Topically Applied Bacteriophage to Control Multi-Drug Resistant Klebsiella pneumoniae Infected Wound in a Rat Model
Abstract
1. Introduction
2. Results
2.1. Bacterial Characterization
2.2. Antibiotic Sensitivity Profile
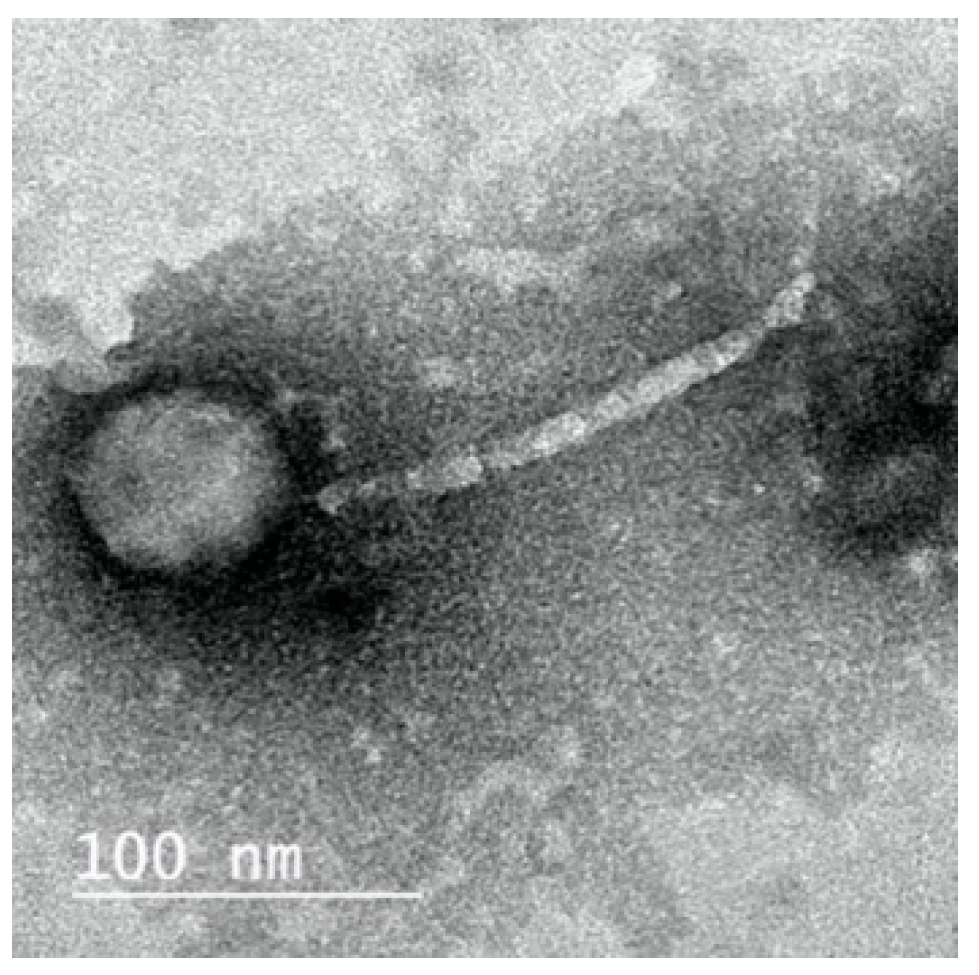
2.3. Morphology of ZCKP8 Phage
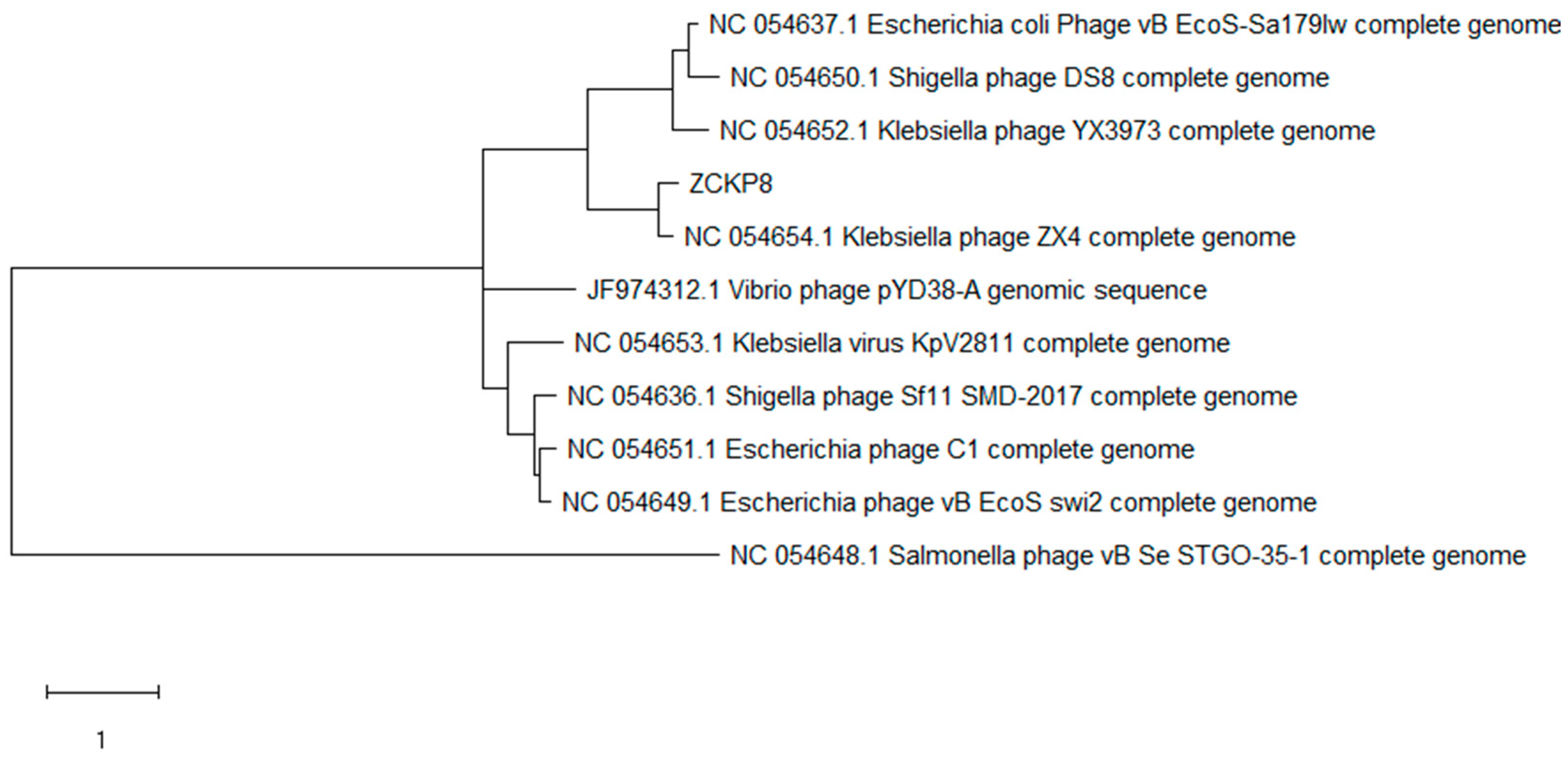
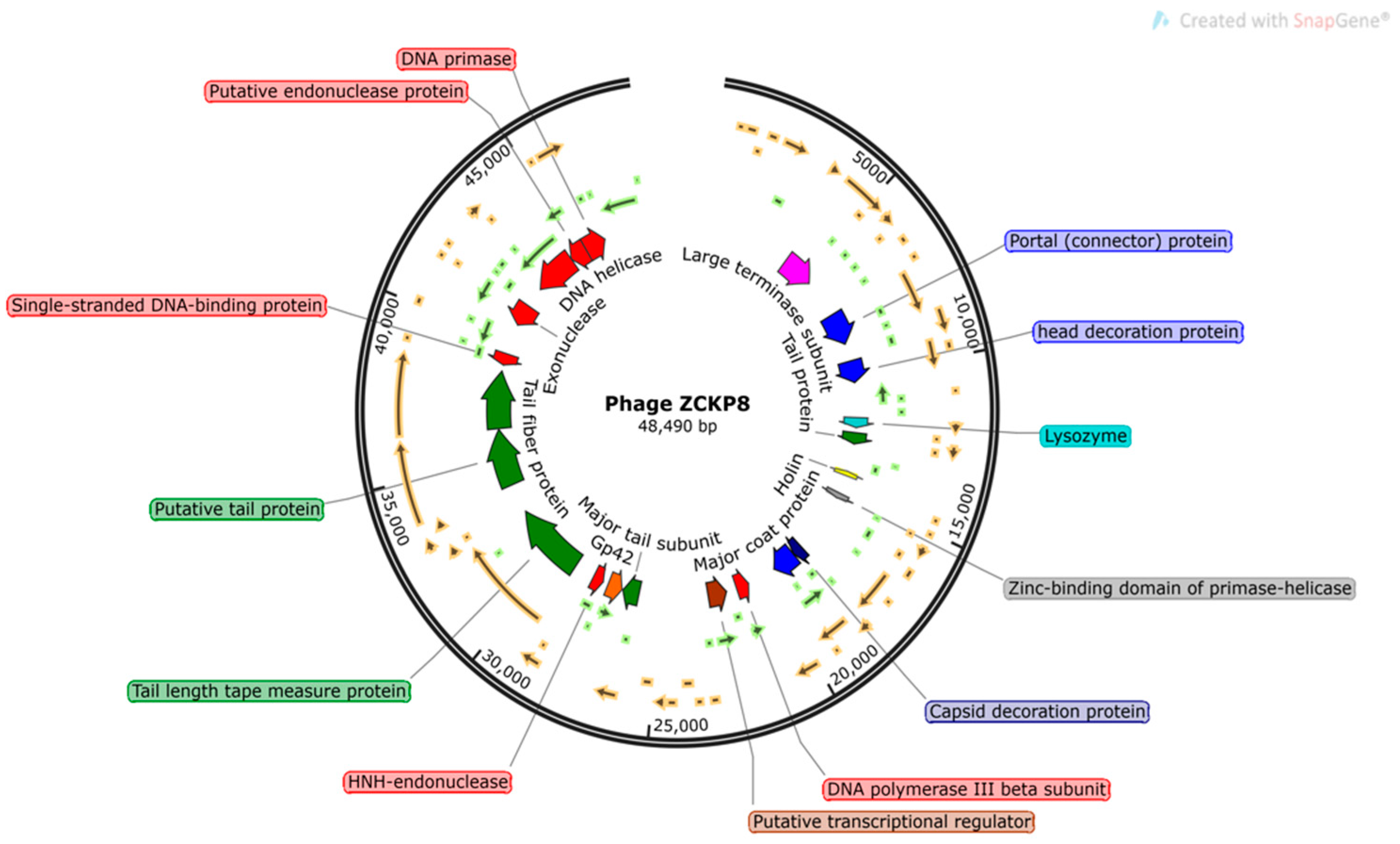
2.4. Phage Genome
2.5. Phage Host Range and Efficiency of Plating (EOP)
2.6. Determination of the Bacteriophage Insensitive Mutant (BIM) Frequency
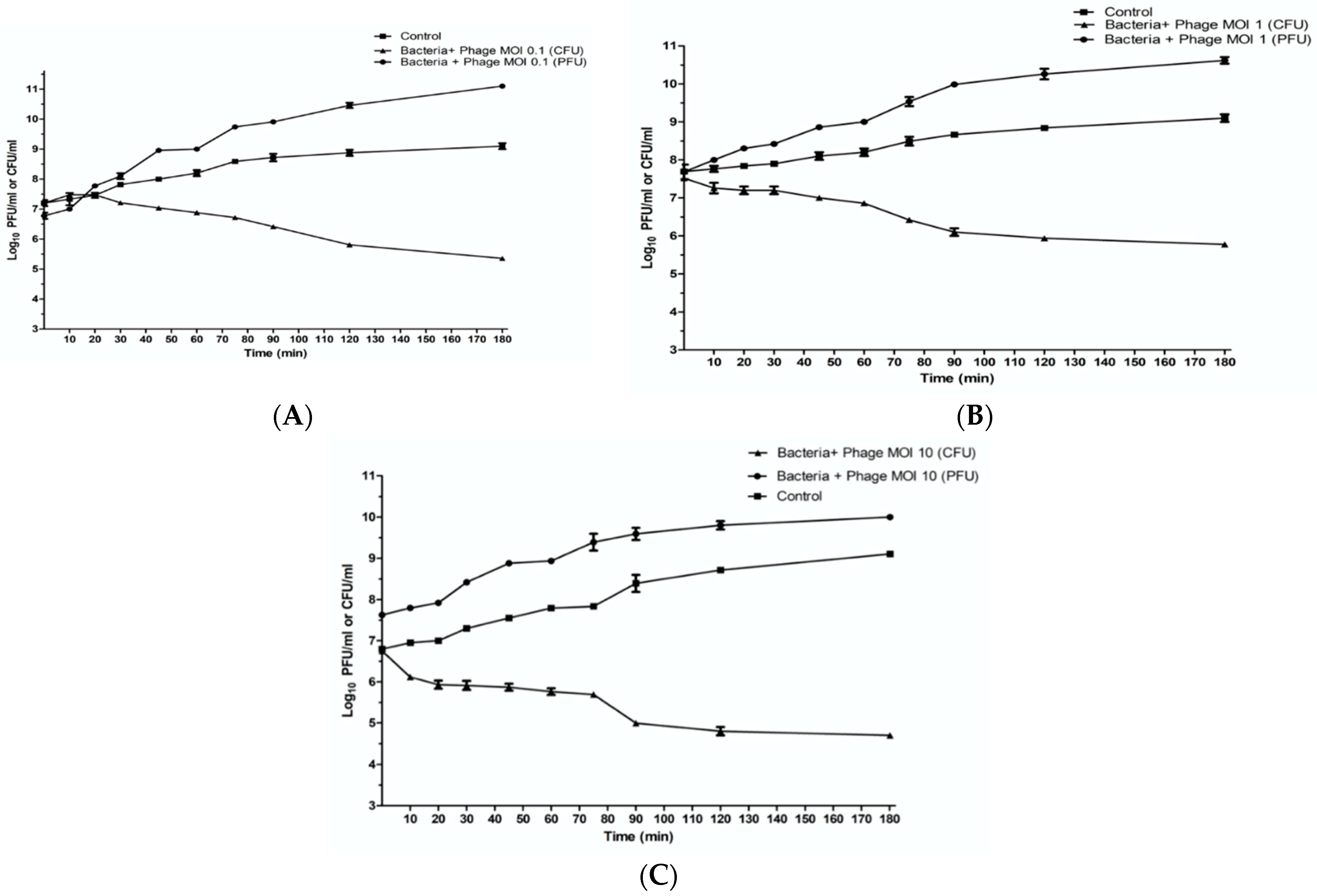
2.7. In Vitro Characterization of Phage ZCKP8
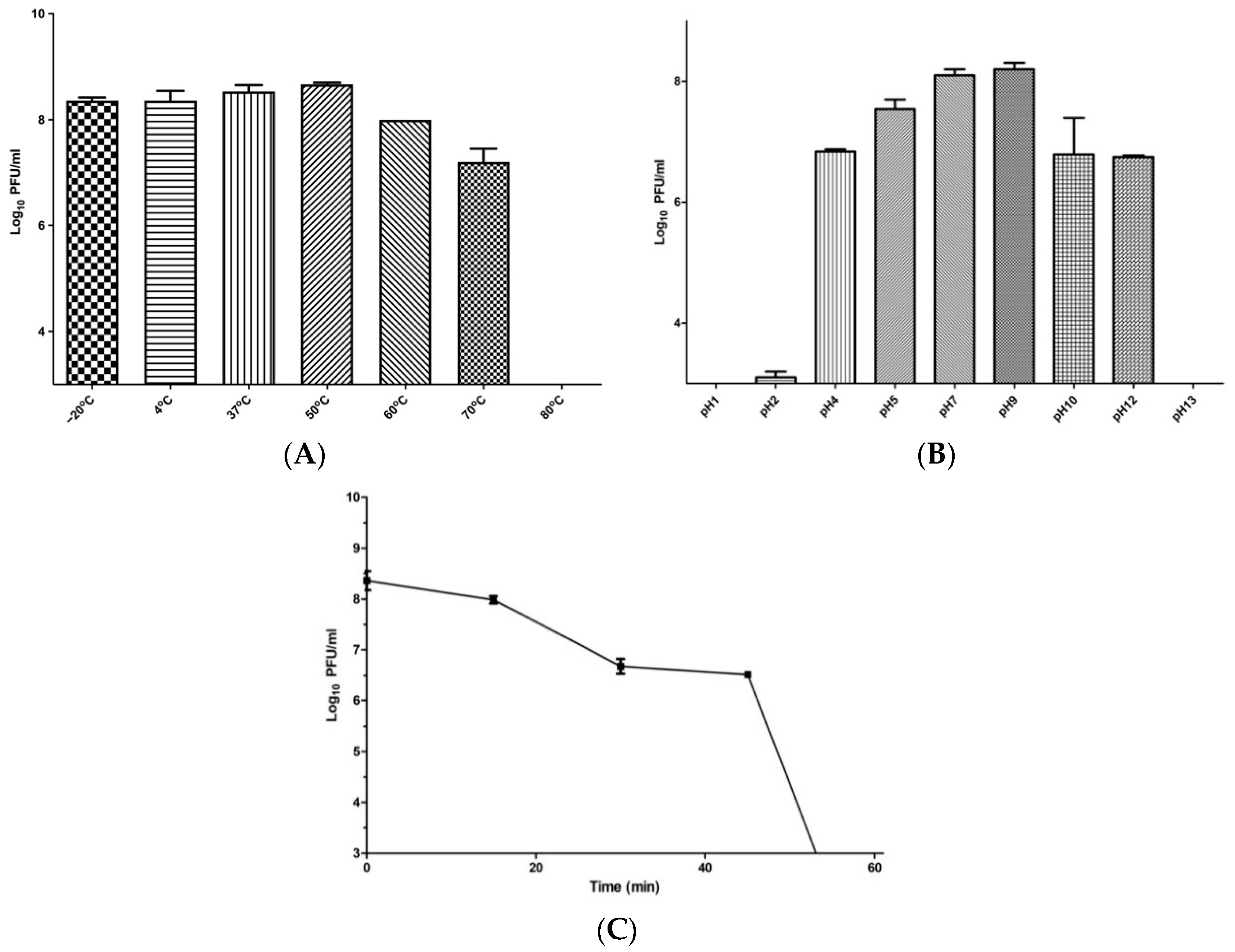
2.8. Phage Temperature, pH, and UV Stability
2.9. In Vivo Wound Healing
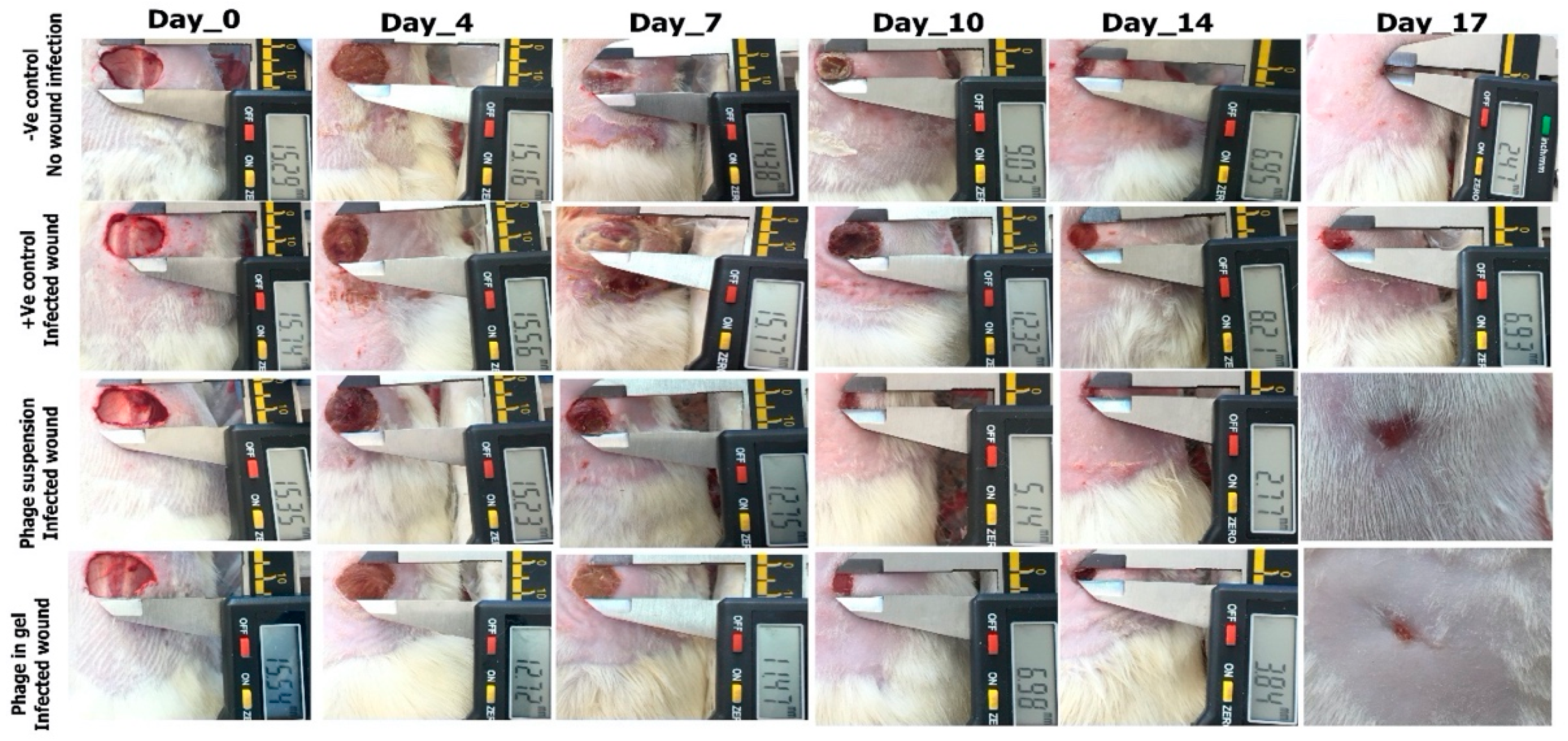
2.9.1. Photodocumentary Analysis and Wound Healing Percentage
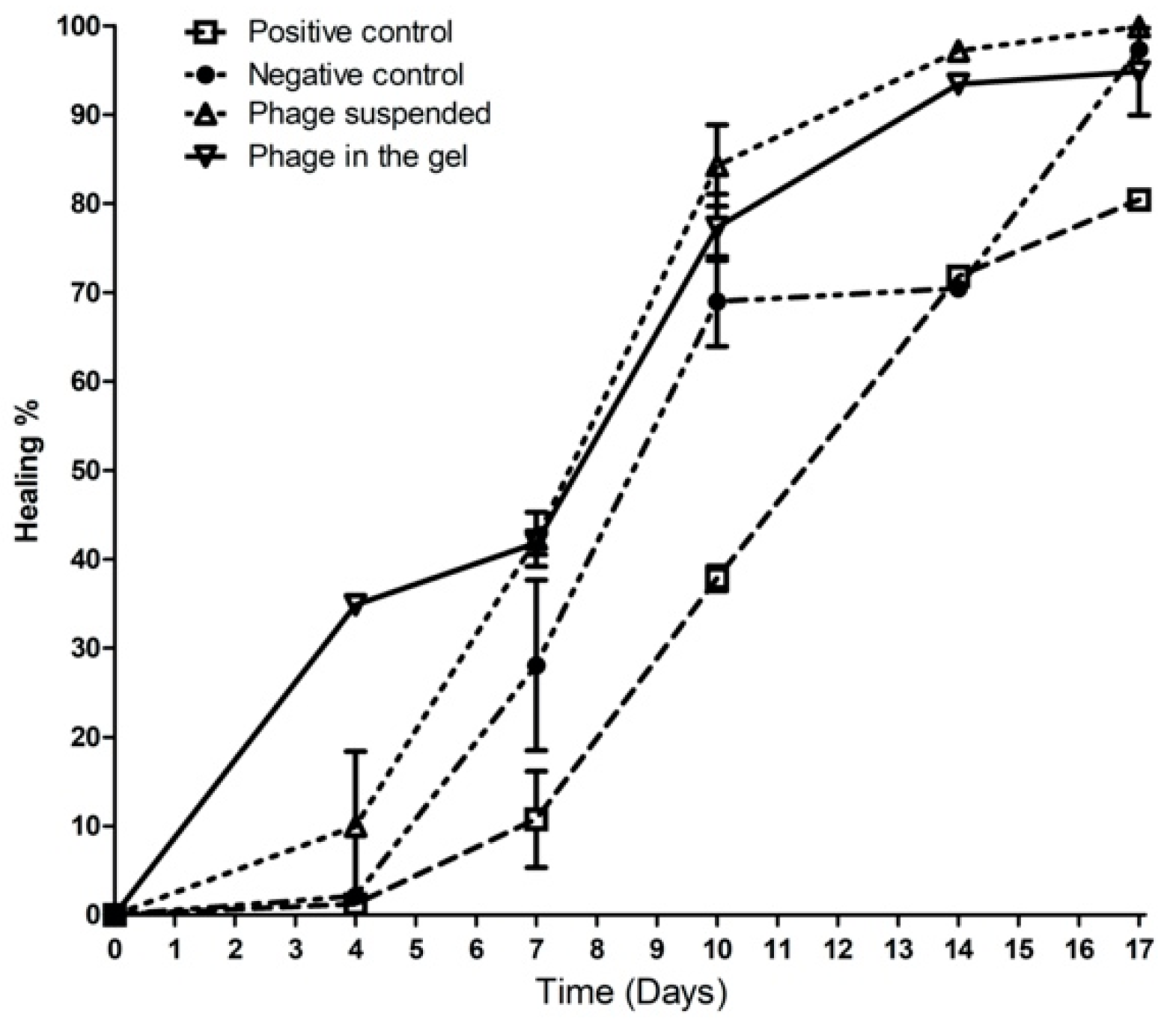
2.9.2. Wound Healing Percentage
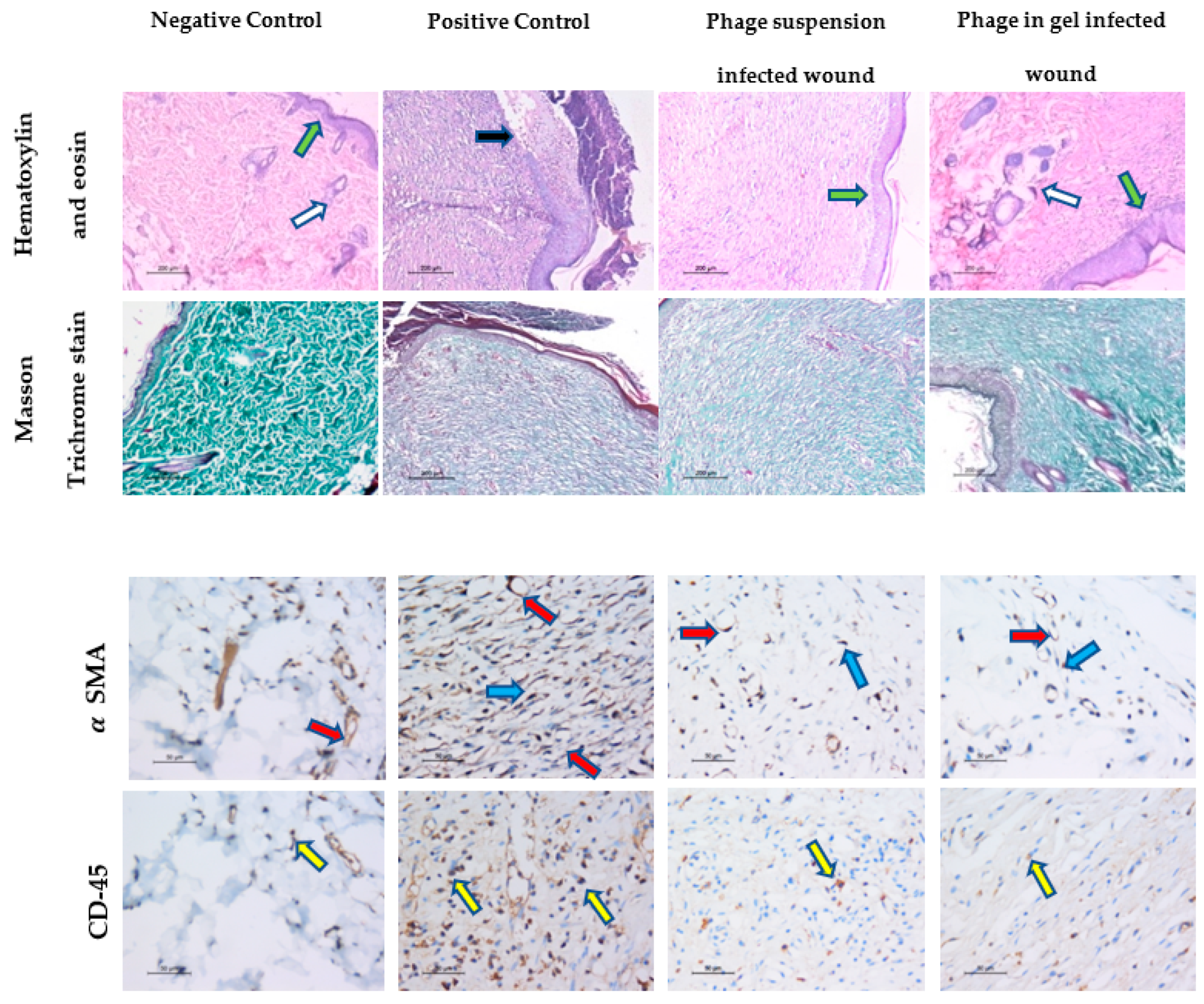
2.9.3. Histopathological Analysis of Wound Healing
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains Isolation and Stocking
4.2. Bacterial Identification and Confirmation by PCR
4.3. 16S rRNA Gene Sequencing
4.4. Antibiotic Sensitivity Test
4.5. Phage Selection, Isolation, Purification, and Amplification
4.6. Characterization of the Isolated Phage
4.6.1. Pulsed Field Gel Electrophoresis (PFGE)
4.6.2. Examination of Phage Morphology by Electron Microscopy (TEM)
4.6.3. Host Range Determination
4.6.4. Phage DNA Sequencing
4.6.5. In Vitro Characterization of Phage ZCKP8
4.6.6. Efficiency of Plating (EOP)
4.6.7. Determination of the Frequency of BIM
4.6.8. Phage pH, Temperature, and UV Stability
4.7. In Vivo Wound Healing Efficiency Using ZCKP8 Phage on a K. pneumoniae Infected Wound on a Rat Model
4.7.1. Surgical Procedures of Full-Thickness Wound Model
4.7.2. Histological Examination
4.7.3. IHC Staining and Interpretation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Das, A.; Acharya, S.; Behera, B.K.; Paria, P.; Bhowmick, S.; Parida, P.; Das, B. Isolation, identification and characterization of Klebsiella pneumoniae from infected farmed Indian Major Carp Labeo rohita (Hamilton 1822) in West Bengal, India. Aquaculture 2018, 482, 111–116. [Google Scholar] [CrossRef]
- Anand, T.; Virmani, N.; Kumar, S.; Mohanty, A.K.; Pavulraj, S.; Bera, B.C.; Vaid, R.K.; Ahlawat, U.; Tripathi, B. Phage therapy for treatment of virulent Klebsiella pneumoniae infection in a mouse model. J. Glob. Antimicrob. Resist. 2020, 21, 34–41. [Google Scholar] [CrossRef]
- Cao, F.; Wang, X.; Wang, L.; Li, Z.; Che, J.; Wang, L.; Li, X.; Cao, Z.; Zhang, J.; Jin, L.; et al. Evaluation of the Efficacy of a Bacteriophage in the Treatment of Pneumonia Induced by Multidrug ResistanceKlebsiella pneumoniaein Mice. BioMed Res. Int. 2015, 2015, 1–9. [Google Scholar] [CrossRef]
- Ramos-Castañeda, J.A.; Ruano-Ravina, A.; Barbosa-Lorenzo, R.; Paillier-Gonzalez, J.E.; Saldaña-Campos, J.C.; Salinas, D.F.; Lemos-Luengas, E.V. Mortality due to KPC carbapenemase-producing Klebsiella pneumoniae infections: Systematic review and meta-analysis. J. Infect. 2018, 76, 438–448. [Google Scholar] [CrossRef]
- Cryz, S.J. Progress in immunization againstKlebsiella infections. Eur. J. Clin. Microbiol. Infect. Dis. 1983, 2, 523–528. [Google Scholar] [CrossRef]
- Chung, P.Y. The emerging problems ofKlebsiella pneumoniaeinfections: Carbapenem resistance and biofilm formation. FEMS Microbiol. Lett. 2016, 363, 219. [Google Scholar] [CrossRef]
- Shoma, S.; Kamruzzaman, M.; Ginn, A.N.; Iredell, J.R.; Partridge, S.R. Characterization of multidrug-resistant Klebsiella pneumoniae from Australia carrying blaNDM-1. Diagn. Microbiol. Infect. Dis. 2014, 78, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Moghadam, M.T.; Khoshbayan, A.; Chegini, Z.; Farahani, I.; Shariati, A. Bacteriophages, a New Therapeutic Solution for Inhibiting Multidrug-Resistant Bacteria Causing Wound Infection: Lesson from Animal Models and Clinical Trials. Drug Des. Dev. Ther. 2020, 14, 1867–1883. [Google Scholar] [CrossRef]
- Simões, D.; Miguel, S.A.P.; Ribeiro, M.; Coutinho, P.; Mendonça, A.; Correia, I.J. Recent advances on antimicrobial wound dressing: A review. Eur. J. Pharm. Biopharm. 2018, 127, 130–141. [Google Scholar] [CrossRef]
- Bowler, P.G.; Duerden, B.I.; Armstrong, D.G. Wound Microbiology and Associated Approaches to Wound Management. Clin. Microbiol. Rev. 2001, 14, 244–269. [Google Scholar] [CrossRef]
- Stearns-Kurosawa, D.J.; Osuchowski, M.F.; Valentine, C.; Kurosawa, S.; Remick, D.G. The Pathogenesis of Sepsis. Annu. Rev. Pathol. Mech. Dis. 2011, 6, 19–48. [Google Scholar] [CrossRef] [PubMed]
- Shariati, A.; Moradabadi, A.; Azimi, T.; Ghaznavi-Rad, E. Wound healing properties and antimicrobial activity of platelet-derived biomaterials. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cienfuegos-Gallet, A.V.; de Los Ríos, A.M.; Viana, P.S.; Brinez, F.R.; Castro, C.R.; Villamil, G.R.; del Corral Londoño, H.; Jiménez, J.N. Risk factors and survival of patients infected with carbapenem-resistant Klebsiella pneumoniae in a KPC endemic setting: A case-control and cohort study. BMC Infect. Dis. 2019, 19, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Scalise, A.; Bianchi, A.; Tartaglione, C.; Bolletta, E.; Pierangeli, M.; Torresetti, M.; Marazzi, M.; Di Benedetto, G. Microenvironment and microbiology of skin wounds: The role of bacterial biofilms and related factors. Semin. Vasc. Surg. 2015, 28, 151–159. [Google Scholar] [CrossRef]
- Fijan, S.; Frauwallner, A.; Langerholc, T.; Krebs, B.; ter Haar née Younes, J.A.; Heschl, A.; Mičetić Turk, D.; Rogelj, I. Efficacy of Using Probiotics with Antagonistic Activity against Pathogens of Wound Infections: An Integrative Review of Literature. BioMed Res. Int. 2019, 2019, 1–21. [Google Scholar] [CrossRef]
- Mihai, M.M.; Preda, M.; Lungu, I.; Gestal, M.C.; Popa, M.I.; Holban, A.M. Nanocoatings for Chronic Wound Repair—Modulation of Microbial Colonization and Biofilm Formation. Int. J. Mol. Sci. 2018, 19, 1179. [Google Scholar] [CrossRef]
- Nakamura, Y.; Daya, M. Use of Appropriate Antimicrobials in Wound Management. Emerg. Med. Clin. N. Am. 2007, 25, 159–176. [Google Scholar] [CrossRef]
- Chegini, Z.; Khoshbayan, A.; Vesal, S.; Moradabadi, A.; Hashemi, A.; Shariati, A. Bacteriophage therapy for inhibition of multi drug-resistant uropathogenic bacteria: A narrative review. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 1–13. [Google Scholar] [CrossRef]
- Azimi, T.; Mosadegh, M.; Nasiri, M.J.; Sabour, S.; Karimaei, S.; Nasser, A. Phage therapy as a renewed therapeutic approach to mycobacterial infections: A comprehensive review. Infect. Drug Resist. 2019, 12, 2943–2959. [Google Scholar] [CrossRef]
- Abdelsattar, A.; Nofal, R.; Makky, S.; Safwat, A.; Taha, A.; El-Shibiny, A. The Synergistic Effect of Biosynthesized Silver Nanoparticles and Phage ZCSE2 as a Novel Approach to Combat Multidrug-Resistant Salmonella enterica. Antibiot. 2021, 10, 678. [Google Scholar] [CrossRef]
- Harada, L.K.; Silva, E.C.; Campos, W.F.; Fiol, F.D.S.D.; Vila, M.; Dąbrowska, K.; Krylov, V.N.; Balcão, V.M. Biotechnological applications of bacteriophages: State of the art. Microbiol. Res. 2018, 212–213, 38–58. [Google Scholar] [CrossRef] [PubMed]
- Beceiro, A.; Tomás, M.; Bou, G. Antimicrobial Resistance and Virulence: A Successful or Deleterious Association in the Bacterial World? Clin. Microbiol. Rev. 2013, 26, 185–230. [Google Scholar] [CrossRef]
- Alsaadi, A.; Beamud, B.; Easwaran, M.; Abdelrahman, F.; El-Shibiny, A.; Alghoribi, M.F.; Domingo-Calap, P. Learning From Mistakes: The Role of Phages in Pandemics. Front. Microbiol. 2021, 12, 653107. [Google Scholar] [CrossRef]
- Kęsik-Szeloch, A.; Drulis-Kawa, Z.; Weber-Dąbrowska, B.; Kassner, J.; Majkowska-Skrobek, G.; Augustyniak, D.; Łusiak-Szelachowska, M.; Żaczek, M.; Górski, A.; Kropinski, A.M. Characterising the biology of novel lytic bacteriophages infecting multidrug resistant Klebsiella pneumoniae. Virol. J. 2013, 10, 100. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.P.; Oliveira, H.; Melo, L.; Sillankorva, S.; Azeredo, J. Bacteriophage-encoded depolymerases: Their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 2016, 100, 2141–2151. [Google Scholar] [CrossRef] [PubMed]
- Hyman, P.; Abedon, S.T. Practical Methods for Determining Phage Growth Parameters. Methods Mol. Biol. 2009, 501, 175–202. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.; Ward, S.; Hyman, P. More Is Better: Selecting for Broad Host Range Bacteriophages. Front. Microbiol. 2016, 7, 1352. [Google Scholar] [CrossRef]
- Verma, V.; Harjai, K.; Chhibber, S. Characterization of a T7-Like Lytic Bacteriophage of Klebsiella pneumoniae B5055: A Potential Therapeutic Agent. Curr. Microbiol. 2009, 59, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Shi, H.; Zhang, Z.; Han, F.; Li, J.; Sun, Y. Isolation and characterization of a lytic bacteriophage φKp-lyy15 of Klebsiella pneumoniae. Virol. Sin. 2015, 30, 66–68. [Google Scholar] [CrossRef]
- Park, E.-A.; Kim, Y.-T.; Cho, J.-H.; Ryu, S.; Lee, J.-H. Characterization and genome analysis of novel bacteriophages infecting the opportunistic human pathogens Klebsiella oxytoca and K. pneumoniae. Arch. Virol. 2016, 162, 1129–1139. [Google Scholar] [CrossRef]
- Karumidze, N.; Kusradze, I.; Rigvava, S.; Goderdzishvili, M.; Rajakumar, K.; Alavidze, Z. Isolation and Characterisation of Lytic Bacteriophages of Klebsiella pneumoniae and Klebsiella oxytoca. Curr. Microbiol. 2013, 66, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Kusradze, I.; Karumidze, N.; Rigvava, S.; Dvalidze, T.; Katsitadze, M.; Amiranashvili, I.; Goderdzishvili, M. Characterization and Testing the Efficiency of Acinetobacter baumannii Phage vB-GEC_Ab-M-G7 as an Antibacterial Agent. Front. Microbiol. 2016, 7, 1590. [Google Scholar] [CrossRef] [PubMed]
- Wintachai, P.; Naknaen, A.; Thammaphet, J.; Pomwised, R.; Phaonakrop, N.; Roytrakul, S.; Smith, D.R. Characterization of extended-spectrum-β-lactamase producing Klebsiella pneumoniae phage KP1801 and evaluation of therapeutic efficacy in vitro and in vivo. Sci. Rep. 2020, 10, 1–18. [Google Scholar] [CrossRef]
- Horváth, M.; Kovács, T.; Koderivalappil, S.; Ábrahám, H.; Rákhely, G.; Schneider, G. Identification of a newly isolated lytic bacteriophage against K24 capsular type, carbapenem resistant Klebsiella pneumoniae isolates. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Jamal, M.; Hussain, T.; Das, C.R.; Andleeb, S. Characterization of Siphoviridae phage Z and studying its efficacy against multidrug-resistant Klebsiella pneumoniae planktonic cells and biofilm. J. Med. Microbiol. 2015, 64, 454–462. [Google Scholar] [CrossRef]
- Chhibber, S.; Nag, D.; Bansal, S. Inhibiting biofilm formation by Klebsiella pneumoniae B5055 using an iron antagonizing molecule and a bacteriophage. BMC Microbiol. 2013, 13, 174. [Google Scholar] [CrossRef]
- Penadés, J.R.; Chen, J.; Quiles-Puchalt, N.; Carpena, N.; Novick, R. Bacteriophage-mediated spread of bacterial virulence genes. Curr. Opin. Microbiol. 2015, 23, 171–178. [Google Scholar] [CrossRef]
- Colomer-Lluch, M.; Jofre, J.; Muniesa, M. Antibiotic Resistance Genes in the Bacteriophage DNA Fraction of Environmental Samples. PLoS ONE 2011, 6, e17549. [Google Scholar] [CrossRef]
- Takeo, M.; Lee, W.; Ito, M. Wound Healing and Skin Regeneration. Cold Spring Harb. Perspect. Med. 2015, 5, a023267. [Google Scholar] [CrossRef]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 162–173. [Google Scholar] [CrossRef]
- Harper, D.R.; Parracho, H.M.R.T.; Walker, J.; Sharp, R.J.; Hughes, G.; Werthén, M.; Lehman, S.M.; Morales, S. Bacteriophages and Biofilms. Antibiotics 2014, 3, 270–284. [Google Scholar] [CrossRef]
- Derakhshan, S.; Peerayeh, S.N.; Bakhshi, B. Association Between Presence of Virulence Genes and Antibiotic Resistance in ClinicalKlebsiella PneumoniaeIsolates. Lab. Med. 2016, 47, 306–311. [Google Scholar] [CrossRef]
- Srivastava, S.; Singh, V.; Kumar, V.; Verma, P.C.; Srivastava, R.; Basu, V.; Gupta, V.; Rawat, A.K. Identification of regulatory elements in 16S rRNA gene of Acinetobacter species isolated from water sample. Bioinformation 2008, 3, 173–176. [Google Scholar] [CrossRef][Green Version]
- Abraham, O.-S.J.; Miguel, T.-S.; Inocencio, H.-C.; Blondy, C.-C. A quick and effective in-house method of DNA purification from agarose gel, suitable for sequencing. 3 Biotech 2017, 7, 180. [Google Scholar] [CrossRef] [PubMed]
- Treves, D.S. Review of Three DNA Analysis Applications for Use in the Microbiology or Genetics Classroom. J. Microbiol. Biol. Educ. 2010, 11, 186–187. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Townsend, E.M.; Kelly, L.; Gannon, L.; Muscatt, G.; Dunstan, R.; Michniewski, S.; Sapkota, H.; Kiljunen, S.J.; Kolsi, A.; Skurnik, M.; et al. Isolation and Characterization of Klebsiella Phages for Phage Therapy. PHAGE 2021, 2, 26–42. [Google Scholar] [CrossRef] [PubMed]
- Bourdin, G.; Schmitt, B.; Guy, L.M.; Germond, J.-E.; Zuber, S.; Michot, L.; Reuteler, G.; Brüssow, H. Amplification and Purification of T4-Like Escherichia coli Phages for Phage Therapy: From Laboratory to Pilot Scale. Appl. Environ. Microbiol. 2013, 80, 1469–1476. [Google Scholar] [CrossRef]
- Pallavali, R.R.; Degati, V.L.; Lomada, D.; Reddy, M.; Durbaka, V.R.P. Isolation and in vitro evaluation of bacteriophages against MDR-bacterial isolates from septic wound infections. PLoS ONE 2017, 12, e0179245. [Google Scholar] [CrossRef] [PubMed]
- Abdelsattar, A.S.; Abdelrahman, F.; Dawoud, A.; Connerton, I.F.; El-Shibiny, A. Encapsulation of E. coli phage ZCEC5 in chitosan–alginate beads as a delivery system in phage therapy. AMB Express 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Islam, M.; Zhou, Y.; Liang, L.; Nime, I.; Liu, K.; Yan, T.; Wang, X.; Li, J. Application of a Phage Cocktail for Control of Salmonella in Foods and Reducing Biofilms. Viruses 2019, 11, 841. [Google Scholar] [CrossRef]
- Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of Bacteriophages by the Direct Plating Plaque Assay. In Advanced Structural Safety Studies; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2009; Volume 501, pp. 77–80. [Google Scholar]
- Pertics, B.; Cox, A.; Nyúl, A.; Szamek, N.; Kovács, T.; Schneider, G. Isolation and Characterization of a Novel Lytic Bacteriophage against the K2 Capsule-Expressing Hypervirulent Klebsiella pneumoniae Strain 52145, and Identification of Its Functional Depolymerase. Microorganisms 2021, 9, 650. [Google Scholar] [CrossRef]
- Senczek, D.; Stephan, R.; Untermann, F. Pulsed-field gel electrophoresis (PFGE) typing of Listeria strains isolated from a meat processing plant over a 2-year period. Int. J. Food Microbiol. 2000, 62, 155–159. [Google Scholar] [CrossRef]
- Taha, O.A.; Connerton, P.L.; Connerton, I.; El-Shibiny, A. Bacteriophage ZCKP1: A Potential Treatment for Klebsiella pneumoniae Isolated From Diabetic Foot Patients. Front. Microbiol. 2018, 9, 2127. [Google Scholar] [CrossRef] [PubMed]
- LaMar, D. FastQC. 2015. Available online: https://qubeshub.org/resources/fastqc (accessed on 8 July 2021).
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Furuno, M.; Kasukawa, T.; Saito, R.; Adachi, J.; Suzuki, H.; Baldarelli, R.; Hayashizaki, Y.; Okazaki, Y. CDS Annotation in Full-Length cDNA Sequence. Genome Res. 2003, 13, 1478–1487. [Google Scholar] [CrossRef]
- Armon, R.; Kott, Y. A simple, rapid and sensitive presence/absence detection test for bacteriophage in drinking water. J. Appl. Bacteriol. 1993, 74, 490–496. [Google Scholar] [CrossRef] [PubMed]
- Viazis, S.; Akhtar, M.; Feirtag, J.; Brabban, A.; Diez-Gonzalez, F. Isolation and characterization of lytic bacteriophages against enterohaemorrhagic Escherichia coli. J. Appl. Microbiol. 2011, 110, 1323–1331. [Google Scholar] [CrossRef]
- O’Flynn, G.; Ross, R.P.; Fitzgerald, G.F.; Coffey, A. Evaluation of a Cocktail of Three Bacteriophages for Biocontrol of Escherichia coli O157:H7. Appl. Environ. Microbiol. 2004, 70, 3417–3424. [Google Scholar] [CrossRef]
- Capra, M.; Quiberoni, A.; Reinheimer, J. Thermal and chemical resistance of Lactobacillus casei and Lactobacillus paracasei bacteriophages. Lett. Appl. Microbiol. 2004, 38, 499–504. [Google Scholar] [CrossRef]
- Hammerl, J.A.; Jäckel, C.; Alter, T.; Janzcyk, P.; Stingl, K.; Knüver, M.T.; Hertwig, S. Reduction of Campylobacter jejuni in Broiler Chicken by Successive Application of Group II and Group III Phages. PLoS ONE 2014, 9, e114785. [Google Scholar] [CrossRef] [PubMed]
- Silva, Y.J.; Costa, L.; Pereira, C.; Cunha, A.; Calado, R.; Gomes, N.; Almeida, A. Influence of environmental variables in the efficiency of phage therapy in aquaculture. Microb. Biotechnol. 2014, 7, 401–413. [Google Scholar] [CrossRef] [PubMed]
- El-Aassar, M.R.; El-Beheri, N.G.; Agwa, M.M.; Eltaher, H.M.; Alseqely, M.; Sadik, W.S.; El-Khordagui, L. Antibiotic-free combinational hyaluronic acid blend nanofibers for wound healing enhancement. Int. J. Biol. Macromol. 2021, 167, 1552–1563. [Google Scholar] [CrossRef]
- Shalaby, M.; Agwa, M.; Saeed, H.; Khedr, S.M.; Morsy, O.; El-Demellawy, M.A. Fish Scale Collagen Preparation, Characterization and Its Application in Wound Healing. J. Polym. Environ. 2020, 28, 166–178. [Google Scholar] [CrossRef]
- Kifelew, L.G.; Warner, M.S.; Morales, S.; Vaughan, L.; Woodman, R.; Fitridge, R.; Mitchell, J.G.; Speck, P. Efficacy of phage cocktail AB-SA01 therapy in diabetic mouse wound infections caused by multidrug-resistant Staphylococcus aureus. BMC Microbiol. 2020, 20, 1–10. [Google Scholar] [CrossRef]
- Capparelli, R.; Parlato, M.; Borriello, G.; Salvatore, P.; Iannelli, D. Experimental Phage Therapy against Staphylococcus aureus in Mice. Antimicrob. Agents Chemother. 2007, 51, 2765–2773. [Google Scholar] [CrossRef]
- Merabishvili, M.; Monserez, R.; Van Belleghem, J.; Rose, T.; Jennes, S.; De Vos, D.; Verbeken, G.; Vaneechoutte, M.; Pirnay, J.-P. Stability of bacteriophages in burn wound care products. PLoS ONE 2017, 12, e0182121. [Google Scholar] [CrossRef]
- Dunn, L.; Prosser, H.C.G.; Tan, M.-J.; Vanags, L.Z.; Ng, M.K.C.; Bursill, C. Murine Model of Wound Healing. J. Vis. Exp. 2013, 75, e50265. [Google Scholar] [CrossRef] [PubMed]
- Santos, T.; Santos, I.; Pereira-Filho, R.; Gomes, S.; Lima-Verde, I.; Marques, M.; Cardoso, J.; Severino, P.; Souto, E.; Albuquerque-Júnior, R. Histological Evidence of Wound Healing Improvement in Rats Treated with Oral Administration of Hydroalcoholic Extract of Vitis labrusca. Curr. Issues Mol. Biol. 2021, 43, 335–352. [Google Scholar] [CrossRef]
- Park, I.-S.; Chung, P.-S.; Ahn, J.C. Enhancement of Ischemic Wound Healing by Spheroid Grafting of Human Adipose-Derived Stem Cells Treated with Low-Level Light Irradiation. PLoS ONE 2015, 10, e0122776. [Google Scholar] [CrossRef] [PubMed]
| Antibiotics | AK 30 | ATM 30 | CIP 5 | CAZ 30 | Fep 30 | CN 10 | IPM 10 | LEV 5 | SXT 25 | TGC 15 | TZP 110 | LZD 30 | MAR Index |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KP/1 | S | R | R | R | R | S | I | I | R | S | I | R | 0.5 |
| KP/2 | S | R | R | R | R | S | S | I | R | R | R | R | 0.7 |
| KP/3 | S | S | R | I | I | S | S | R | R | R | I | R | 0.4 |
| KP/4 | S | S | I | I | I | S | I | I | R | R | I | R | 0.3 |
| KP/5 | R | S | R | R | R | S | R | R | R | R | R | R | 0.8 |
| KP/6 | R | R | R | R | R | S | S | I | R | R | R | R | 0.8 |
| KP/7 | I | S | S | S | S | S | I | S | R | R | I | R | 0.3 |
| KP/8 | R | R | R | R | R | R | R | I | R | R | R | R | 0.9 |
| KP/9 | R | R | R | R | R | R | R | R | R | S | I | R | 0.8 |
| KP/10 | R | R | R | R | R | R | R | R | R | R | R | R | 1 |
| KP/11 | R | R | R | R | R | R | R | R | R | R | I | R | 0.9 |
| KP/12 | S | R | R | R | R | R | S | R | R | R | I | R | 0.8 |
| KP/13 | R | R | R | R | R | R | I | R | R | R | R | R | 0.9 |
| KP/14 | S | I | R | I | I | S | S | R | R | I | I | R | 0.3 |
| KP/15 | I | R | R | R | R | S | S | I | R | R | R | R | 0.7 |
| KP/16 | R | R | R | I | S | S | R | R | R | S | R | R | 0.7 |
| KP/17 | S | I | R | I | I | S | S | R | R | S | I | R | 0.3 |
| KP/18 | S | R | I | S | R | S | S | S | R | S | S | R | 0.3 |
| KP/19 | S | R | I | I | R | S | I | I | R | I | S | R | 0.3 |
| KP/20 | S | S | S | R | I | S | S | R | R | I | R | R | 0.4 |
| Total R% | 8(40%) | 13(65%) | 15(75%) | 12(60%) | 13(65%) | 6(30%) | 6(30%) | 11(55%) | 20(100%) | 12(60%) | 9(45%) | 20(100%) |
| Bacterial Species | K. pneumoniae (n = 8) |
|---|---|
| EOP 1 | 4 |
| EOP 1 | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fayez, M.S.; Hakim, T.A.; Agwa, M.M.; Abdelmoteleb, M.; Aly, R.G.; Montaser, N.N.; Abdelsattar, A.S.; Rezk, N.; El-Shibiny, A. Topically Applied Bacteriophage to Control Multi-Drug Resistant Klebsiella pneumoniae Infected Wound in a Rat Model. Antibiotics 2021, 10, 1048. https://doi.org/10.3390/antibiotics10091048
Fayez MS, Hakim TA, Agwa MM, Abdelmoteleb M, Aly RG, Montaser NN, Abdelsattar AS, Rezk N, El-Shibiny A. Topically Applied Bacteriophage to Control Multi-Drug Resistant Klebsiella pneumoniae Infected Wound in a Rat Model. Antibiotics. 2021; 10(9):1048. https://doi.org/10.3390/antibiotics10091048
Chicago/Turabian StyleFayez, Mohamed S., Toka A. Hakim, Mona M. Agwa, Mohamed Abdelmoteleb, Rania G. Aly, Nada N. Montaser, Abdallah S. Abdelsattar, Nouran Rezk, and Ayman El-Shibiny. 2021. "Topically Applied Bacteriophage to Control Multi-Drug Resistant Klebsiella pneumoniae Infected Wound in a Rat Model" Antibiotics 10, no. 9: 1048. https://doi.org/10.3390/antibiotics10091048
APA StyleFayez, M. S., Hakim, T. A., Agwa, M. M., Abdelmoteleb, M., Aly, R. G., Montaser, N. N., Abdelsattar, A. S., Rezk, N., & El-Shibiny, A. (2021). Topically Applied Bacteriophage to Control Multi-Drug Resistant Klebsiella pneumoniae Infected Wound in a Rat Model. Antibiotics, 10(9), 1048. https://doi.org/10.3390/antibiotics10091048








