Antibiotic Biosynthesis Pathways from Endophytic Streptomyces SUK 48 through Metabolomics and Genomics Approaches
Abstract
:1. Introduction
2. Materials and Methods
2.1. Brasilia sp. Collection
2.2. Chemicals
2.3. Genomic DNA Extraction and Whole Genome Sequencing
2.4. Bioinformatic Analysis
2.5. Liquid Chromatography-Mass Spectrometry (LC-MS) Analysis
2.6. Mass Spectrometry (MS) Data Handling
2.7. Metabolite Identification
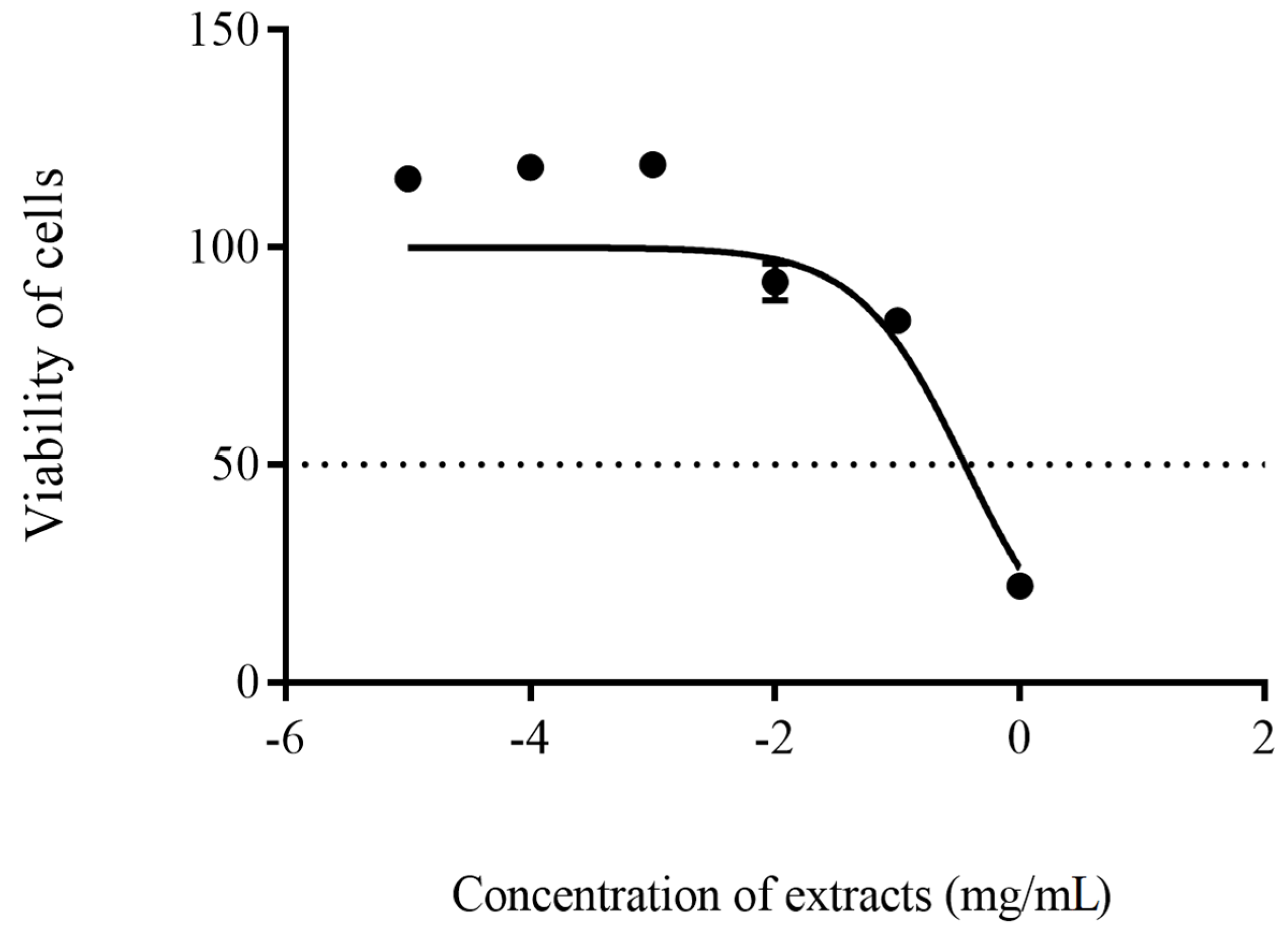
2.8. In-Vitro Cytotoxicity Test
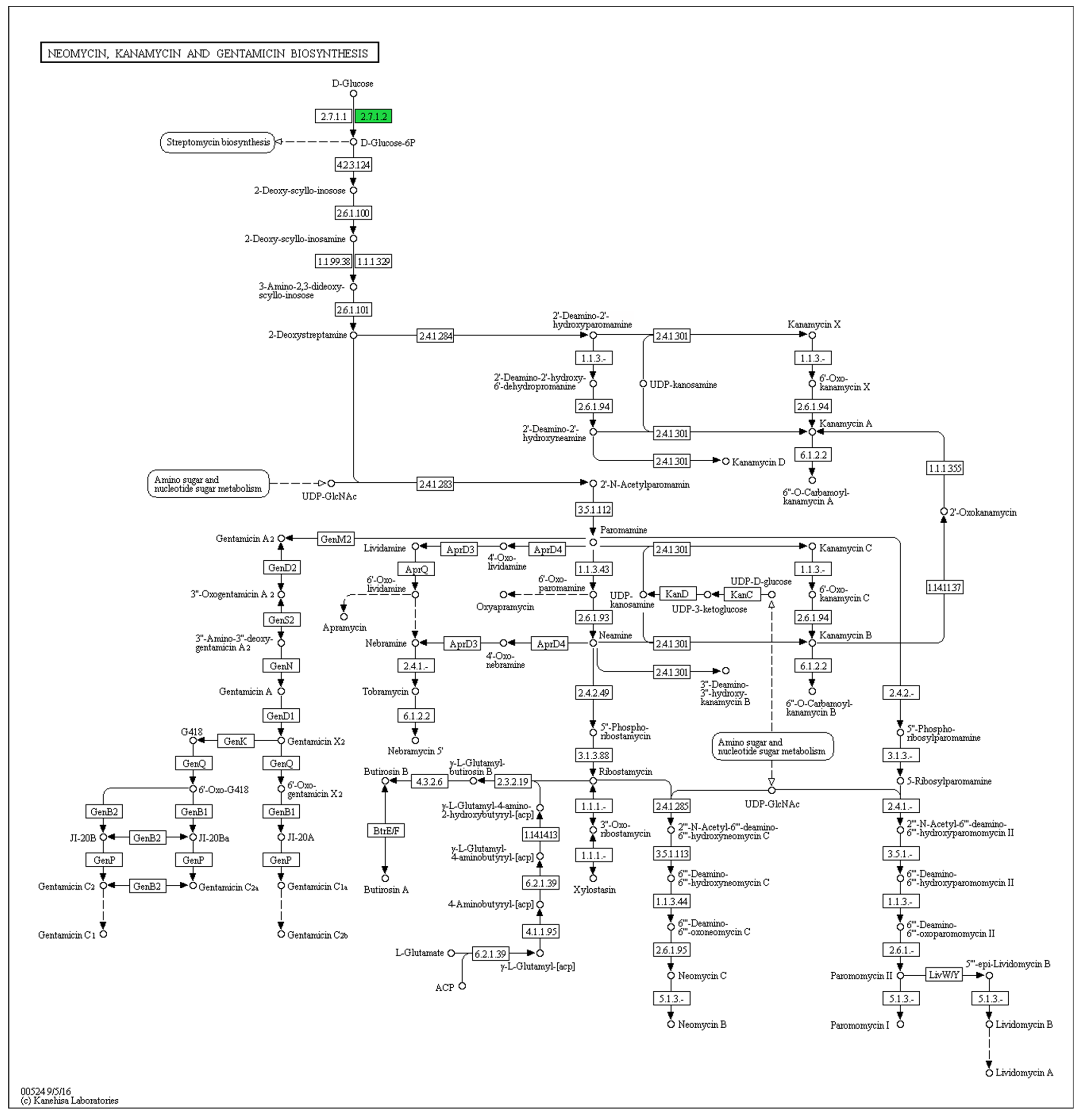
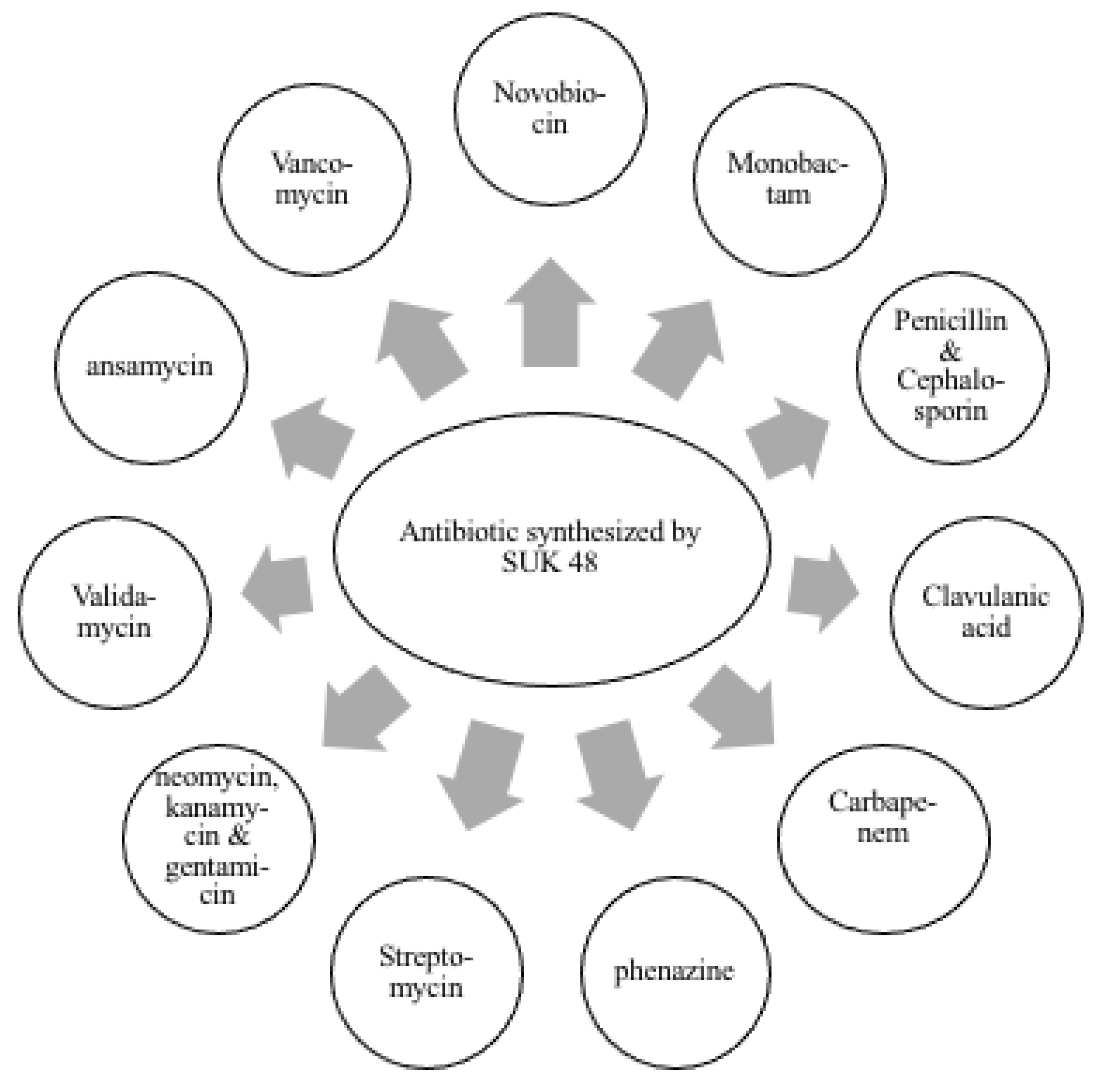
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bérdy, J. Bioactive Microbial Metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirling, E.B.; Gottlieb, D. Cooperative Description of Type Strains of Streptomyces. Int. J. Syst. Bacteriol. 1972, 22, 265–394. [Google Scholar] [CrossRef] [Green Version]
- Shirling, E.B.; Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 1966, 16, 313–340. [Google Scholar] [CrossRef] [Green Version]
- Ezra, D.; Castillo, U.F.; Strobel, G.A.; Hess, W.M.; Porter, H.; Jensen, J.B.; Condron, M.A.M.; Teplow, D.B.; Sears, J.; Maranta, M.; et al. Coronamycins, peptide antibiotics produced by a verticillate Streptomyces sp. (MSU-2110) endophytic on Monstera sp. Microbiology 2004, 150, 785–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castillo, U.F.; Strobel, G.A.; Ford, E.J.; Hess, W.M.; Porter, H.; Jensen, J.B.; Albert, H.; Robison, R.; Condron, M.A.M.; Teplow, D.B.; et al. Munumbicins, wide-spectrum antibiotics produced by Streptomyces NRRL 30562, endophytic on Kennedia nigriscans. Microbiology 2002, 148, 2675–2685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doroghazi, J.R.; Metcalf, W.W. Comparative genomics of Actinomycetes with a focus on natural product biosynthetic genes. BMC Genom. 2013, 14, 611. [Google Scholar] [CrossRef] [Green Version]
- Remali, J.; Zin, N.M.; Ng, C.L.; Aizat, W.M.; Tiong, J.J.L. Fenazin sebagai Potensi Antibiotik Baru daripada Streptomyces kebangsaanensis. Sains Malays. 2019, 48, 543–553. [Google Scholar] [CrossRef]
- Sarmin, N.I.M.; Tan, G.Y.A.; Franco, C.M.M.; Edrada-Ebel, R.; Latip, J.; Zin, N.M. Streptomyces kebangsaanensis sp. nov., an endophytic actinomycete isolated from an ethnomedicinal plant, which produces phenazine-1-carboxylic acid. Int. J. Syst. Bacteriol. 2013, 63, 3733–3738. [Google Scholar] [CrossRef] [Green Version]
- Abdi, S.N.; Ghotaslou, R.; Ganbarov, K.; Mobed, A.; Tanomand, A.; Yousefi, M.; Asgharzadeh, M.; Kafil, H.S. Acinetobacter baumannii efflux pumps and antibiotic resistance. Infect. Drug Resist. 2020, 13, 423–434. [Google Scholar] [CrossRef] [Green Version]
- Coyne, S.; Courvalin, P.; Périchon, B. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob. Agents Chemother. 2011, 55, 947–953. [Google Scholar] [CrossRef] [Green Version]
- Antunes, L.C.S.; Visca, P.; Towner, K.J. Acinetobacter baumannii: Evolution of a global pathogen. Pathog. Dis. 2014, 71, 292–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeom, J.; Shin, J.H.; Yang, J.Y.; Kim, J.; Hwang, G.S. 1H NMR-Based Metabolite Profiling of Planktonic and Biofilm Cells in Acinetobacter baumannii 1656-2. PLoS ONE 2013, 8, 57730. [Google Scholar] [CrossRef] [Green Version]
- Rice, L.B. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- Kyriakidis, I.; Vasileiou, E.; Pana, Z.D.; Tragiannidis, A. Acinetobacter baumannii antibiotic resistance mechanisms. Pathogens 2021, 10, 373. [Google Scholar] [CrossRef]
- Arias, C.A.; Murray, B.E. A New Antibiotic and the Evolution of Resistance. N. Engl. J. Med. 2015, 372, 1168–1170. [Google Scholar] [CrossRef]
- Kon, K.V.; Rai, M.K. Plant essential oils and their constituents in coping with multidrug-resistant bacteria. Expert Rev. Anti-Infect. 2012, 10, 775–790. [Google Scholar] [CrossRef]
- Roemer, T.; Boone, C. Systems-level antimicrobial drug and drug synergy discovery. Nat. Chem. Biol. 2013, 9, 222–231. [Google Scholar] [CrossRef]
- Tang, Y.W. Non-genomic Omic Techniques. In Molecular Medical Microbiology, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2014; Volume 1–3, pp. 399–406. [Google Scholar]
- Nambiar, P.R.; Gupta, R.R.; Misra, V. An “Omics” based survey of human colon cancer. Mutat. Res. Fundam. Mol. Mech. Mutagenesis 2010, 693, 3–18. [Google Scholar] [CrossRef]
- dos Santos, B.S.; da Silva, L.C.N.; da Silva, T.D.; Rodrigues, J.F.S.; Grisotto, M.A.G.; Correia, M.T.d.S.; Napoleão, T.H.; da Silva, M.V.; Paiva, P.M.G. Application of omics technologies for evaluation of antibacterial mechanisms of action of plant-derived products. Front. Microbiol. 2016, 7, 1466. [Google Scholar] [CrossRef] [PubMed]
- Lankadurai, B.P.; Nagato, E.G.; Simpson, M.J. Environmental metabolomics: An emerging approach to study organism responses to environmental stressors. Environ. Rev. 2013, 21, 180–205. [Google Scholar] [CrossRef]
- Aliferis, K.A.; Jabaji, S. Metabolomics—A robust bioanalytical approach for the discovery of the modes-of-action of pesticides: A review. Pestic. Biochem. Phys. 2011, 100, 105–117. [Google Scholar] [CrossRef]
- Yu, Y.; Yi, Z.-B.; Liang, Y.-Z. Main antimicrobial components of Tinospora capillipes, and their mode of action against Staphylococcus aureus. FEBS Lett. 2007, 581, 4179–4183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birkenstock, T.; Liebeke, M.; Winstel, V.; Krismer, B.; Gekeler, C.; Niemiec, M.J.; Bisswanger, H.; Lalk, M.; Peschel, A. Exometabolome analysis identifies pyruvate dehydrogenase as a target for the antibiotic triphenylbismuthdichloride in multiresistant bacterial pathogens. J. Biol. Chem. 2012, 287, 2887–2895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zin, N.M.; Nur Faizah, A.B.; Aishah, I.; Baba, M.S.; Dan Sidik, N.M. Pencirian dan aktiviti antibakteria endofit streptomyces sp. dari hutan simpan penyelidikan UKM Bangi. Malays. Appl. Biol. 2015, 44, 107–117. [Google Scholar]
- Eid, J.; Fehr, A.; Gray, J.; Luong, K.; Lyle, J.; Otto, G.; Peluso, P.; Rank, D.; Baybayan, P.; Bettman, B.; et al. Real-time DNA sequencing from single polymerase molecules. Science 2009, 323, 133–138. [Google Scholar] [CrossRef]
- Chin, C.-S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E.; et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [Green Version]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Medema, M.H.; Blin, K.; Cimermancic, P.; De Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. AntiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef]
- Blin, K.; Pascal Andreu, V.; De Los Santos, E.L.C.; Del Carratore, F.; Lee, S.Y.; Medema, M.H.; Weber, T. The antiSMASH database version 2: A comprehensive resource on secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2019, 47, D625–D630. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Rosli, M.A.F.; Azizan, K.A.; Baharum, S.N.; Goh, H.H. Mass spectrometry data of metabolomics analysis of Nepenthes pitchers. Data Brief 2017, 14, 295–297. [Google Scholar] [CrossRef] [PubMed]
- Mamat, S.F.; Azizan, K.A.; Baharum, S.N.; Noor, N.M.; Aizat, W.M. Metabolomics analysis of mangosteen (Garcinia mangostana Linn.) fruit pericarp using different extraction methods and GC-MS. Plant. OMICS 2018, 11, 89–97. [Google Scholar] [CrossRef]
- Mamat, S.F.; Azizan, K.A.; Baharum, S.N.; Noor, N.M.; Aizat, W.M. ESI-LC-MS based-metabolomics data of mangosteen (Garcinia mangostana Linn.) fruit pericarp, aril and seed at different ripening stages. Data Brief 2018, 17, 1074–1077. [Google Scholar] [CrossRef] [PubMed]
- Veeramohan, R.; Azizan, K.A.; Aizat, W.M.; Goh, H.H.; Mansor, S.M.; Yusof, N.S.M.; Baharum, S.N.; Ng, C.L. Metabolomics data of Mitragyna speciosa leaf using LC-ESI-TOF-MS. Data Brief 2018, 18, 1212–1216. [Google Scholar] [CrossRef]
- Smith, C.A.; O’Maille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. METLIN: A metabolite mass spectral database. Ther. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Macintyre, L.; Zhang, T.; Viegelmann, C.; Martinez, I.J.; Cheng, C.; Dowdells, C.; Abdelmohsen, U.R.; Gernert, C.; Hentschel, U.; Edrada-Ebel, R.A. Metabolomic tools for secondary metabolite discovery from marine microbial symbionts. Mar. Drugs 2014, 12, 3416–3448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azmi, F.; Elliott, A.G.; Marasini, N.; Ramu, S.; Ziora, Z.; Kavanagh, A.M.; Blaskovich, M.A.T.; Cooper, M.A.; Skwarczynski, M.; Toth, I. Short cationic lipopeptides as effective antibacterial agents: Design, physicochemical properties and biological evaluation. Bioorgan. Med. Chem. 2016, 24, 2235–2241. [Google Scholar] [CrossRef]
- Ahmad, S.J.; Zin, N.M. Data on DNA-seq analysis of Endophytic Streptomyces sp. SUK 48. Data Brief 2021, 35, 106768. [Google Scholar] [CrossRef]
- Ahmad, S.J.; Zin, N.M.; Mazlan, N.W.; Baharum, S.N.; Baba, M.S.; Lau, Y.L. Metabolite Profiling of Endophytic Streptomyces spp. and its Antiplasmodial Potential. PeerJ. 2021, 9, e10816. [Google Scholar] [CrossRef]
- Chopra, I.; Roberts, M. Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghannoum, M.A.; Rice, L.B. Antifungal agents: Mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin. Microbiol. Rev. 1999, 12, 501–517. [Google Scholar] [CrossRef] [Green Version]
- Risdian, C.; Mozef, T.; Wink, J. Biosynthesis of polyketides in Streptomyces. Microorganisms 2019, 7, 124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, Y.J.; Kim, E.S.; Hwang, Y.S.; Choi, C.Y. Avermectin: Biochemical and molecular basis of its biosynthesis and regulation. Appl. Microbiol. 2004, 63, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, S.; Taguchi, T.; Ochi, K.; Ichinose, K. Biosynthesis of Actinorhodin and Related Antibiotics: Discovery of Alternative Routes for Quinone Formation Encoded in the act Gene Cluster. Chem. Biol. 2009, 16, 226–236. [Google Scholar] [CrossRef] [Green Version]
- Umezawa, H.; Ueda, M.; Maeda, K.; Yagishita, K.; Kondo, S.; Okami, Y.; Utahara, R.; Osato, Y.; Nitta, K.; Takeuchi, T. Production and isolation of a new antibiotic: Kanamycin. Antibiotics 1957, 10, 181–188. [Google Scholar] [CrossRef]
- Wolf, H.; Zähner, H. Metabolic products of microorganisms—99. Kirromycin. Arch. Microbiol. 1972, 83, 147–154. [Google Scholar] [CrossRef]
- Lin, H.C.; Chiou, G.; Chooi, Y.H.; McMahon, T.C.; Xu, W.; Garg, N.K.; Tang, Y. Elucidation of the concise biosynthetic pathway of the communesin indole alkaloids. Angew. Chem. Int. Ed. 2015, 54, 3004–3007. [Google Scholar] [CrossRef]
- Chen, K.L.; Lai, C.Y.; Pham, M.T.; Chein, R.J.; Tang, Y.; Lin, H.C. Enzyme-Catalyzed Azepinoindole Formation in Clavine Alkaloid Biosynthesis. Org. Lett. 2020, 22, 3302–3306. [Google Scholar] [CrossRef] [PubMed]
- Qader, S.W.; Abdulla, M.A.; Chua, L.S.; Najim, N.; Zain, M.M.; Hamdan, S. Antioxidant, total phenolic content and cytotoxicity evaluation of selected Malaysian plants. Molecules 2011, 16, 3433–3443. [Google Scholar] [CrossRef] [PubMed]
- López-García, J.; Lehocký, M.; Humpolíček, P.; Sáha, P. HaCaT Keratinocytes Response on Antimicrobial Atelocollagen Substrates: Extent of Cytotoxicity, Cell Viability and Proliferation. J. Funct. Biomater. 2014, 5, 43–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baba, M.S.; Mohamad Zin, N.; Ahmad, S.J.; Mazlan, N.W.; Baharum, S.N.; Ahmad, N.; Azmi, F. Antibiotic Biosynthesis Pathways from Endophytic Streptomyces SUK 48 through Metabolomics and Genomics Approaches. Antibiotics 2021, 10, 969. https://doi.org/10.3390/antibiotics10080969
Baba MS, Mohamad Zin N, Ahmad SJ, Mazlan NW, Baharum SN, Ahmad N, Azmi F. Antibiotic Biosynthesis Pathways from Endophytic Streptomyces SUK 48 through Metabolomics and Genomics Approaches. Antibiotics. 2021; 10(8):969. https://doi.org/10.3390/antibiotics10080969
Chicago/Turabian StyleBaba, Mohd Shukri, Noraziah Mohamad Zin, Siti Junaidah Ahmad, Noor Wini Mazlan, Syarul Nataqain Baharum, Nuraziemah Ahmad, and Fazren Azmi. 2021. "Antibiotic Biosynthesis Pathways from Endophytic Streptomyces SUK 48 through Metabolomics and Genomics Approaches" Antibiotics 10, no. 8: 969. https://doi.org/10.3390/antibiotics10080969
APA StyleBaba, M. S., Mohamad Zin, N., Ahmad, S. J., Mazlan, N. W., Baharum, S. N., Ahmad, N., & Azmi, F. (2021). Antibiotic Biosynthesis Pathways from Endophytic Streptomyces SUK 48 through Metabolomics and Genomics Approaches. Antibiotics, 10(8), 969. https://doi.org/10.3390/antibiotics10080969





