xu, Z.; Wang, L.; Zhou, M.; Wu, Y.; Chen, T.; Shaw, C. Assessment of antimicrobial and wound healing effects of Brevinin-2Ta against the bacterium Klebsiella pneumoniae in dermally-wounded rats. Oncotarget 2017, 8, 111369–111385. [Google Scholar] [CrossRef]
- Etienne, O.; Picart, C.; Taddei, C.; Haikel, Y.; Dimarcq, J.L.; Schaaf, P.; Voegel, J.C.; Ogier, J.A.; Egles, C. Multilayer Polyelectrolyte Films Functionalized by Insertion of Defensin: A New Approach to Protection of Implants from Bacterial Colonization. Antimicrob. Agents Chemother. 2004, 48, 3662–3669. [Google Scholar] [CrossRef]
- Gomes, A.; Mano, J.; Queiroz, J.; Gouveia, I. Incorporation of antimicrobial peptides on functionalized cotton gauzes for medical applications. Carbohydr. Polym. 2015, 127, 451–461. [Google Scholar] [CrossRef]
- Webber, J.L.; Namivandi-Zangeneh, R.; Drozdek, S.; Wilk, K.A.; Boyer, C.; Wong, E.H.H.; Bradshaw-Hajek, B.H.; Krasowska, M.; Beattie, D.A. Incorporation and antimicrobial activity of nisin Z within carrageenan/chitosan multilayers. Sci. Rep. 2021, 11, 1–15. [Google Scholar] [CrossRef]
- Conn, C.; Drummond, C. Nanostructured bicontinuous cubic lipid self-assembly materials as matrices for protein encapsulation. Soft Matter 2013, 9, 3449–3464. [Google Scholar] [CrossRef]
- Meikle, T.; Zabara, A.; Waddington, L.J.; Separovic, F.; Drummond, C.J.; Conn, C.E. Incorporation of antimicrobial peptides in nanostructured lipid membrane mimetic bilayer cubosomes. Colloids Surf. B Biointerfaces 2017, 152, 143–151. [Google Scholar] [CrossRef]
- Rodríguez-Vázquez, M.; Vega-Ruiz, B.; Ramos-Zúñiga, R.; Saldaña-Koppel, D.A.; Quiñones-Olvera, L.F. Chitosan and its potential use as a scaffold for tissue engineering in regenerative medicine. BioMed Res. Int. 2015. [Google Scholar] [CrossRef]
- Lee, E.Y.; Zhang, C.; Di Domizio, J.; Jin, F.; Connell, W.; Hung, M.; Malkoff, N.; Veksler, V.; Gilliet, M.; Ren, P.; et al. Helical antimicrobial peptides assemble into protofibril scaffolds that present ordered dsDNA to TLR9. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Zhu, X.; Mutreja, I.; Boda, S.K.; Fischer, N.G.; Zhang, A.; Lui, C.; Qi, Y.; Aparicio, C. Biomimetic mineralized hybrid scaffolds with antimicrobial peptides. Bioact. Mater. 2021, 6, 2250–2260. [Google Scholar] [CrossRef] [PubMed]
- Rabanal, F.; Grau, A.; Vila-Farrés, X.; Gonzalez-Linares, J.; Borràs, M.; Vila, J.; Manresa, A.; Cajal, Y. A bioinspired peptide scaffold with high antibiotic activity and low in vivo toxicity. Sci. Rep. 2015, 5, 10558. [Google Scholar] [CrossRef] [PubMed]
- Williams, G.R.; Raimi-Abraham, B.T.; Luo, C. Nanofibres in Drug Delivery; UCL Press: London, UK, 2018. [Google Scholar]
- Zare, M.; Dziemidowicz, K.; Williams, G.; Ramakrishna, S. Encapsulation of Pharmaceutical and Nutraceutical Active Ingredients Using Electrospinning Processes. Nanomaterials 2021, 11, 1968. [Google Scholar] [CrossRef]
- Kluin, J.; Talacua, H.; Smits, A.; Emmert, M.Y.; Brugmans, M.C.; Fioretta, E.; Dijkman, P.E.; Söntjens, S.; Duijvelshoff, R.; Dekker, S.; et al. In situ heart valve tissue engineering using a bioresorbable elastomeric implant—From material design to 12 months follow-up in sheep. Biomaterials 2017, 125, 101–117. [Google Scholar] [CrossRef]
- Riool, M.; De Breij, A.; Drijfhout, J.W.; Nibbering, P.H.; Zaat, S.A.J. Antimicrobial Peptides in Biomedical Device Manufacturing. Front. Chem. 2017, 5, 63. [Google Scholar] [CrossRef]
- López-Abarrategui, C.; Alba, A.; Lima, L.A.; Neto, S.M.; Vasconcelos, I.M.; Oliveira, J.T.A.; Dias, S.C.; Otero-Gonzalez, A.J.; Franco, O.L. Screening of Antimicrobials from Caribbean Sea Animals and Isolation of Bactericidal Proteins from the Littoral Mollusk Cenchritis muricatus. Curr. Microbiol. 2012, 64, 501–505. [Google Scholar] [CrossRef]
- Viana, J.F.C.; Carrijo, J.; Freitas, C.G.; Paul, A.; Alcaraz, J.; Lacorte, C.C.; Migliolo, L.; de Andrade, C.A.S.; Falcão, R.; Santos, N.; et al. Antifungal nanofibers made by controlled release of sea animal derived peptide. Nanoscale 2015, 7, 6238–6246. [Google Scholar] [CrossRef]
- Wang, X.; Yue, T.; Lee, T.-C. Development of Pleurocidin-poly(vinyl alcohol) electrospun antimicrobial nanofibers to retain antimicrobial activity in food system application. Food Control 2015, 54, 150–157. [Google Scholar] [CrossRef]
- Avila, E.E. Functions of Antimicrobial Peptides in Vertebrates. Curr. Protein Pept. Sci. 2017, 18, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T.; Selsted, M.E.; Lehrer, R.I. Defensins. Eur. J. Haematol. 1990, 44, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Selsted, M.E.; Ouellette, A.J. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 2005, 6, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Lynn, D.J.; Bradley, D.G. Discovery of α-defensins in basal mammals. Dev. Comp. Immunol. 2007, 31, 963–967. [Google Scholar] [CrossRef]
- Selsted, M.E.; Tang, Y.Q.; Morris, W.L.; McGuire, P.A.; Novotny, M.J.; Smith, W.; Henschen, A.H.; Cullor, J.S. Purification, primary structures, and antibacterial activities of beta-defensins, a new family of antimicrobial peptides from bovine neutrophils. J. Biol. Chem. 1993, 268, 6641–6648. [Google Scholar] [CrossRef]
- Tran, D.; Tran, P.A.; Tang, Y.Q.; Yuan, J.; Cole, T.; Selsted, M.E. Homodimeric θ-Defensins from Rhesus macaqueLeukocytes: Isolation, synthesis, antimicrobial activities, and bacterial binding properties of the cyclic peptides. J. Biol. Chem. 2002, 277, 3079–3084. [Google Scholar] [CrossRef]
- Bals, R.; Wilson, J.M. Cathelicidins-a family of multifunctional antimicrobial peptides. Cell. Mol. Life Sci. CMLS 2003, 60, 711–720. [Google Scholar] [CrossRef]
- Zanetti, M.; Gennaro, R.; Romeo, D. Cathelicidins: A novel protein family with a common proregion and a variable C-terminal antimicrobial domain. FEBS Lett. 1995, 374, 1–5. [Google Scholar] [CrossRef]
- Agerberth, B.; Lee, J.-Y.; Bergman, T.; Carlquist, M.; Boman, H.G.; Mutt, V.; Jornvall, H. Amino acid sequence of PR-39. Isolation from pig intestine of a new member of the family of proline-arginine-rich antibacterial peptides. Eur. J. Biochem. 1991, 202, 849–854. [Google Scholar] [CrossRef]
- Gennaro, R.; Skerlavaj, B.; Romeo, D. Purification, composition, and activity of two bactenecins, antibacterial peptides of bovine neutrophils. Infect. Immun. 1989, 57, 3142–3146. [Google Scholar] [CrossRef] [PubMed]
- Wang, G. Human Antimicrobial Peptides and Proteins. Pharmaceuticals 2014, 7, 545–594. [Google Scholar] [CrossRef] [PubMed]
- Shaat, R.M.; El Meadawy, S.; Rizk, E.M.; Elgawad, M.S.A.; Elsaid, T.O. The significance of α-defensins 1-3 in Behcet’s disease: A case-control study among Egyptian patients. Egypt. Rheumatol. Rehabil. 2020, 47, 1–8. [Google Scholar] [CrossRef]
- Shukla, P.K.; Meena, A.S.; Rao, V.; Rao, R.G.; Balazs, L.; Rao, R. Human Defensin-5 Blocks Ethanol and Colitis-Induced Dysbiosis, Tight Junction Disruption and Inflammation in Mouse Intestine. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Taha-Abdelaziz, K.; Perez-Casal, J.; Schott, C.; Hsiao, J.; Attah-Poku, S.; Slavić, D.; Caswell, J.L. Bactericidal activity of tracheal antimicrobial peptide against respiratory pathogens of cattle. Vet. Immunol. Immunopathol. 2013, 152, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Coretti, L.; Natale, A.; Cuomo, M.; Florio, E.; Keller, S.; Lembo, F.; Chiariotti, L.; Pero, R. The Interplay between Defensins and Microbiota in Crohn’s Disease. Mediat. Inflamm. 2017, 2017, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Jiang, H.; Chen, R.; Zhou, J.; Xiao, Y.; Zhang, Y.; Yan, F. Human β-defensin 3 gene modification promotes the osteogenic differentiation of human periodontal ligament cells and bone repair in periodontitis. Int. J. Oral Sci. 2020, 12, 1–10. [Google Scholar] [CrossRef]
- Cole, A.M.; Hong, T.; Boo, L.M.; Nguyen, T.; Zhao, C.; Bristol, G.; Zack, J.A.; Waring, A.J.; Yang, O.O.; Lehrer, R.I. Retrocyclin: A primate peptide that protects cells from infection by T- and M-tropic strains of HIV-1. Proc. Natl. Acad. Sci. USA 2002, 99, 1813–1818. [Google Scholar] [CrossRef] [PubMed]
- Schaal, J.B.; Tran, D.; Tran, P.; Ösapay, G.; Trinh, K.; Roberts, K.D.; Brasky, K.M.; Tongaonkar, P.; Ouellette, A.J.; Selsted, M.E. Rhesus Macaque Theta Defensins Suppress Inflammatory Cytokines and Enhance Survival in Mouse Models of Bacteremic Sepsis. PLoS ONE 2012, 7, e51337. [Google Scholar] [CrossRef]
- YYasin, B.; Wang, W.; Pang, M.; Cheshenko, N.; Hong, T.; Waring, A.J.; Herold, B.C.; Wagar, E.A.; Lehrer, R.I. θ Defensins Protect Cells from Infection by Herpes Simplex Virus by Inhibiting Viral Adhesion and Entry. J. Virol. 2004, 78, 5147–5156. [Google Scholar] [CrossRef]
- Liang, Q.-L.; Zhou, K.; He, H.-X. Retrocyclin 2: A new therapy against avian influenza H5N1 virus in vivo and vitro. Biotechnol. Lett. 2009, 32, 387–392. [Google Scholar] [CrossRef]
- Rekha, R.S.; Muvva, S.S.V.J.R.; Wan, M.; Raqib, R.; Bergman, P.; Brighenti, S.; Gudmundsson, G.H.; Agerberth, B. Phenylbutyrate induces LL-37-dependent autophagy and intracellular killing of Mycobacterium tuberculosis in human macrophages. Autophagy 2015, 11, 1688–1699. [Google Scholar] [CrossRef]
- Van Eijk, M.; Boerefijn, S.; Cen, L.; Rosa, M.; Morren, M.J.; Van Der Ent, C.K.; Kraak, B.; Dijksterhuis, J.; Valdes, I.D.; Haagsman, H.P.; et al. Cathelicidin-inspired antimicrobial peptides as novel antifungal compounds. Med. Mycol. 2020, 58, 1073–1784. [Google Scholar] [CrossRef]
- Tonk, M.; Cabezas-Cruz, A.; Valdés, J.J.; Rego, R.O.; Chrudimská, T.; Strnad, M.; Šíma, R.; Bell-Sakyi, L.; Franta, Z.; Vilcinskas, A.; et al. Defensins from the tick Ixodes scapularis are effective against phytopathogenic fungi and the human bacterial pathogen Listeria grayi. Parasites Vectors 2014, 7, 1–8. [Google Scholar] [CrossRef]
- Dolezal, T.; Krejcova, G.; Bajgar, A.; Nedbalova, P.; Strasser, P. Molecular regulations of metabolism during immune response in insects. Insect Biochem. Mol. Biol. 2019, 109, 31–42. [Google Scholar] [CrossRef]
- Tonk, M.; Vilcinskas, A.; Rahnamaeian, M. Insect antimicrobial peptides: Potential tools for the prevention of skin cancer. Appl. Microbiol. Biotechnol. 2016, 100, 7397–7405. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.R.; Kim, H.; Nam, S.H.; Yun, E.Y.; Kim, S.R.; Ahn, M.Y.; Chang, J.S.; Hwang, J.S. CopA3 peptide from Copris tripartitus induces apoptosis in human leukemia cells via a caspase-independent pathway. BMB Rep. 2012, 45, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Zhang, H.; Niu, L.; Wang, X. Efficient Screening of a Novel Antimicrobial Peptide from Jatropha curcas by Cell Membrane Affinity Chromatography. J. Agric. Food Chem. 2011, 59, 1145–1151. [Google Scholar] [CrossRef]
- Li, J.; Hu, S.; Jian, W.; Xie, C.; Yang, X. Plant antimicrobial peptides: Structures, functions, and applications. Bot. Stud. 2021, 62, 1–15. [Google Scholar] [CrossRef]
- Salimi, A.; Noorbakhsh, A.; Mamkhezri, H.; Ghavami, R. Electrocatalytic Reduction of H2O2 and Oxygen on the Surface of Thionin Incorporated onto MWCNTs Modified Glassy Carbon Electrode: Application to Glucose Detection. Electroanalysis 2007, 19, 1100–1108. [Google Scholar] [CrossRef]
- Zhang, D.; Li, W.; Wang, H.; Ma, Z. A novel immunoprobe composed of reduced graphene oxide-hemin-thionin-Au nanohybrid for ultrasensitive detection of tumor marker. Sens. Actuators B Chem. 2018, 258, 141-14. [Google Scholar] [CrossRef]
- Hu, E.; Wang, D.; Chen, J.; Tao, X. Novel cyclotides from Hedyotis diffusa induce apoptosis and inhibit proliferation and migration of prostate cancer cells. Int. J. Clin. Exp. Med. 2015, 8, 4059. [Google Scholar]
- Thery, T.; Arendt, E.K. Antifungal activity of synthetic cowpea defensin Cp-thionin II and its application in dough. Food MicroBiol. 2018, 73, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Loo, S.; Kam, A.; Xiao, T.; Nguyen, G.K.T.; Liu, C.F.; Tam, J.P. Identification and Characterization of Roseltide, a Knottin-type Neutrophil Elastase Inhibitor Derived from Hibiscus sabdariffa. Sci. Rep. 2016, 6, 39401. [Google Scholar] [CrossRef] [PubMed]
- Islam, K.T.; Velivelli, S.; Berg, R.H.; Oakley, B.; Shah, D.M. A novel bi-domain plant defensin MtDef5 with potent broad-spectrum antifungal activity binds to multiple phospholipids and forms oligomers. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Egan, K.; Field, D.; Rea, M.C.; Ross, R.; Hill, C.; Cotter, P.D. Bacteriocins: Novel Solutions to Age Old Spore-Related Problems? Front. Microbiol. 2016, 7, 461. [Google Scholar] [CrossRef]
- Lubelski, J.; Rink, R.; Khusainov, R.; Moll, G.N.; Kuipers, O.P. Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Experientia 2007, 65, 455–476. [Google Scholar] [CrossRef]
- De Arauz, L.J.; Jozala, A.F.; Mazzola, P.G.; Penna, T.C. Nisin biotechnological production and application: A review. Trends Food Sci. Technol. 2009, 20, 146–154. [Google Scholar] [CrossRef]
- Kamarajan, P.; Hayami, T.; Matte, B.; Liu, Y.; Danciu, T.; Ramamoorthy, A.; Worden, F.; Kapila, S.; Kapila, Y. Nisin ZP, a Bacteriocin and Food Preservative, Inhibits Head and Neck Cancer Tumorigenesis and Prolongs Survival. PLoS ONE 2015, 10, e0131008. [Google Scholar] [CrossRef]
- Preet, S.; Bharati, S.; Panjeta, A.; Tewari, R.; Rishi, P. Effect of nisin and doxorubicin on DMBA-induced skin carcinogenesis—A possible adjunct therapy. Tumor Biol. 2015, 36, 8301–8308. [Google Scholar] [CrossRef]
- Begde, D.; Bundale, S.; Mashitha, P.; Rudra, J.; Nashikkar, N.; Upadhyay, A. Immunomodulatory efficacy of nisin-a bacterial lantibiotic peptide. J. Pept. Sci. 2011, 17, 438–444. [Google Scholar] [CrossRef]
- Al Atya, A.K.; Belguesmia, Y.; Chataigne, G.; Ravallec, R.; Vachée, A.; Szunerits, S.; Boukherroub, R.; Drider, D. Anti-MRSA activities of enterocins DD28 and DD93 and evidences on their role in the inhibition of biofilm formation. Front. Microbiol. 2016, 7, 817. [Google Scholar] [CrossRef] [PubMed]
- Joo, N.E.; Ritchie, K.; Kamarajan, P.; Miao, D.; Kapila, Y.L. Nisin, an apoptogenic bacteriocin and food preservative, attenuates HNSCC tumorigenesis via CHAC 1. Cancer Med. 2012, 1, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Field, D.; Begley, M.; O’Connor, P.M.; Daly, K.M.; Hugenholtz, F.; Cotter, P.D.; Hill, C.; Ross, R. Bioengineered Nisin A Derivatives with Enhanced Activity against Both Gram Positive and Gram Negative Pathogens. PLoS ONE 2012, 7, e46884. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.M.; Ateia, I.; Paulus, J.R.; Liu, H.; Fenno, J.C.; Rickard, A.H.; Kapila, Y.L. Antimicrobial nisin acts against saliva derived multi-species biofilms without cytotoxicity to human oral cells. Front. Microbiol. 2015, 6, 617. [Google Scholar] [CrossRef] [PubMed]
- Tallet, L.; Gribova, V.; Ploux, L.; Vrana, N.E.; LaValle, P. New Smart Antimicrobial Hydrogels, Nanomaterials, and Coatings: Earlier Action, More Specific, Better Dosing? Adv. Healthc. Mater. 2020, 10, e2001199. [Google Scholar] [CrossRef]
- Alvarez-Lorenzo, C.; Concheiro, A. Intelligent drug delivery systems: Polymeric micelles and hydrogels. Mini Rev. Med. Chem. 2008, 8, 1065–1074. [Google Scholar] [CrossRef]
- Sousa, M.G.; Rezende, T.M.; Franco, O.L. Nanofibers as drug-delivery systems for antimicrobial peptides. Drug Discov. Today 2021, 26, 2064–2074. [Google Scholar] [CrossRef]
- Greber, K.E.; Dawgul, M. Antimicrobial peptides under clinical trials. Curr. Top. Med. Chem. 2017, 17, 620. [Google Scholar] [CrossRef]
- Azharuddin, M.; Zhu, G.H.; Das, D.; Ozgur, E.; Uzun, L.; Turner, A.P.F.; Patra, H.K. A repertoire of biomedical applications of noble metal nanoparticles. Chem. Commun. 2019, 55, 6964. [Google Scholar] [CrossRef]
- Kaur, J.; Gill, G.S.; Jeet, K. Applications of Carbon Nanotubes in Drug Delivery: A Comprehensive Review. In Characterization and Biology of Nanomaterials for Drug Delivery: Nanoscience and Nanotechnology in Drug Delivery, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2019; p. 133. [Google Scholar]
- Weiner, N.; Martin, F.; Riaz, M. Liposomes as a drug delivery system. Drug Dev. Ind. Pharm. 1989, 15, 1523. [Google Scholar] [CrossRef]
- Lancelot, A.; Sierra, T.; Serrano, J.L. Nanostructured liquid-crystalline particles for drug delivery. Expert Opin. Drug Deliv. 2014, 11, 547. [Google Scholar] [CrossRef]
- Sherje, A.P.; Jadhav, M.; Dravyakar, B.R.; Kadam, D. Dendrimers: A versatile nanocarrier for drug delivery and targeting. Int. J. Pharm. 2018, 548, 707. [Google Scholar] [CrossRef] [PubMed]
- Ray, P.; White, R.R. Aptamers for targeted drug delivery. Pharmaceuticals 2010, 3, 1761–1778. [Google Scholar] [CrossRef] [PubMed]
- Hoare, T.R.; Kohane, D.S. Hydrogels in drug delivery: Progress and challenges. Polymers 2008, 49, 1993. [Google Scholar] [CrossRef]
- Joseph, B.; George, A.; Gopi, S.; Kalarikkal, N.; Thomas, S. Polymer sutures for simultaneous wound healing and drug delivery—A review. Int. J. Pharm. 2017, 524, 454. [Google Scholar] [CrossRef] [PubMed]
- Rajewski, R.A.; Stella, V.J. Pharmaceutical applications of cyclodextrins. 2. In vivo drug delivery. J. Pharm. Sci. 1996, 85, 1142. [Google Scholar]
- Matthews, J.A.; Wnek, G.E.; Simpson, D.G.; Bowlin, G.L. Electrospinning of Collagen Nanofibers. Biomacromolecules 2002, 3, 232–238. [Google Scholar] [CrossRef]
- Li, J.; Fan, C.; Pei, H.; Shi, J.; Huang, Q. Smart Drug Delivery Nanocarriers with Self-Assembled DNA Nanostructures. Adv. Mater. 2013, 25, 4386–4396. [Google Scholar] [CrossRef]
- Wang, Q.; Uzunoglu, E.; Wu, Y.; Libera, M. Self-Assembled Poly(ethylene glycol)-co-Acrylic Acid Microgels to Inhibit Bacterial Colonization of Synthetic Surfaces. ACS Appl. Mater. Interfaces 2012, 4, 2498–2506. [Google Scholar] [CrossRef] [PubMed]
- Bennet, D.; Kim, S. Polymer nanoparticles for smart drug delivery. In Application of Nanotechnology in Drug Delivery; InTech Open: London, UK, 2014. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).