Whole-Genome Sequencing for Molecular Characterization of Carbapenem-Resistant Enterobacteriaceae Causing Lower Urinary Tract Infection among Pediatric Patients
Abstract
1. Introduction
2. Results
2.1. Demographics and Etiology
2.2. Phenotypic Resistance Profiles of the CRE Isolates
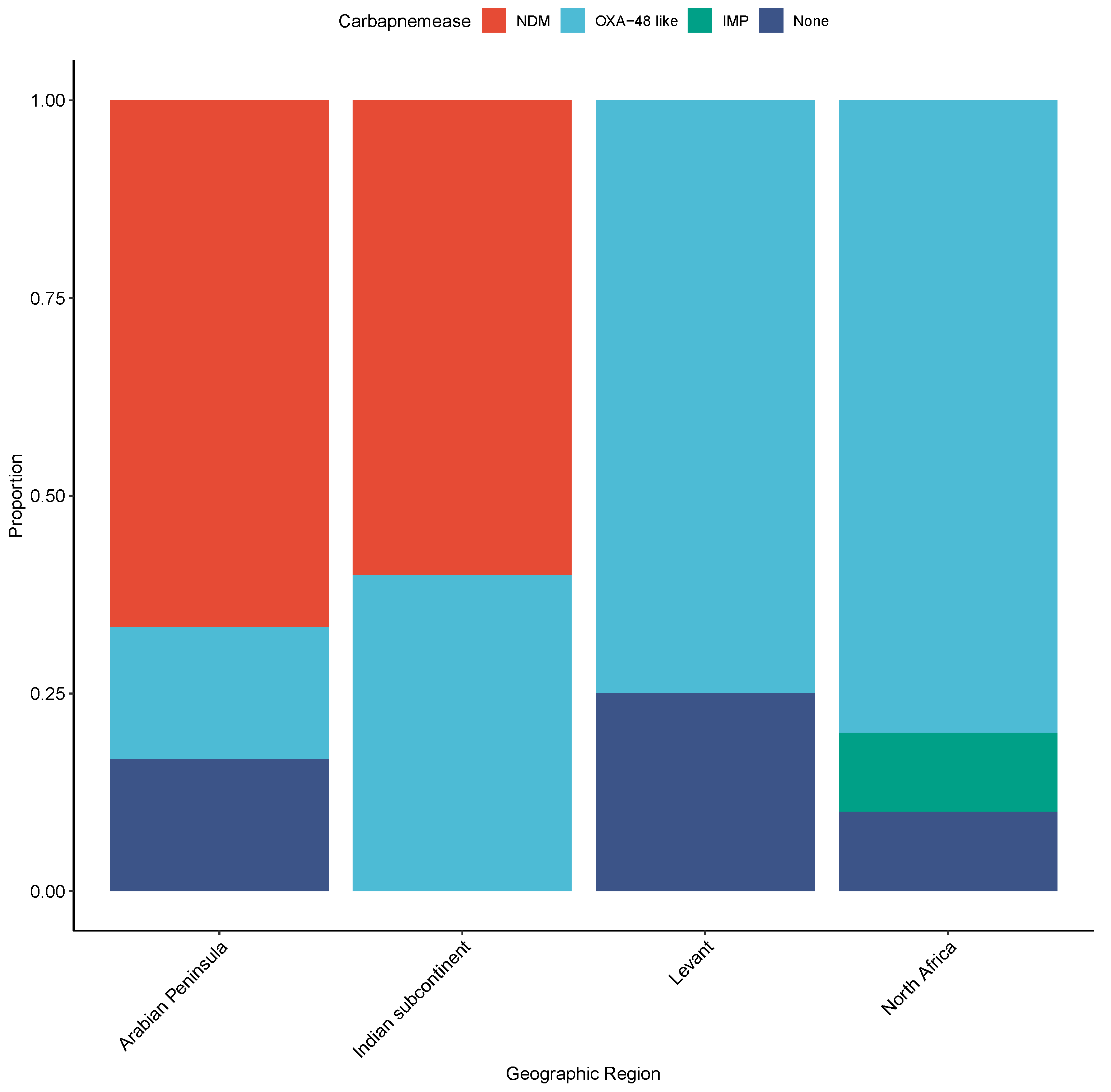
2.3. Molecular Genotyping Profile of CRE Isolates
3. Discussion
4. Materials and Methods
4.1. Clinical Isolates and Control Strains
4.2. Carbapenemase Phenotypic Confirmation
4.3. Molecular Characterization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2013; US Department of Health and Human Services, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2013. [Google Scholar]
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2019; US Department of Health and Human Services, Centers for Disease Control and Prevention: Atlanta, GA, USA, 2019. [Google Scholar]
- Smith, H.Z.; Kendall, B. Carbapenem-Resistant Enterobacteriaceae (CRE); StatPearls: Treasure Islanfe, FL, USA, 2019. [Google Scholar]
- Tacconelli, E.; Magrini, N.; Kahlmeter, G.; Singh, N. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. Lancet Infect. Dis. 2017, 27, 318–327. [Google Scholar]
- Vardakas, K.Z.; Tansarli, G.S.; Rafailidis, P.I.; Falagas, M.E. Carbapenems versus alternative antibiotics for the treatment of bacteraemia due to Enterobacteriaceae producing extended-spectrum β-lactamases: A systematic review and meta-analysis. J. Antimicrob. Chemother. 2012, 67, 2793–2803. [Google Scholar] [CrossRef] [PubMed]
- Codjoe, F.S.; Donkor, E.S. Carbapenem Resistance: A Review. Med. Sci. 2017, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Marsik, F.J.; Nambiar, S. Review of carbapenemases and AmpC-beta lactamases. Pediatr. Infect. Dis. J. 2011, 30, 1094–1095. [Google Scholar] [CrossRef]
- Patel, G.; Bonomo, R. “Stormy waters ahead”: Global emergence of carbapenemases. Front. Microbiol. 2013, 4, 48. [Google Scholar] [CrossRef] [PubMed]
- Walsh, F. The multiple roles of antibiotics and antibiotic resistance in nature. Front. Microbiol. 2013, 4, 255. [Google Scholar] [CrossRef] [PubMed]
- Gasink, L.B.; Edelstein, P.H.; Lautenbach, E.; Synnestvedt, M.; Fishman, N.O. Risk factors and clinical impact of Klebsiella pneumoniae carbapenemase–producing K. pneumoniae. Infect. Control Hosp. Epidemiol. 2009, 30, 1180. [Google Scholar] [CrossRef]
- Arnold, R.S.; Thom, K.A.; Sharma, S.; Phillips, M.; Johnson, J.K.; Morgan, D.J. Emergence of Kl1–3ebsiella pneumoniae carbapenemase (KPC)-producing bacteria. South Med. J. 2011, 104, 40–45. [Google Scholar] [CrossRef]
- Tzouvelekis, L.; Markogiannakis, A.; Psichogiou, M.; Tassios, P.; Daikos, G. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: An evolving crisis of global dimensions. Clin. Microbiol. Rev. 2012, 25, 682–707. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Dortet, L.; Poirel, L. Carbapenem resistance in Enterobacteriaceae: Here is the storm! Trends Mol. Med. 2012, 18, 263–272. [Google Scholar] [CrossRef]
- Zorc, J.J.; Kiddoo, D.A.; Shaw, K.N. Diagnosis and management of pediatric urinary tract infections. Clin. Microbiol. Rev. 2005, 18, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Logan, L.K.; Weinstein, R.A. The epidemiology of carbapenem-resistant Enterobacteriaceae: The impact and evolution of a global menace. J. Infect. Dis. 2017, 215 (Suppl. 1), S28–S36. [Google Scholar] [CrossRef] [PubMed]
- Touati, A.; Mairi, A. Epidemiology of Carbapenemase-producing Enterobacterales in the Middle East: A systematic review. Expert Rev. Anti-Infect. Ther. 2020, 18, 241–250. [Google Scholar] [CrossRef]
- Chiotos, K.; Han, J.H.; Tamma, P.D. Carbapenem-resistant Enterobacteriaceae infections in children. Curr. Infect. Dis. Rep. 2016, 18, 2. [Google Scholar] [CrossRef]
- Abid, F.B.; Tsui, C.K.M.; Doi, Y.; Deshmukh, A.; McElheny, C.L.; Bachman, W.C.; Fowler, E.L.; Albishawi, A.; Mushtaq, K.; Ibrahim, E.B.; et al. Molecular characterization of clinical carbapenem-resistant Enterobacterales from Qatar. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2021, 40, 1779–1785. [Google Scholar] [CrossRef] [PubMed]
- Queenan, A.M.; Bush, K. Carbapenemases: The Versatile β-Lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef] [PubMed]
- Zahedi-Bialvaei, A.; Samadi-Kafil, H.; Ebrahimzadeh-Leylabadlo, H.; Asgharzadeh, M.; Aghazadeh, M. Dissemination of carbapenemases producing Gram negative bacteria in the Middle East. Iran. J. Microbiol. 2015, 7, 226–246. [Google Scholar] [PubMed]
- Eltai, N.O.; Yassine, H.M.; al Thani, A.A.; Madi, M.A.A.; Ismail, A.; Ibrahim, E.; Alali, W.Q. Prevalence of antibiotic resistant Escherichia coli isolates from fecal samples of food handlers in Qatar. Antimicrob. Resist. Infect. Control 2018, 7, 78. [Google Scholar] [CrossRef]
- Walsh, T.R. New Delhi metallo-β-lactamase-1: Detection and prevention. CMAJ Can. Med. Assoc. J. 2011, 183, 1240–1241. [Google Scholar] [CrossRef][Green Version]
- Poirel, L.; Potron, A.; Nordmann, P. OXA-48-like carbapenemases: The phantom menace. J. Antimicrob. Chemother. 2012, 67, 1597–1606. [Google Scholar] [CrossRef]
- Sonnevend, Á.; Ghazawi, A.A.; Hashmey, R.; Jamal, W.; Rotimi, V.O.; Shibl, A.M.; Al-Jardani, A.; Al-Abri, S.S.; Tariq, W.U.Z.; Weber, S.; et al. Characterization of Carbapenem-Resistant Enterobacteriaceae with High Rate of Autochthonous Transmission in the Arabian Peninsula. PLoS ONE 2015, 10, e0131372. [Google Scholar] [CrossRef]
- Oteo, J.; Hernández, J.M.; Espasa, M.; Fleites, A.; Sáez, D.; Bautista, V.; Pérez-Vázquez, M.; Fernández-García, M.D.; Delgado-Iribarren, A.; Sánchez-Romero, I. Emergence of OXA-48-producing Klebsiella pneumoniae and the novel carbapenemases OXA-244 and OXA-245 in Spain. J. Antimicrob. Chemother. 2013, 68, 317–321. [Google Scholar] [CrossRef]
- Potron, A.; Poirel, L.; Dortet, L.; Nordmann, P. Characterisation of OXA-244, a chromosomally-encoded OXA-48-like β-lactamase from Escherichia coli. Int. J. Antimicrob. Agents 2016, 47, 102–103. [Google Scholar] [CrossRef]
- Masseron, A.; Poirel, L.; Falgenhauer, L.; Imirzalioglu, C.; Kessler, J.; Chakraborty, T.; Nordmann, P. Ongoing dissemination of OXA-244 carbapenemase-producing Escherichia coli in Switzerland and their detection. Diagn. Microbiol. Infect. Dis. 2020, 97, 115059. [Google Scholar] [CrossRef] [PubMed]
- Hoyos-Mallecot, Y.; Naas, T.; Bonnin, R.A.; Patino, R.; Glaser, P.; Fortineau, N.; Dortet, L. OXA-244-Producing Escherichia coli Isolates, a Challenge for Clinical Microbiology Laboratories. Antimicrob. Agents Chemother. 2017, 61, e00818-17. [Google Scholar] [CrossRef]
- Doménech-Sánchez, A.; Hernández-Allés, S.; Martínez-Martínez, L.; Benedí, V.J.; Albertí, S. Identification and characterization of a new porin gene of Klebsiella pneumoniae: Its role in beta-lactam antibiotic resistance. J. Bacteriol. 1999, 181, 2726–2732. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, E.; Kojima, S.; Nikaido, H. Klebsiella pneumoniae Major Porins OmpK35 and OmpK36 Allow More Efficient Diffusion of β-Lactams than Their Escherichia coli Homologs OmpF and OmpC. J. Bacteriol. 2016, 198, 3200–3208. [Google Scholar] [CrossRef]
- Wong, J.L.C.; Romano, M.; Kerry, L.E.; Kwong, H.O.; Low, W.E.; Brett, S.J.; Clements, A.; Beis, K.; Frankel, G. OmpK36-mediated Carbapenem resistance attenuates ST258 Klebsiella pneumoniae in vivo. Nat. Commun. 2019, 10, 3957. [Google Scholar] [CrossRef]
- David, S.; Romano, M.; Kerry, L.E.; Kwong, H.O.; Low, W.E.; Brett, S.J.; Clements, A.; Beis, K.; Frankel, G. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat. Microbiol. 2019, 4, 1919–1929. [Google Scholar] [CrossRef]
- McMurry, L.M.; George, A.M.; Levy, S.B. Active efflux of chloramphenicol in susceptible Escherichia coli strains and in multiple-antibiotic-resistant (Mar) mutants. Antimicrob. Agents Chemother. 1994, 38, 542–546. [Google Scholar] [CrossRef]
- White, D.G.; Goldman, J.D.; Demple, B.; Levy, S.B. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J. Bacteriol. 1997, 179, 6122–6126. [Google Scholar] [CrossRef] [PubMed]
- Aono, R.; Tsukagoshi, N.; Yamamoto, M. Involvement of outer membrane protein TolC, a possible member of the mar-sox regulon, in maintenance and improvement of organic solvent tolerance of Escherichia coli K-12. J. Bacteriol. 1998, 180, 938–944. [Google Scholar] [CrossRef]
- Chetri, S.; Choudhury, A.; Zhang, S.; Garst, A.D.; Song, X.; Liu, X.; Chen, T.; Gill, R.T.; Wang, Z. Transcriptional response of mar, sox and rob regulon against concentration gradient carbapenem stress within Escherichia coli isolated from hospital acquired infection. BMC Res. 2020, 13, 168. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.H.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2020, 76, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Arredondo-Alonso, S.; Rogers, M.R.; Braat, J.C.; Verschuuren, T.D.; Top, J.; Corander, J.; Willems, R.J.L.; Schürch, A.C. Mlplasmids: A user-friendly tool to predict plasmid-and chromosome-derived sequences for single species. Microb. Genom. 2018, 4, e000224. [Google Scholar] [CrossRef]

| Gender | Total Number (%) | Nationality | |
|---|---|---|---|
| Qatari | Non-Qatari | ||
| Male | 8 (27.6%) | 0 (0%) | 8 (27.6%) |
| Female | 21 (72.4%) | 4 (13.8%) | 17 (58.6%) |
| Total | 29 (100%) | 4 (13.8%) | 25 (86.2%) |
| Age | |||
| <2 | 5 (17.2%) | 0 (0%) | 5 (17.2%) |
| 2–5 | 13 (44.8%) | 1 (3.4%) | 12 (41.4%) |
| 6–15 | 11 (37.9%) | 3 (10.3%) | 8 (27.6%) |
| Total | 29 (100%) | 4 (13.8%) | 25 (86.2%) |
| Isolate ID | Collection Date (Month-Year) | Sequence Type (ST) | Carbapenem Resistance Genes (Mlplasmid Posterior Probability for Carbapenemases) |
|---|---|---|---|
| Escherechia coli | |||
| EC-QU-7 | June-2015 | 162 | blaNDM-1 (0.126) |
| EC-QU-5 | November-2015 | 38 | blaOXA-48 (0.971) |
| EC-QU-14 | November-2015 | 448 | blaNDM-5 (0.63) |
| EC-QU-1 | April-2016 | 11,021 | blaNDM-4 (0.937) |
| EC-QU-10 | December-2016 | 11,021 | marA mutation |
| EC-QU-16 | January-2017 | 131 | blaIMP-26 (0.126) |
| EC-QU-6 | March-2017 | 11,021 | blaNDM-4 (0.764) |
| EC-QU-19 | September-2018 | 2083 | blaNDM-5 (0.65) |
| EC-QU-21 | October-2018 | 162 | blaNDM-5 (0.79) |
| EC-QU-23 | January-2019 | 38 | blaOXA-244 (0.852) |
| EC-QU-25 | January-2019 | 38 | blaOXA-244 (0.972) |
| EC-QU-26 | May-2019 | 38 | blaOXA-244 (0.80) |
| EC-QU-27 | July-2019 | 95 | blaOXA-181 (0.84) |
| EC-QU-31 | August-2019 | 410 | blaOXA-484 (0.78) |
| EC-QU-30 | September-2019 | 227 | blaOXA-48 (0.94) |
| EC-QU-29 | September-2019 | 38 | blaOXA-244 (0.991) |
| EC-QU-28 | September-2018 | 131 | blaOXA-244 (0.94) |
| EC-QU-33 | September-2019 | 2346 | blaOXA-244 (0.983) |
| EC-QU-35 | October-2019 | 10 | blaOXA-244 (0.731) |
| Klebsiella pneumonaie | |||
| KPN-QU-9 | October-2015 | 45 | ompK37 mutation |
| KPN-QU-15 | January-2017 | 218 | ompK37 and ompK36 mutations |
| KPN-QU-17 | March-2017 | 101 | blaNDM-1 (0.884) |
| KPN-QU-3 | March-2017 | 196 | blaNDM-1 (0.992) |
| KPN-QU-11 | April-2017 | 987 | ompK37 mutation blaOXA-48 (0.947) |
| KPN-QU-20 | April-2018 | 147 | blaNDM-5 (0.942) |
| KPN-QU-22 | January-2019 | 35 | ompK37, ompK36 mutations blaOXA-181 (0.913) |
| KPN-QU-37 | August-2019 | 3712 | ompK37 mutation bla OXA-181 (0.97) |
| KPN-QU-36 | October-2019 | 870 | ompK37 and ompK36 mutations blaNDM-1 (0.982) |
| Enterobacter hormaechi | |||
| EBH-QU-2 | October-2016 | 269 | blaNDM-7 a |
| Enterobacter cloacae | |||
| EBH-QU-4 | December-2016 | 171 | blaNDM-1 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al Mana, H.; Sundararaju, S.; Tsui, C.K.M.; Perez-Lopez, A.; Yassine, H.; Al Thani, A.; Al-Ansari, K.; Eltai, N.O. Whole-Genome Sequencing for Molecular Characterization of Carbapenem-Resistant Enterobacteriaceae Causing Lower Urinary Tract Infection among Pediatric Patients. Antibiotics 2021, 10, 972. https://doi.org/10.3390/antibiotics10080972
Al Mana H, Sundararaju S, Tsui CKM, Perez-Lopez A, Yassine H, Al Thani A, Al-Ansari K, Eltai NO. Whole-Genome Sequencing for Molecular Characterization of Carbapenem-Resistant Enterobacteriaceae Causing Lower Urinary Tract Infection among Pediatric Patients. Antibiotics. 2021; 10(8):972. https://doi.org/10.3390/antibiotics10080972
Chicago/Turabian StyleAl Mana, Hassan, Sathyavathi Sundararaju, Clement K. M. Tsui, Andres Perez-Lopez, Hadi Yassine, Asmaa Al Thani, Khalid Al-Ansari, and Nahla O. Eltai. 2021. "Whole-Genome Sequencing for Molecular Characterization of Carbapenem-Resistant Enterobacteriaceae Causing Lower Urinary Tract Infection among Pediatric Patients" Antibiotics 10, no. 8: 972. https://doi.org/10.3390/antibiotics10080972
APA StyleAl Mana, H., Sundararaju, S., Tsui, C. K. M., Perez-Lopez, A., Yassine, H., Al Thani, A., Al-Ansari, K., & Eltai, N. O. (2021). Whole-Genome Sequencing for Molecular Characterization of Carbapenem-Resistant Enterobacteriaceae Causing Lower Urinary Tract Infection among Pediatric Patients. Antibiotics, 10(8), 972. https://doi.org/10.3390/antibiotics10080972







