Cationic Peptidomimetic Amphiphiles Having a N-Aryl- or N-Naphthyl-1,2,3-Triazole Core Structure Targeting Clostridioides (Clostridium) difficile: Synthesis, Antibacterial Evaluation, and an In Vivo C. difficile Infection Model
Abstract
:1. Introduction
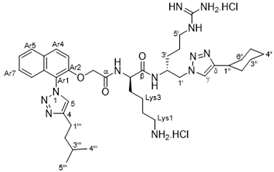
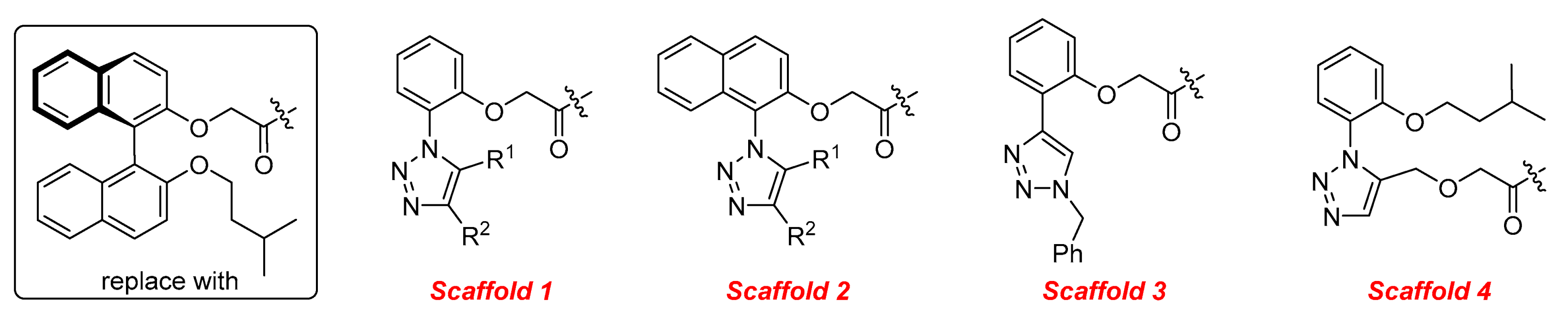
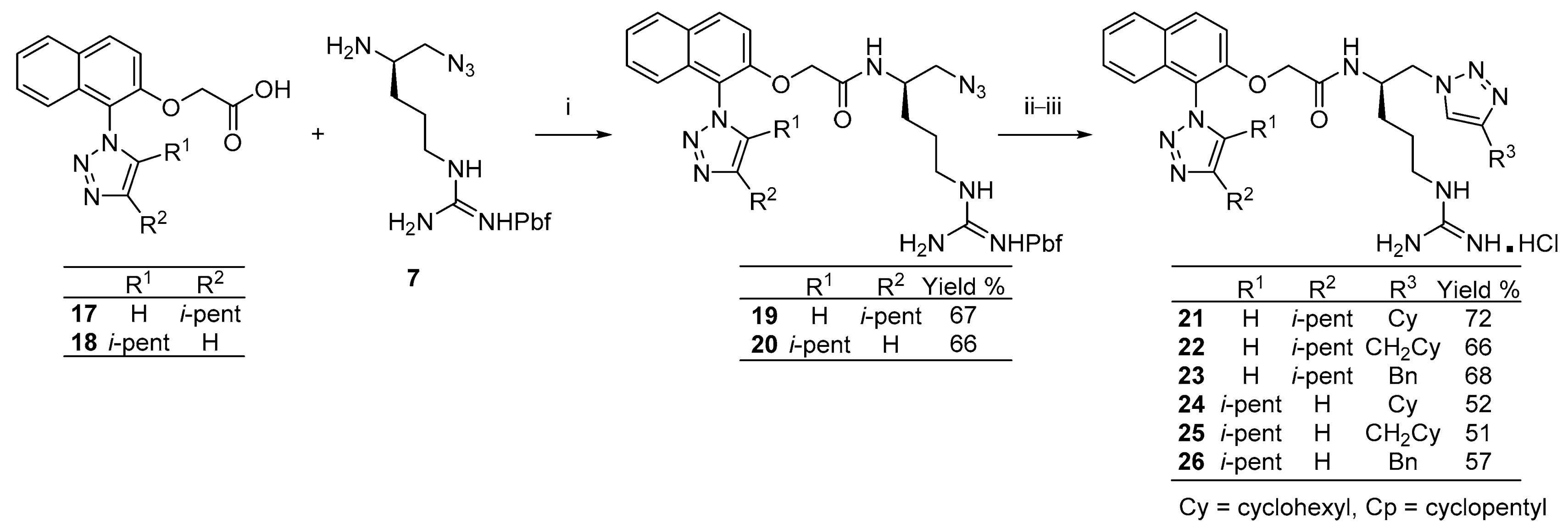
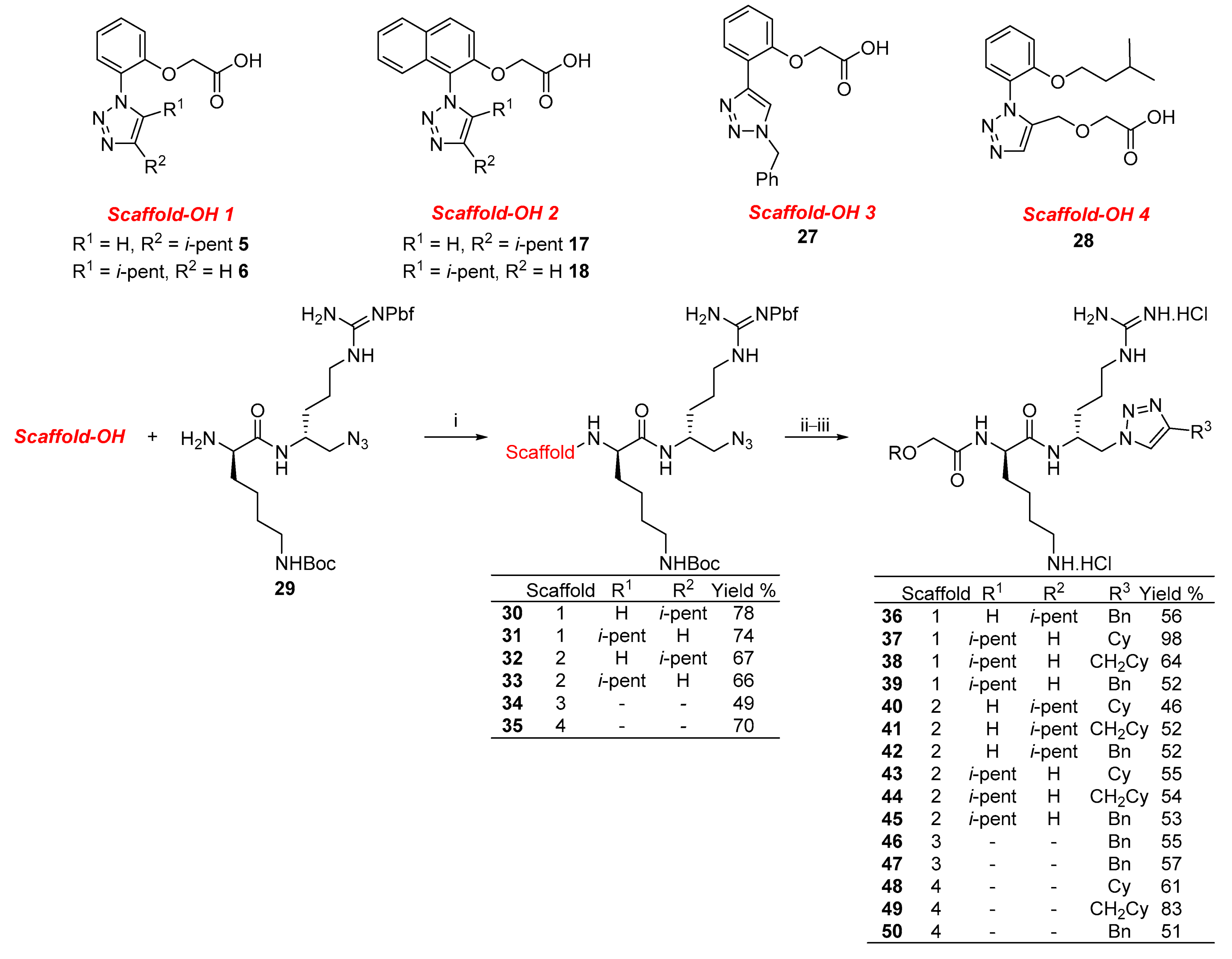
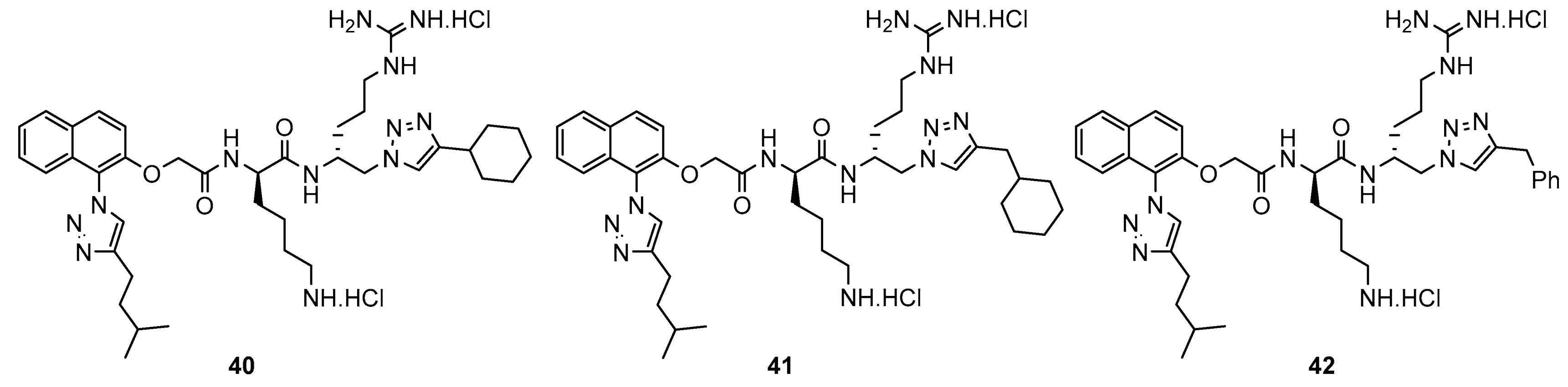
2. Results and Discussion
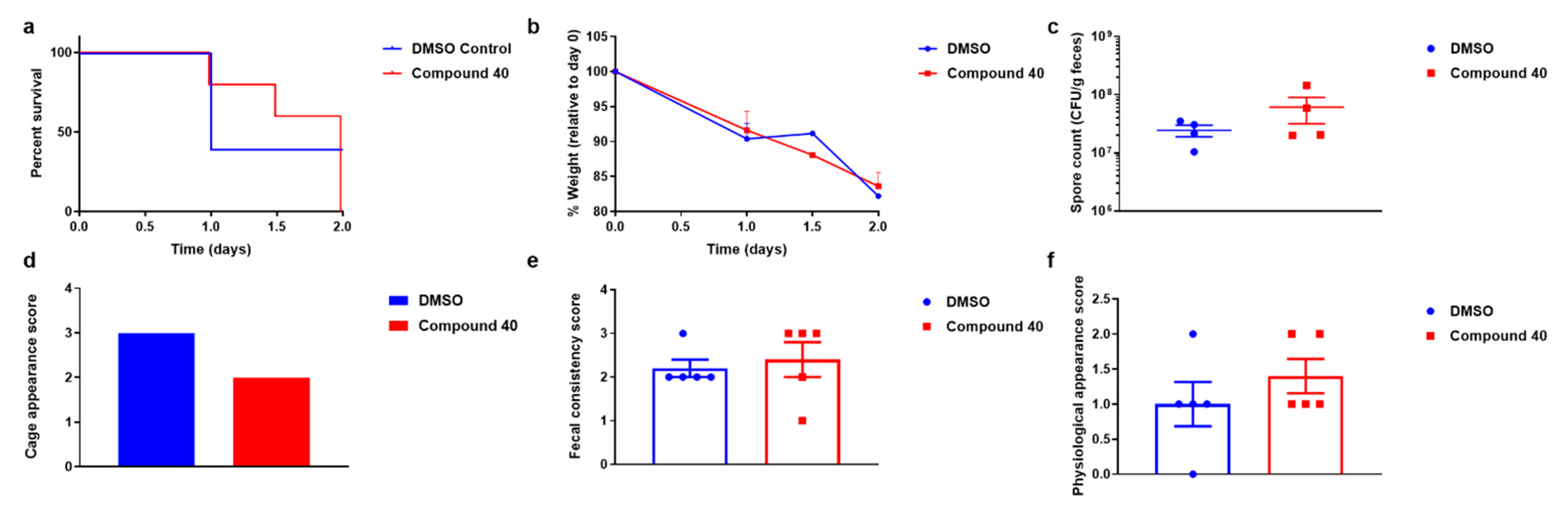
3. In Vivo Assay: Murine Model of CDI
4. Materials and Methods
4.1. General Synthesis Procedures
4.1.1. General Procedure I: Alkylation of Phenols (with Ethyl Bromoacetate)
4.1.2. General Procedure II: Ester Hydrolysis
4.1.3. General Procedure III: Amide Coupling
4.1.4. General Procedure IV: Copper-Catalyzed Azide-Alkyne Cycloaddition
4.1.5. General Procedure VII: Amine Deprotection (N-Boc and/or N-Pbf Removal)
4.2. Representative Synthesis of Compound 40
4.2.1. Ethyl 2-((1-iodonaphthalen-2-yl)oxy)acetate

4.2.2. Ethyl 2-((1-(4-isopentyl-1H-1,2,3-triazol-1-yl)naphthalen-2-yl)oxy)acetate
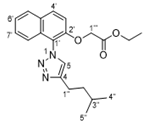
4.2.3. 2-((1-(4-Isopentyl-1H-1,2,3-triazol-1-yl)naphthalen-2-yl)oxy)acetic acid (17)
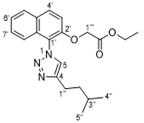
4.2.4. (9H-Fluoren-9-yl)methyl tert-butyl ((R)-6-(((R)-1-azido-5-(2-((2,2-dimethyl-2,3-dihydro benzofuran-5-yl)sulfonyl)guanidino)pentan-2-yl)amino)-6-oxohexane-1,5-diyl) dicarbamate
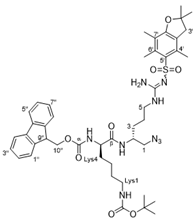
4.2.5. Tert-butyl ((R)-5-amino-6-(((R)-1-azido-5-(2-((2,2-dimethyl-2,3-dihydrobenzofuran-5-yl)sulfonyl)guanidino)pentan-2-yl)amino)-6-oxohexyl)carbamate (31)

4.2.6. Tert-butyl ((R)-6-(((R)-1-azido-5-(2-((2,2,4,6,7-pentamethyl-2,3-dihydrobenzofuran-5-yl)sulfonyl)guanidino)pentan-2-yl)amino)-5-(2-((1-(4-isopentyl-1H-1,2,3-triazol-1-yl)naphthalen-2-yl)oxy)acetamido)-6-oxohexyl)carbamate (32)
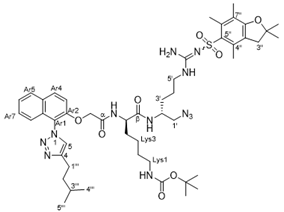
4.2.7. (R)-6-Amino-N-((R)-1-(4-cyclohexyl-1H-1,2,3-triazol-1-yl)-5-guanidinopentan-2-yl)-2-(2-((1-(4-isopentyl-1H-1,2,3-triazol-1-yl)naphthalen-2-yl)oxy)acetamido)hexanamide dihydrochloride (40)
4.3. Microbiological Assays
4.4. In Vivo Murine Model of CDI Treatment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leffler, D.A.; Lamont, J.T. Treatment of Clostridium difficile-Associated Disease. Gastroenterology 2009, 136, 1899–1912. [Google Scholar] [CrossRef]
- Knight, D.R.; Elliott, B.; Chang, B.J.; Perkins, T.T.; Riley, T.V. Diversity and Evolution in the Genome of Clostridium difficile. Clin. Microbiol. Rev. 2015, 28, 721–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eaton, S.R.; Mazuski, J.E. Overview of Severe Clostridium difficile Infection. Crit. Care Clin. 2013, 29, 827–839. [Google Scholar] [CrossRef] [PubMed]
- Di Bella, S.; Ascenzi, P.; Siarakas, S.; Petrosillo, N.; Di Masi, A. Clostridium difficile Toxins A and B: Insights into Pathogenic Properties and Extraintestinal Effects. Toxins 2016, 8, 134. [Google Scholar] [CrossRef] [Green Version]
- Chandrasekaran, R.; Lacy, D.B. The role of toxins in Clostridium difficile infection. FEMS Microbiol. Rev. 2017, 41, 723–750. [Google Scholar] [CrossRef] [Green Version]
- Johnson, A.P. New antibiotics for selective treatment of gastrointestinal infection caused by Clostridium difficile. Expert Opin. Ther. Pat. 2010, 20, 1389–1399. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Clostridium Difficile Update. 2019. Available online: https://www.cdc.gov/drugresistance/pdf/threats-report/CRE-508.pdf (accessed on 26 June 2021).
- Stanley, J.D.; Bartlett, J.G.; Dart, B.W.; Ashcraft, J. Clostridium difficile infection. Curr. Probl. Surg. 2013, 50, 302–337. [Google Scholar] [CrossRef] [PubMed]
- Ritter, A.S.; Petri, W.A. New developments in chemotherapeutic options for Clostridium difficile colitis. Curr. Opin. Infect. Dis. 2013, 26, 461–470. [Google Scholar] [CrossRef]
- Cornely, O.A.; Miller, M.A.; Louie, T.J.; Crook, D.W.; Gorbach, S.L. Treatment of First Recurrence of Clostridium difficile Infection: Fidaxomicin Versus Vancomycin. Clin. Infect. Dis. 2012, 55, S154–S161. [Google Scholar] [CrossRef]
- Hostler, C.J.; Chen, L.F. Fidaxomicin for treatment of Clostridium difficile-associated diarrhea and its potential role for prophylaxis. Expert Opin. Pharmacother. 2013, 14, 1529–1536. [Google Scholar] [CrossRef]
- Cho, J.M.; Pardi, D.S.; Khanna, S. Update on Treatment of Clostridioides difficile Infection. Mayo Clin Proc. 2020, 95, 758–769. [Google Scholar] [CrossRef]
- Tague, A.J.; Putsathit, P.; Hammer, K.A.; Wales, S.M.; Knight, D.R.; Riley, T.V.; Keller, P.A.; Pyne, S.G. Cationic biaryl 1,2,3-triazolyl peptidomimetic amphiphiles targeting Clostridioides (Clostridium) difficile: Synthesis, antibacterial evaluation and an in vivo C. difficile infection model. Eur. J. Med. Chem. 2019, 170, 203–224. [Google Scholar] [CrossRef] [PubMed]
- Wales, S.M.; Hammer, K.A.; King, A.M.; Tague, A.J.; Lyras, D.; Riley, T.V.; Keller, P.A.; Pyne, S.G. Binaphthyl-1,2,3-triazole peptidomimetics with activity against Clostridium difficile and other pathogenic bacteria. Org. Biomol. Chem. 2015, 13, 5743–5756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tague, A.J.; Putsathit, P.; Riley, T.V.; Keller, P.A.; Pyne, S.G. Positional Isomers of Biphenyl Antimicrobial Peptidomimetic Amphiphiles. ACS Med. Chem. Lett. 2021, 12, 413–419. [Google Scholar] [CrossRef]
- Zhang, S.J.; Yang, Q.; Xu, L.; Chang, J.; Sun, X. Synthesis and antibacterial activity against Clostridium difficile of novel demethylvancomycin derivatives. Bioorg. Med. Chem. Lett. 2012, 22, 4942–4945. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.M.; Williams, J.D.; Peet, N.P.; Moir, D.T.; Panchal, R.G.; Bavari, S.; Shinabarger, D.L.; Bowlin, T.L. Comparative In Vitro Activity Profiles of Novel Bis-Indole Antibacterials against Gram-Positive and Gram-Negative Clinical Isolates. Antimicrob. Agents Chemother. 2010, 54, 3974–3977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dvoskin, S.; Xu, W.-C.; Brown, N.C.; Yanachkov, I.B.; Yanachkova, M.; Wright, G.E. A Novel Agent Effective against Clostridium difficile Infection. Antimicrob. Agents Chemother. 2012, 56, 1624–1626. [Google Scholar] [CrossRef] [Green Version]
- Ueda, C.; Tateda, K.; Horikawa, M.; Kimura, S.; Ishii, Y.; Nomura, K.; Yamada, K.; Suematsu, T.; Inoue, Y.; Ishiguro, M.; et al. Anti-Clostridium difficile Potential of Tetramic Acid Derivatives from Pseudomonas aeruginosa Quorum-Sensing Autoinducers. Antimicrob. Agents Chemother. 2010, 54, 683–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ballard, T.E.; Wang, X.; Olekhnovich, I.; Koerner, T.; Seymour, C.; Hoffman, P.S.; Macdonald, T.L. Biological Activity of Modified and Exchanged 2-Amino-5-Nitrothiazole Amide Analogues of Nitazoxanide. Bioorg. Med. Chem. Lett. 2010, 20, 3537–3539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirst, H.A.; Toth, J.E.; Debono, M.; Willard, K.E.; Truedell, B.A.; Ott, J.L.; Counter, F.T.; Felty-Duckworth, A.M.; Pekarek, R.S. Synthesis and evaluation of tylosin-related macrolides modified at the aldehyde function: A new series of orally effective antibiotics. J. Med. Chem. 1988, 31, 1631–1641. [Google Scholar] [CrossRef]
- Liu, R.; Suárez, J.M.; Weisblum, B.; Gellman, S.H.; McBride, S.M. Synthetic Polymers Active against Clostridium difficile Vegetative Cell Growth and Spore Outgrowth. J. Am. Chem. Soc. 2014, 136, 14498–14504. [Google Scholar] [CrossRef] [Green Version]
- Jarrad, A.M.; Karoli, T.; Blaskovich, M.A.T.; Lyras, D.; Cooper, M.A. Clostridium difficile Drug Pipeline: Challenges in Discovery and Development of New Agents. J. Med. Chem. 2015, 58, 5164–5185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowes, R. FDA Approves Zinplava for Preventing Return of C. difficile. Available online: https://www.medscape.com/viewarticle/870887 (accessed on 26 June 2021).
- Bremner, J.B.; Keller, P.A.; Pyne, S.G.; Boyle, T.P.; Brkic, Z.; David, D.M.; Garas, A.; Morgan, J.; Robertson, M.; Somphol, K.; et al. Binaphthyl-Based Dicationic Peptoids with Therapeutic Potential. Angew. Chem. Int. Ed. 2010, 49, 537–540. [Google Scholar] [CrossRef] [PubMed]
- Bremner, J.B.; Keller, P.A.; Pyne, S.G.; Boyle, T.P.; Brkic, Z.; David, D.M.; Robertson, M.; Somphol, K.; Baylis, D.; Coates, J.A.; et al. Synthesis and antibacterial studies of binaphthyl-based tripeptoids. Part 1. Bioorg. Med. Chem. 2010, 18, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Bremner, J.B.; Keller, P.A.; Pyne, S.G.; Boyle, T.P.; Brkic, Z.; Morgan, J.; Somphol, K.; Coates, J.A.; Deadman, J.; Rhodes, D.I. Synthesis and antibacterial studies of binaphthyl-based tripeptoids. Part 2. Bioorg. Med. Chem. 2010, 18, 4793–4800. [Google Scholar] [CrossRef]
- Mahadari, M.K.; Tague, A.J.; Keller, P.A.; Pyne, S.G. Synthesis of sterically congested 1,5-disubstituted-1,2,3-Triazoles using chloromagnesium acetylides and hindered 1-naphthyl azides. Tetrahedron 2021, 81, 131916. [Google Scholar] [CrossRef]
- Blaskovich, M.A.T.; Zuegg, J.; Elliott, A.G.; Cooper, M.A. Helping chemists discover new antibiotics. ACS Infect. Dis. 2015, 1, 285–287. [Google Scholar] [CrossRef]
- Wales, S.M.; Hammer, K.A.; Somphol, K.; Kemker, I.; Schröder, D.C.; Tague, A.J.; Brkic, Z.; King, A.M.; Lyras, D.; Riley, T.V.; et al. Synthesis and antimicrobial activity of binaphthylbased, functionalized oxazole and thiazole peptidomimetics. Org. Biomol. Chem. 2019, 13, 10813–10824. [Google Scholar] [CrossRef]
- Tague, A.J.; Putsathit, P.; Hammer, K.A.; Wales, S.M.; Knight, D.R.; Riley, T.V.; Keller, P.A.; Pyne, S.G. Cationic biaryl 1,2,3-triazolyl peptidomimetic amphiphiles: Synthesis, antibacterial evaluation and preliminary mechanism of action studies. Eur. J. Med. Chem. 2019, 168, 386–404. [Google Scholar] [CrossRef]
- Zhu, D.; Ma, J.; Luo, K.; Fu, H.; Zhang, L.; Zhu, S. Enantioselective Intramolecular C-H Insertion of Donor and Donor/Donor Carbenes by a Nondiazo Approach. Angew. Chem. Int. Ed. 2016, 55, 8452–8456. [Google Scholar] [CrossRef]
- Maehr, H.; Smallheer, J. Total syntheses of rivularins D1 and D3. J. Am. Chem. Soc. 1985, 107, 2943–2945. [Google Scholar] [CrossRef]
- Gamble, A.B.; Garner, J.; Gordon, C.P.; O’Conner, S.M.J.; Keller, P.A. Aryl Nitro Reduction with Iron Powder or Stannous Chloride under Ultrasonic Irradiation. Synth. Commun. 2007, 37, 2777–2786. [Google Scholar] [CrossRef]
- Zilla, M.K.; Nayak, D.; Vishwakarma, R.A.; Sharma, P.R.; Goswami, A.; Ali, A. A convergent synthesis of alkyne-azide cycloaddition derivatives of 4-α,β-2-propyne podophyllotoxin depicting potent cytotoxic activity. Eur. J. Med. Chem. 2014, 77, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, 9th ed.; CLSI Document M07-A10; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015. [Google Scholar]
- Clinical and Laboratory Standards Institute. Methods for Antimicrobial Susceptibility Testing of Anaerobic Bacteria, 8th ed.; CLSI Document M11-A8; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Clinical Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; 28th Informational Supplement; CLSI Document M100-S28; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Carter, G.P.; Lyras, D.; Allen, D.L.; Mackin, K.E.; Howarth, P.M.; O’Connor, J.R.; Rood, J.I. Binary toxin production in Clostridium difficile is regulated by CdtR, a LytTR family response regulator. J. Bacteriol. 2007, 189, 7290–7301. [Google Scholar] [CrossRef] [Green Version]
- Hutton, M.L.; Cunningham, B.A.; Mackin, K.E.; Lyon, S.A.; James, M.L.; Rood, J.I.; Lyras, D. Bovine antibodies targeting primary and recurrent Clostridium difficile disease are a potent antibiotic alternative. Sci. Rep. 2017, 7, 3665. [Google Scholar] [CrossRef]
- Lyon, S.A.; Hutton, M.L.; Rood, J.I.; Cheung, J.K.; Lyras, D. CdtR regulates TcdA and TcdB production in Clostridium difficile. PLoS Pathog. 2016, 12, e1005758. [Google Scholar] [CrossRef] [Green Version]
- Awad, M.M.; Hutton, M.L.; Quek, A.J.; Klare, W.P.; Mileto, S.J.; Mackin, K.; Ly, D.; Oorschot, V.; Bosnjak, M.; Jenkin, G.; et al. Human Plasminogen Exacerbates Clostridioides difficile Enteric Disease and Alters the Spore Surface. Gastroenterology 2020, 159, 1431–1443. [Google Scholar] [CrossRef]
- Carter, G.P.; Chakravorty, A.; Pham Nguyen, T.A.; Mileto, S.; Schreiber, F.; Li, L.; Howarth, P.; Clare, S.; Cunningham, B.; Sambol, S.P.; et al. Defining the roles of TcdA and TcdB in localized gastrointestinal disease, systemic organ damage, and the host response during Clostridium difficile infections. mBio 2015, 6, e00551. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Compound | C. difficile ATCC 700057 | C. difficile 132 b (RT027) | S. aureus ATCC 29213 | S. aureus NCTC 10442 c | E. faecalis ATCC 29212 | S. pneumoniae ATCC 49619 | E. coli ATCC 25922 | CC50 d | HC50 e | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 10 | 32 | 64 | 32 | 32 | 32 | 16 | 128 | >32 | >32 |
| 2 | 11 | 32 | 64 | 32 | 32 | 32 | 16 | 128 | >32 | >32 |
| 3 | 12 | 32 | 64 | 32 | 32 | 32 | 16 | 128 | >32 | >32 |
| 4 | 13 | 32 | 64 | 32 | 32 | 32 | 16 | 128 | >32 | >32 |
| 5 | 14 | 128 | 128 | 32 | 32 | 32 | 32 | >128 | >32 | >32 |
| 6 | 15 | 64 | 64 | 16 | 16 | 16 | 16 | 128 | >32 | >32 |
| 7 | 16 | 32 | 32 | 32 | 32 | 32 | 32 | >128 | >32 | >32 |
| 8 | 21 | 128 | >128 | 8 | 8 | 16 | 16 | 128 | >32 | >32 |
| 8 | 22 | 32 | 32 | 4 | 4 | 8 | 8 | 32 | >32 | >32 |
| 9 | 23 | 32 | 32 | 4 | 4 | 4 | 4 | 64 | >32 | >32 |
| 10 | 24 | 32 | 32 | 8 | 8 | 8 | 8 | 32 | 21.9 | >32 |
| 11 | 25 | 32 | 64 | 4 | 4 | 8 | 8 | 16 | >32 | 10.6 |
| 12 | 26 | 64 | >128 | 8 | 8 | 16 | 4 | 64 | 23.5 | >32 |
| 13 | 36 | 128 | 128 | 32 | 32 | 64 | 16 | 128 | >32 | >32 |
| 14 | 37 | 64 | 64 | 16 | 32 | 16 | 8 | 64 | >32 | >32 |
| 15 | 38 | 128 | >128 | 128 | 128 | >128 | 128 | >128 | 32 | 32 |
| 16 | 39 | 64 | 32 | 32 | 64 | 32 | 16 | 128 | >32 | >32 |
| 17 | 40 | 8 | 8 | 16 | 16 | 32 | 16 | 64 | 32 | 32 |
| 18 | 41 | 16 | 16 | 8 | 8 | 8 | 16 | 128 | 16 | 32 |
| 19 | 42 | 8 | 8 | 8 | 8 | 8 | 8 | 32 | 32 | 32 |
| 20 | 43 | 128 | 128 | 16 | 16 | 64 | 4 | 64 | >32 | >32 |
| 21 | 44 | 32 | 32 | 8 | 4 | 16 | 4 | 64 | >32 | >32 |
| 22 | 45 | 128 | 128 | 16 | 16 | 64 | 4 | 128 | >32 | >32 |
| 23 | 46 | 128 | 128 | 32 | 32 | 64 | 16 | 128 | >32 | >32 |
| 24 | 47 | 128 | 128 | 32 | 32 | 64 | 16 | 128 | >32 | >32 |
| 25 | 48 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | 24.5 |
| 26 | 49 | >32 | >32 | 32 | >32 | >32 | >32 | >32 | >32 | 17.1 |
| 27 | 50 | >32 | >32 | 32 | >32 | >32 | >32 | >32 | >32 | 19.8 |
| vanc | 0.5 | 0.5 | 1 | 1 | 4 | 1 | >16 | - | - | |
| colistin | 0.25 | 0.25 | 0.25 | 0.125 | ||||||
| tamoxifen | 13.1 |
 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Compound | C. difficile ATCC 700057 | C. difficile 132 b (RT027) | S. aureus ATCC 29213 | S. aureus ATCC 43300 c | S. aureus NCTC 104422 c,d | E. faecalis ATCC 29212 | S. pneumoniae ATCC 49619 | E. coli ATCC 25922 | CC50 e | HC50 f |
| 40 | 8 | 8 | 16 | 8 | 16 | 32 | 16 | 64 | 32 | 32 |
| 41 | 16 | 16 | 8 | 4 | 8 | 8 | 16 | 128 | 16 | 32 |
| 42 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 32 | 32 | 32 |
| 1 [30] | 8 g | 8 h | 2 | - | - | 2 | 2 | 16 | - | - |
| 2 [31] | - | 32 | 4 | 2 | 4 | 4 | 4 | 8 | 27.4 | 94% i |
| 3 [19] | 8 g | 8 h | 2 | - | 2 | 4 | 8 | >128 | - | - |
| 4 [31] | - | 8 | 8 | 4 | 4 | 8 | 4 | 8 | 14.2 | 23% i |
| vanc | 0.5 | 0.5 | 1 | - | 1 | 4 | 1 | >16 | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahadari, M.K.; Jennepalli, S.; Tague, A.J.; Putsathit, P.; Hutton, M.L.; Hammer, K.A.; Knight, D.R.; Riley, T.V.; Lyras, D.; Keller, P.A.; et al. Cationic Peptidomimetic Amphiphiles Having a N-Aryl- or N-Naphthyl-1,2,3-Triazole Core Structure Targeting Clostridioides (Clostridium) difficile: Synthesis, Antibacterial Evaluation, and an In Vivo C. difficile Infection Model. Antibiotics 2021, 10, 913. https://doi.org/10.3390/antibiotics10080913
Mahadari MK, Jennepalli S, Tague AJ, Putsathit P, Hutton ML, Hammer KA, Knight DR, Riley TV, Lyras D, Keller PA, et al. Cationic Peptidomimetic Amphiphiles Having a N-Aryl- or N-Naphthyl-1,2,3-Triazole Core Structure Targeting Clostridioides (Clostridium) difficile: Synthesis, Antibacterial Evaluation, and an In Vivo C. difficile Infection Model. Antibiotics. 2021; 10(8):913. https://doi.org/10.3390/antibiotics10080913
Chicago/Turabian StyleMahadari, Muni Kumar, Sreenu Jennepalli, Andrew J. Tague, Papanin Putsathit, Melanie L. Hutton, Katherine A. Hammer, Daniel R. Knight, Thomas V. Riley, Dena Lyras, Paul A. Keller, and et al. 2021. "Cationic Peptidomimetic Amphiphiles Having a N-Aryl- or N-Naphthyl-1,2,3-Triazole Core Structure Targeting Clostridioides (Clostridium) difficile: Synthesis, Antibacterial Evaluation, and an In Vivo C. difficile Infection Model" Antibiotics 10, no. 8: 913. https://doi.org/10.3390/antibiotics10080913
APA StyleMahadari, M. K., Jennepalli, S., Tague, A. J., Putsathit, P., Hutton, M. L., Hammer, K. A., Knight, D. R., Riley, T. V., Lyras, D., Keller, P. A., & Pyne, S. G. (2021). Cationic Peptidomimetic Amphiphiles Having a N-Aryl- or N-Naphthyl-1,2,3-Triazole Core Structure Targeting Clostridioides (Clostridium) difficile: Synthesis, Antibacterial Evaluation, and an In Vivo C. difficile Infection Model. Antibiotics, 10(8), 913. https://doi.org/10.3390/antibiotics10080913






