Characterisation of Colistin -Resistant Enterobacterales and Acinetobacter Strains Carrying mcr Genes from Asian Aquaculture Products
Abstract
:1. Introduction
2. Results
2.1. Bacterial Isolates
2.2. Colistin Susceptibility
2.3. Multi-Locus Sequence Typing (MLST)
2.4. Detected mcr Genes
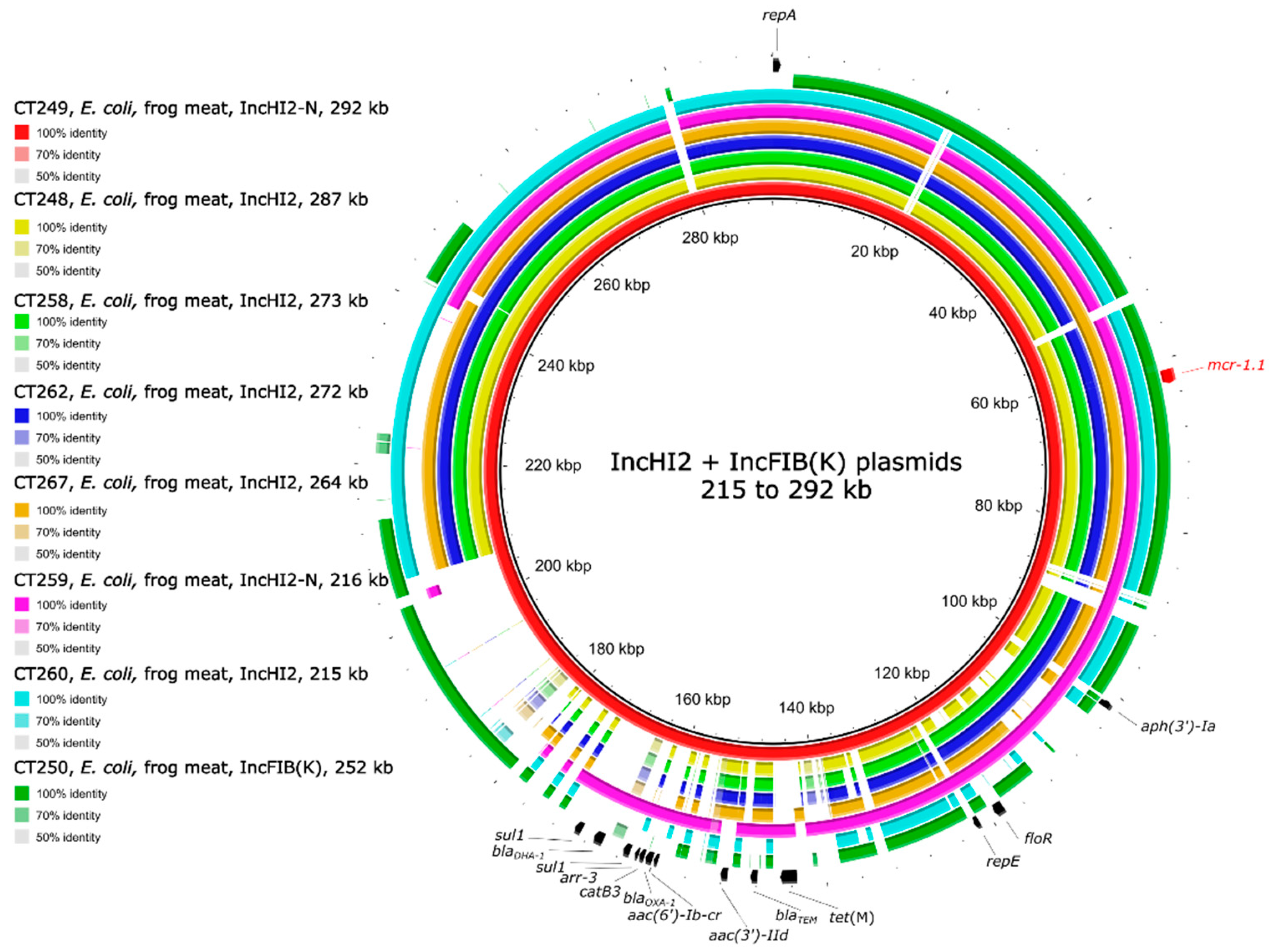
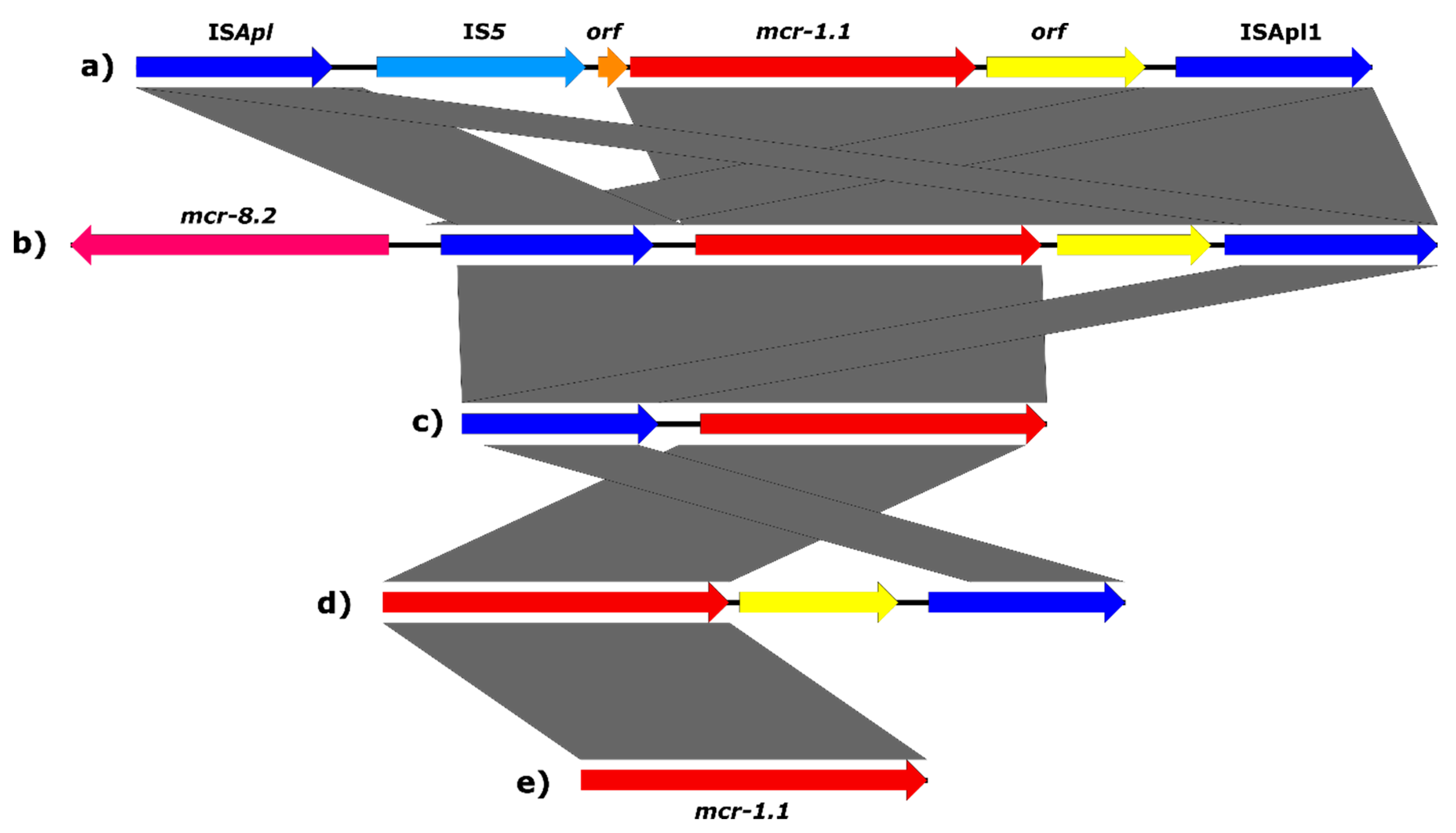
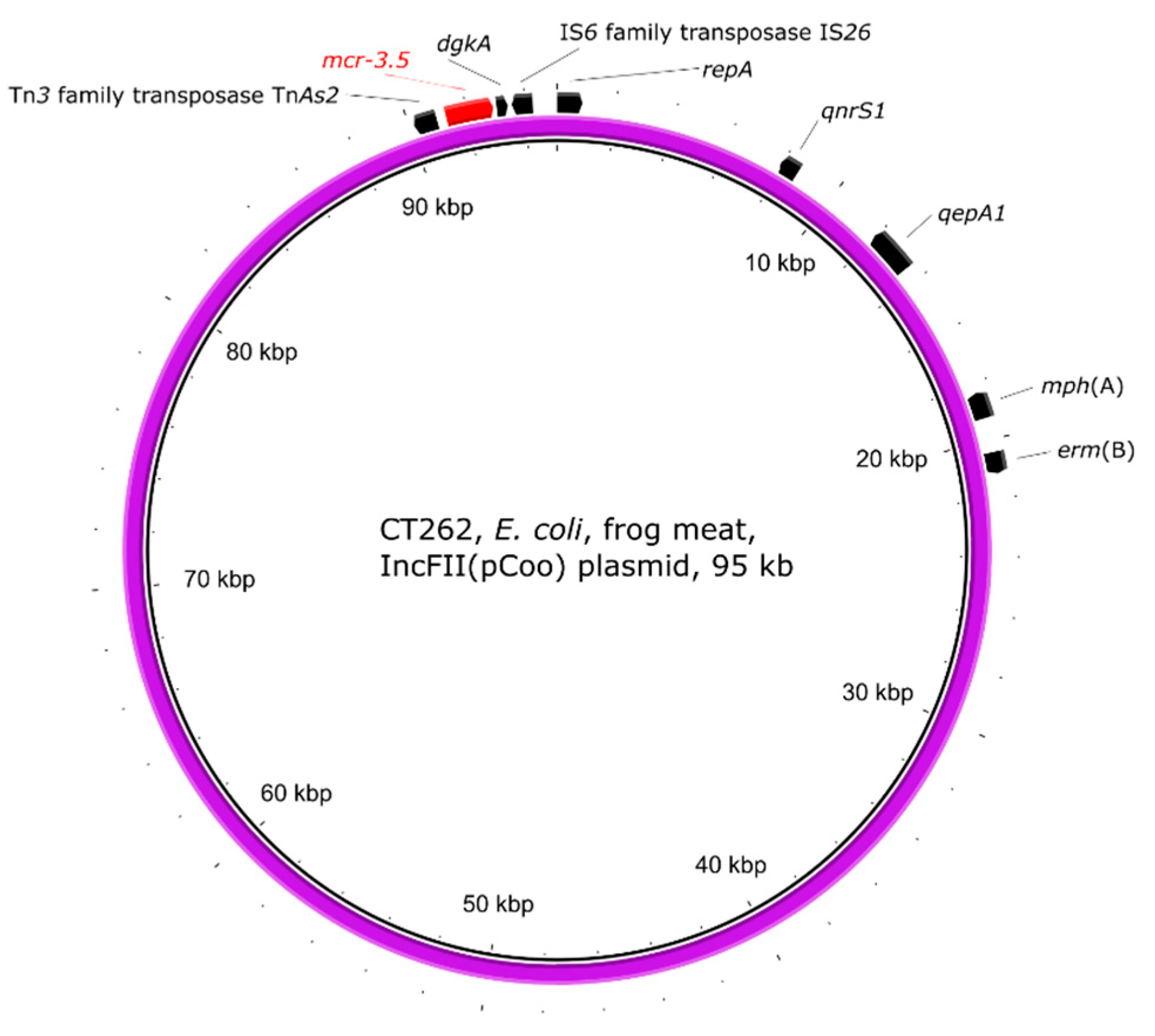
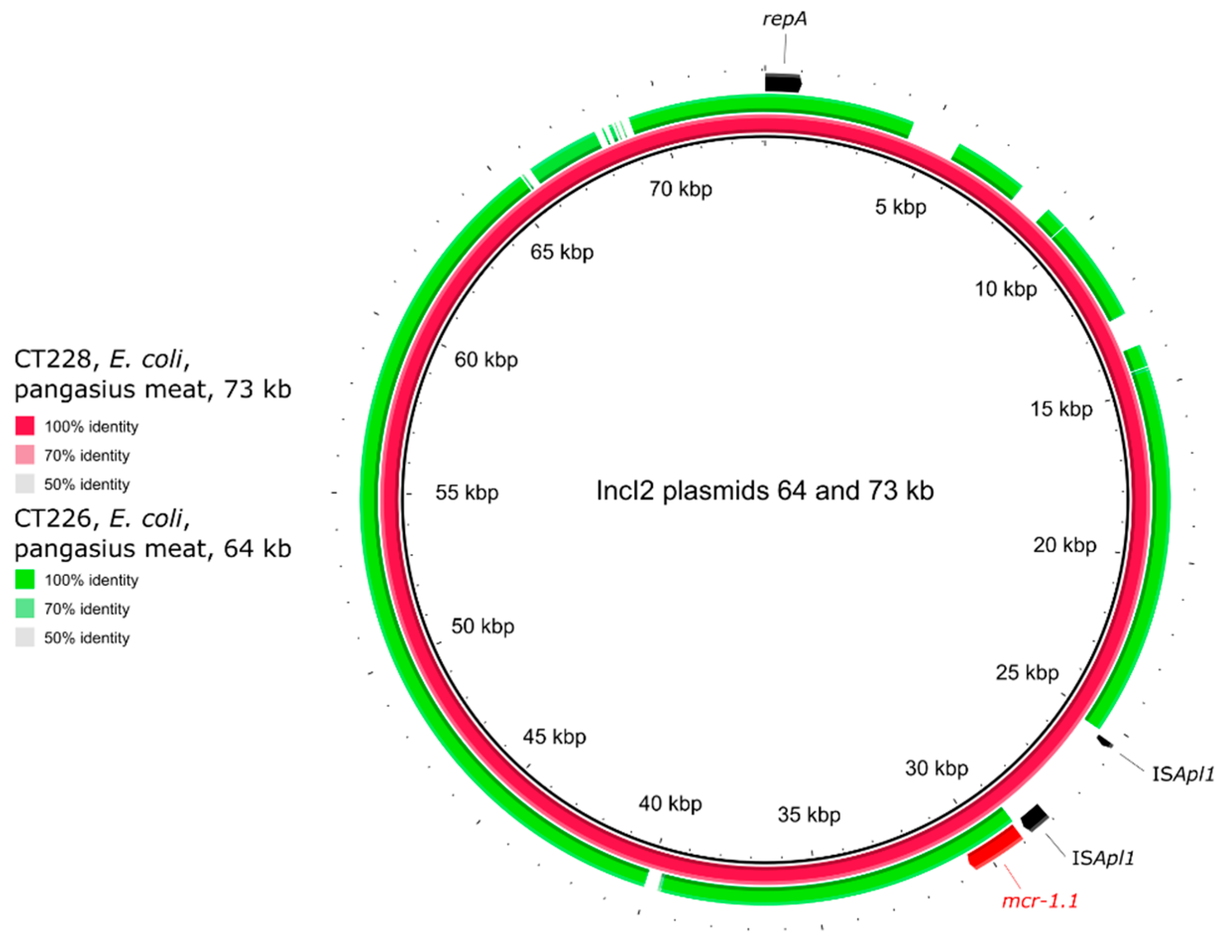
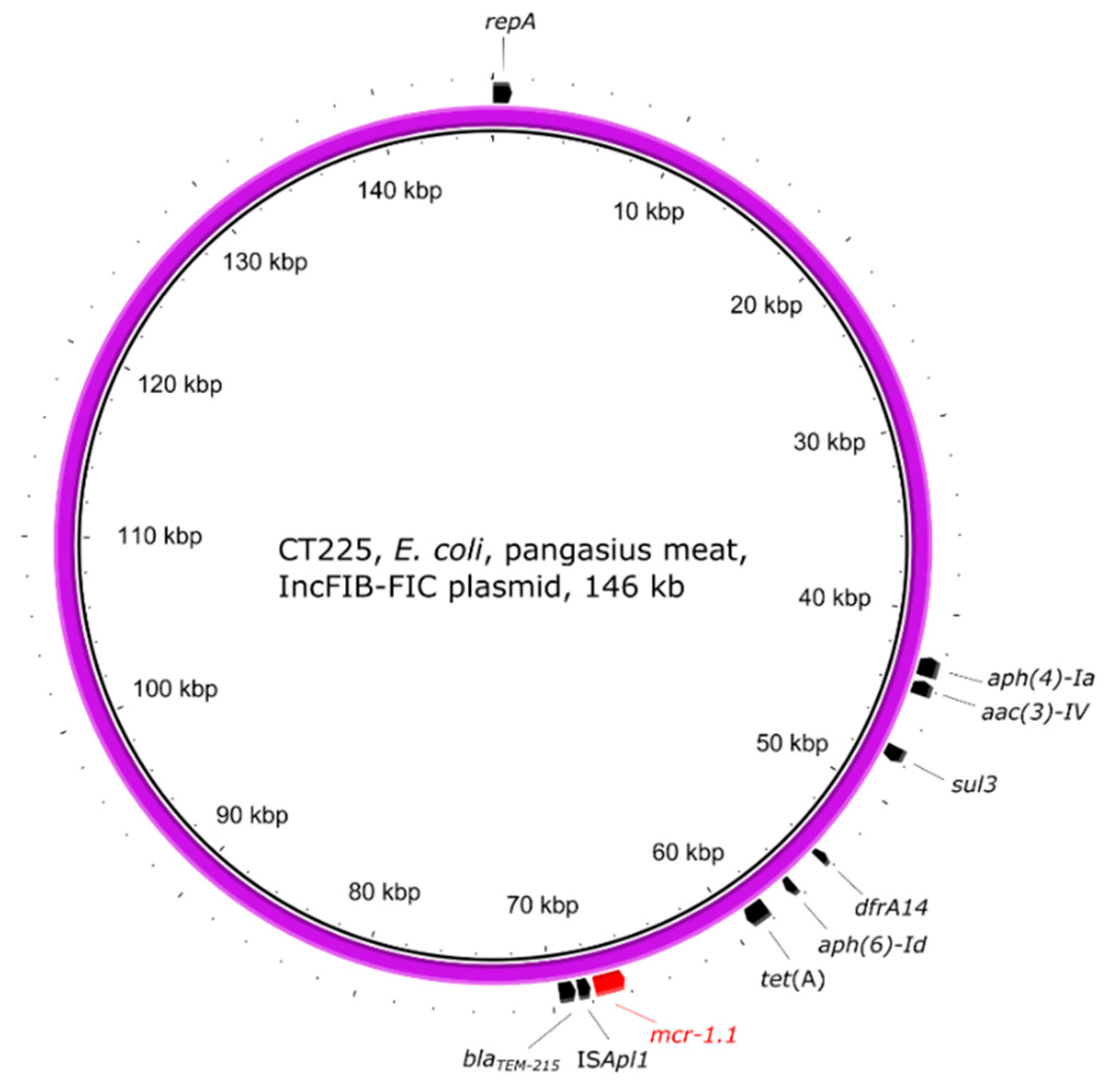
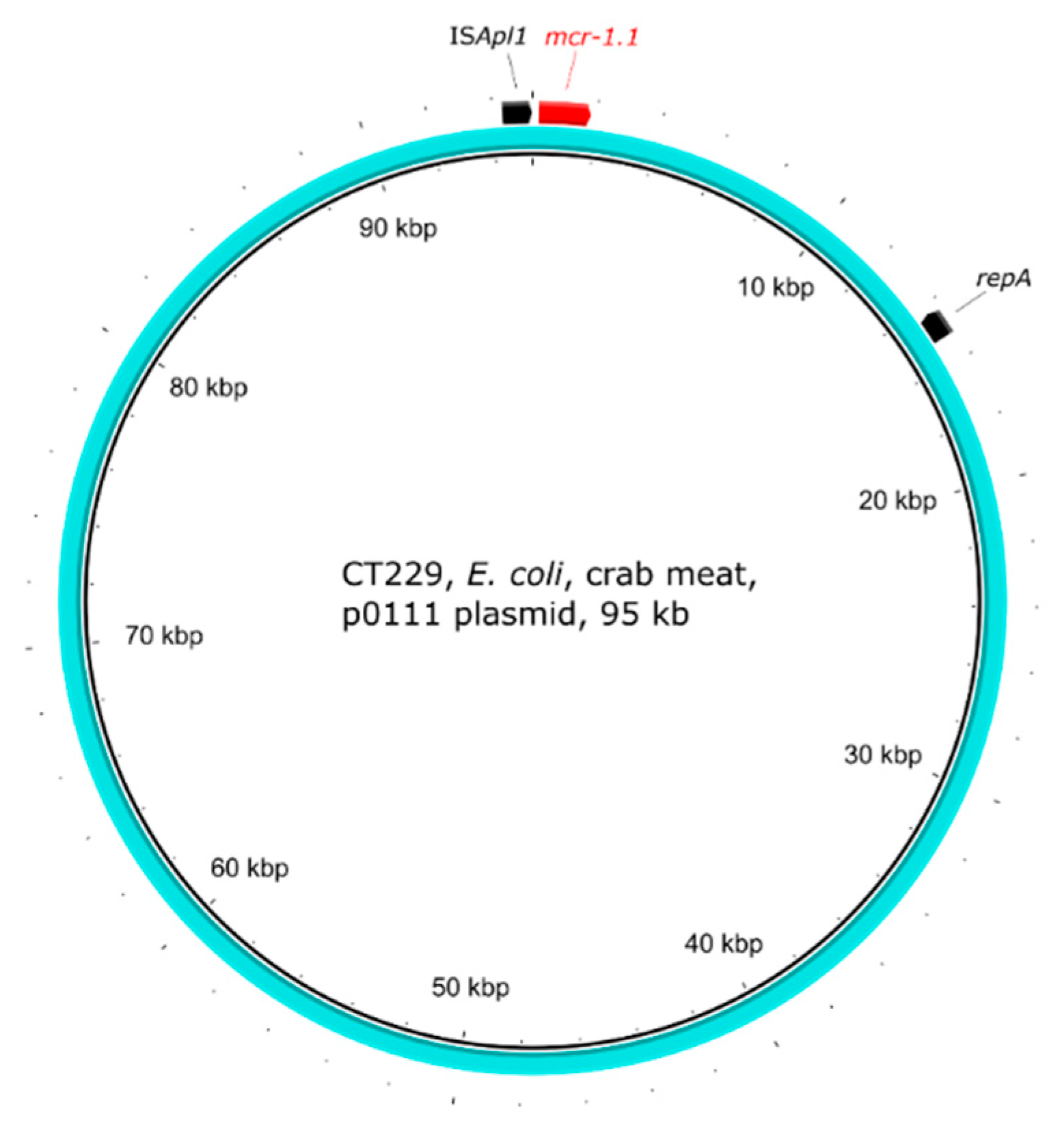
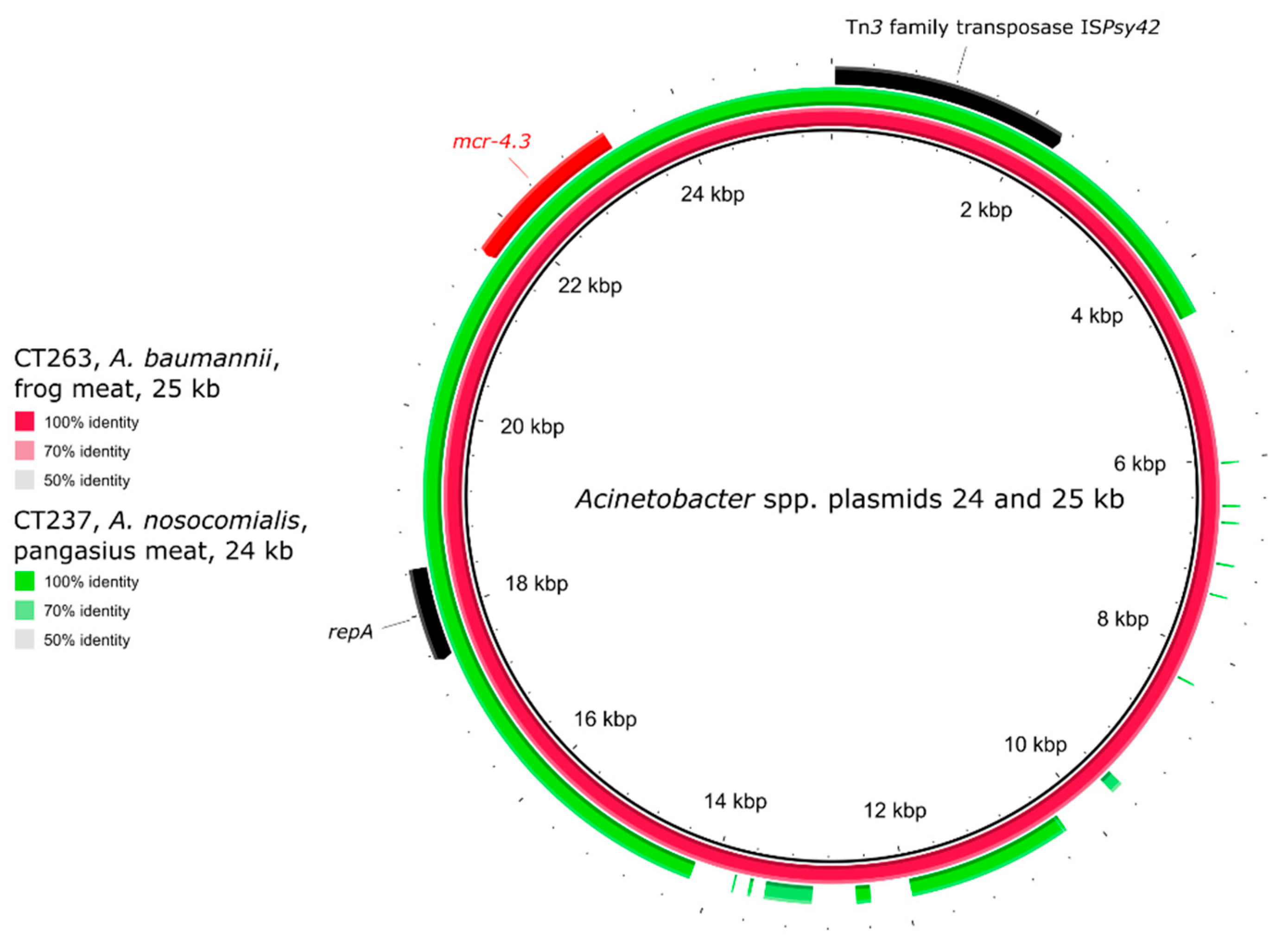
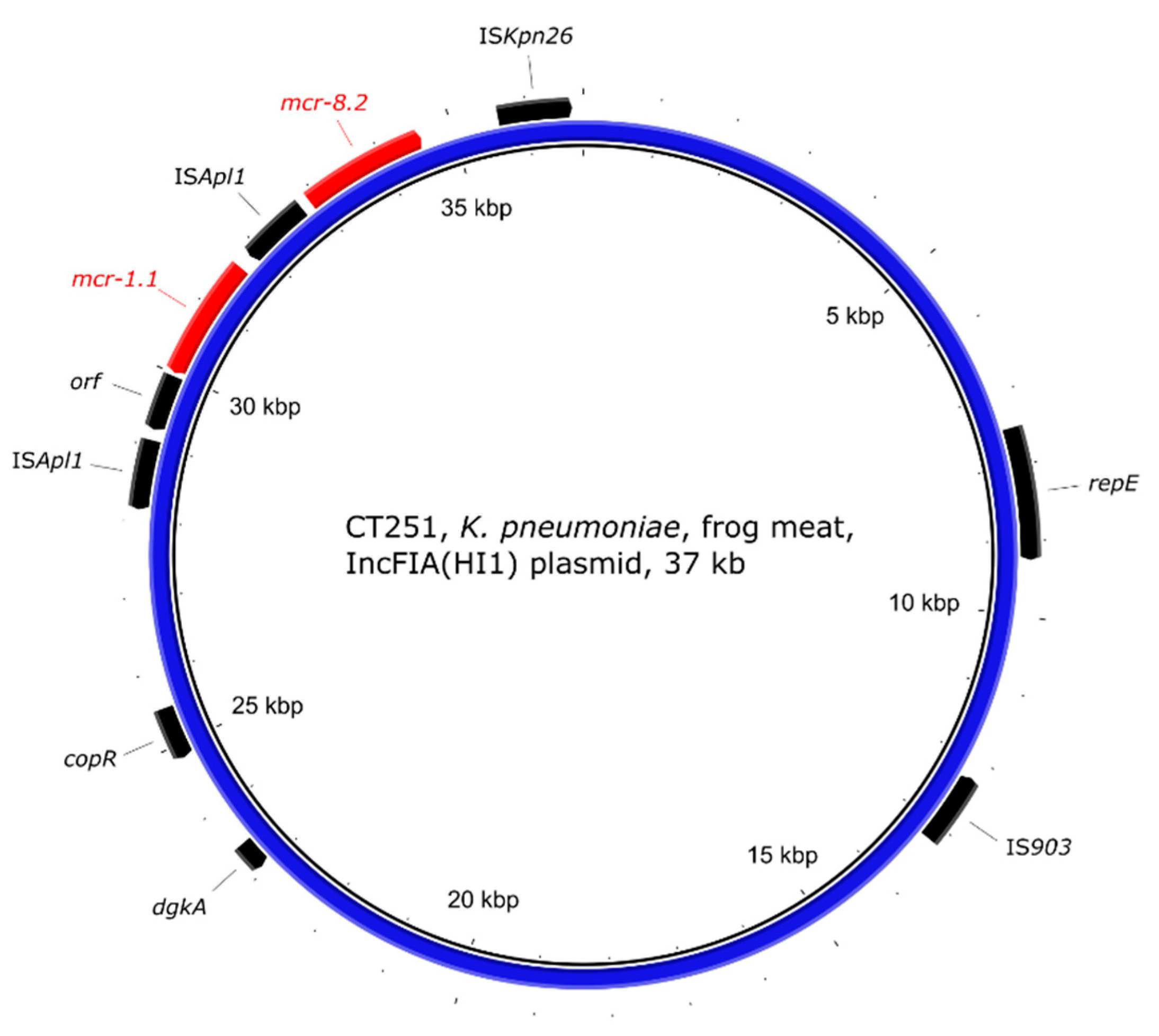
2.5. Genetic Environment of the mcr Genes on Plasmids
2.6. Genetic Surroundings of the mcr-1 Gene in the Chromosome
2.7. Co-Occurrence of Genes Encoding Resistance to Different Classes of Antimicrobials
3. Discussion
4. Materials and Methods
4.1. Bacterial Isolates Collection with Colistin Resistance
4.2. Genomic DNA Extraction, Whole-Genome Sequencing (WGS), and Genome Assembly
4.3. Multilocus Sequence Typing (MLST)
4.4. Genetic Analysis of Plasmids and Antibiotic Resistance Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Henriksson, P.J.G.; Rico, A.; Troell, M.; Klinger, D.H.; Buschmann, A.H.; Saksida, S.; Chadag, M.V.; Zhang, W. Unpacking Factors Influencing Antimicrobial Use in Global Aquaculture and Their Implication for Management: A Review from a Systems Perspective. Sustain. Sci. 2018, 13, 1105–1120. [Google Scholar] [CrossRef] [Green Version]
- Han, Q.F.; Zhao, S.; Zhang, X.R.; Wang, X.L.; Song, C.; Wang, S.G. Distribution, Combined Pollution and Risk Assessment of Antibiotics in Typical Marine Aquaculture Farms Surrounding the Yellow Sea, North China. Environ. Int. 2020, 138, 105551. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Guan, Y.; Cheng, M.; Chen, H.; He, J.; Wang, S.; Wang, Z. Occurrence and Distribution of Antibiotics and Antibiotic Resistance Genes in Ba River, China. Sci. Total Environ. 2018, 642, 1136–1144. [Google Scholar] [CrossRef] [PubMed]
- Catry, B. Use of Colistin-Containing Products within the European Union and European Economic Area (EU/EEA): Development of Resistance in Animals and Possible Impact on Human and Animal Health. Int. J. Antimicrob. Agents 2015, 10, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Luo, Q.; Wang, Y.; Xiao, Y. Prevalence and Transmission of Mobilized Colistin Resistance (Mcr) Gene in Bacteria Common to Animals and Humans. Biosaf. Health 2020, 8, 71–78. [Google Scholar] [CrossRef]
- Xavier, B.B.; Lammens, C.; Ruhal, R.; Kumar-Singh, S.; Butaye, P.; Goossens, H.; Malhotra-Kumar, S. Identification of a Novel Plasmid-Mediated Colistin-Resistance Gene, Mcr-2, in Escherichia Coli, Belgium, June 2016. Euro. Surveill. 2016, 21, 30280. [Google Scholar] [CrossRef]
- Yin, W.; Li, H.; Shen, Y.; Liu, Z.; Wang, S.; Shen, Z.; Zhang, R.; Walsh, T.R.; Shen, J.; Wang, Y. Novel Plasmid-Mediated Colistin Resistance Gene Mcr-3 in Escherichia Coli. mBio 2017, 8, e00543-17. [Google Scholar] [CrossRef] [Green Version]
- Carattoli, A.; Villa, L.; Feudi, C.; Curcio, L.; Orsini, S.; Luppi, A.; Pezzotti, G.; Magistrali, C.F. Novel Plasmid-Mediated Colistin Resistance Mcr-4 Gene in Salmonella and Escherichia Coli, Italy 2013, Spain and Belgium, 2015 to 2016. Eurosurveillance 2017, 22, 30589. [Google Scholar] [CrossRef] [Green Version]
- Borowiak, M.; Fischer, J.; Hammerl, J.A.; Hendriksen, R.S.; Szabo, I.; Malorny, B. Identification of a Novel Transposon-Associated Phosphoethanolamine Transferase Gene, Mcr-5, Conferring Colistin Resistance in d-Tartrate Fermenting Salmonella Enterica Subsp. Enterica Serovar Paratyphi B. J. Antimicrob. Chemother. 2017, 72, 3317–3324. [Google Scholar] [CrossRef] [Green Version]
- AbuOun, M.; Stubberfield, E.J.; Duggett, N.A.; Kirchner, M.; Dormer, L.; Nunez-Garcia, J.; Randall, L.P.; Lemma, F.; Crook, D.W.; Teale, C.; et al. Mcr-1 and Mcr-2 (Mcr-6.1) Variant Genes Identified in Moraxella Species Isolated from Pigs in Great Britain from 2014 to 2015. J. Antimicrob. Chemother. 2017, 72, 2745–2749. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.-Q.; Li, Y.-X.; Lei, C.-W.; Zhang, A.-Y.; Wang, H.-N. Novel Plasmid-Mediated Colistin Resistance Gene Mcr-7.1 in Klebsiella Pneumoniae. J. Antimicrob. Chemother. 2018, 73, 1791–1795. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wang, Y.; Zhou, Y.; Li, J.; Yin, W.; Wang, S.; Zhang, S.; Shen, J.; Shen, Z.; Wang, Y. Emergence of a Novel Mobile Colistin Resistance Gene, Mcr-8, in NDM-Producing Klebsiella Pneumoniae. Emerg. Microbes Infect. 2018, 7, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Carroll, L.M.; Gaballa, A.; Guldimann, C.; Sullivan, G.; Henderson, L.O.; Wiedmann, M. Identification of Novel Mobilized Colistin Resistance Gene Mcr-9 in a Multidrug-Resistant, Colistin-Susceptible Salmonella Enterica Serotype Typhimurium Isolate. mBio 2019, 10, e00853-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Feng, Y.; Liu, L.; Wei, L.; Kang, M.; Zong, Z. Identification of Novel Mobile Colistin Resistance Gene Mcr-10. Emerg. Microbes Infect. 2020, 9, 508–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tkadlec, J.; Kalova, A.; Brajerova, M.; Gelbicova, T.; Karpiskova, R.; Smelikova, E.; Nyc, O.; Drevinek, P.; Krutova, M. The Intestinal Carriage of Plasmid-Mediated Colistin-Resistant Enterobacteriaceae in Tertiary Care Settings. Antibiotics 2021, 10, 258. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-C.; Kuroda, M.; Suzuki, S.; Mu, J.-J. Emergence of the Mcr-1 Colistin Resistance Gene in Extended-Spectrum β-Lactamase-Producing Klebsiella Pneumoniae in Taiwan. J. Global Antimicrob. Resist. 2021, 24, 278–284. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.; Chen, M.; Hu, J.; Zhang, H.; Xiang, Y.; Yang, H.; Qiu, S.; Song, H. Plasmid-Borne Colistin Resistance Gene Mcr-1 in a Multidrug Resistant Salmonella Enterica Serovar Typhimurium Isolate from an Infant with Acute Diarrhea in China. Int. J. Infect. Dis. 2021, 103, 13–18. [Google Scholar] [CrossRef]
- Martins-Sorenson, N.; Snesrud, E.; Xavier, D.E.; Cacci, L.C.; Iavarone, A.T.; McGann, P.; Riley, L.W.; Moreira, B.M. A Novel Plasmid-Encoded Mcr-4.3 Gene in a Colistin-Resistant Acinetobacter Baumannii Clinical Strain. J. Antimicrob. Chemother. 2020, 75, 60–64. [Google Scholar] [CrossRef]
- Rozwandowicz, M.; Brouwer, M.S.M.; Fischer, J.; Wagenaar, J.A.; Gonzalez-Zorn, B.; Guerra, B.; Mevius, D.J.; Hordijk, J. Plasmids Carrying Antimicrobial Resistance Genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 1121–1137. [Google Scholar] [CrossRef] [Green Version]
- Carattoli, A. Resistance Plasmid Families in Enterobacteriaceae. Antimicrob. Agents Chemother. 2009, 53, 2227–2238. [Google Scholar] [CrossRef] [Green Version]
- Salgado-Camargo, A.D.; Castro-Jaimes, S.; Gutierrez-Rios, R.-M.; Lozano, L.F.; Altamirano-Pacheco, L.; Silva-Sanchez, J.; Pérez-Oseguera, Á.; Volkow, P.; Castillo-Ramírez, S.; Cevallos, M.A. Structure and Evolution of Acinetobacter Baumannii Plasmids. Front. Microbiol. 2020, 11, 1283. [Google Scholar] [CrossRef]
- Ma, F.; Shen, C.; Zheng, X.; Liu, Y.; Chen, H.; Zhong, L.; Liang, Y.; Liao, K.; Xia, Y.; Tian, G.-B.; et al. Identification of a Novel Plasmid Carrying Mcr-4.3 in an Acinetobacter Baumannii Strain in China. Antimicrob. Agents Chemother. 2019, 63, e00133-19. [Google Scholar] [CrossRef] [Green Version]
- Bitar, I.; Medvecky, M.; Gelbicova, T.; Jakubu, V.; Hrabak, J.; Zemlickova, H.; Karpiskova, R.; Dolejska, M. Complete Nucleotide Sequences of Mcr-4.3 -Carrying Plasmids in Acinetobacter Baumannii Sequence Type 345 of Human and Food Origin from the Czech Republic, the First Case in Europe. Antimicrob. Agents Chemother. 2019, 63, e01166-19. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Gourbeyre, E.; Chandler, M. Bacterial Insertion Sequences: Their Genomic Impact and Diversity. FEMS Microbiol. Rev. 2014, 38, 865–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aziz, R.K.; Breitbart, M.; Edwards, R.A. Transposases Are the Most Abundant, Most Ubiquitous Genes in Nature. Nucleic Acids Res. 2010, 38, 4207–4217. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, S.; Johnson, A.P. Transferable Resistance to Colistin: A New but Old Threat: Table 1. J. Antimicrob. Chemother. 2016, 71, 2066–2070. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhang, R.; Schwarz, S.; Wu, C.; Shen, J.; Walsh, T.R.; Wang, Y. Farm Animals and Aquaculture: Significant Reservoirs of Mobile Colistin Resistance Genes. Environ. Microbiol. 2020, 22, 2469–2484. [Google Scholar] [CrossRef] [Green Version]
- Lv, L.; Cao, Y.; Yu, P.; Huang, R.; Wang, J.; Wen, Q.; Zhi, C.; Zhang, Q.; Liu, J.-H. Detection of Mcr-1 Gene among Escherichia Coli Isolates from Farmed Fish and Characterization of Mcr-1 -Bearing IncP Plasmids. Antimicrob. Agents Chemother. 2018, 62, e02378-17. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Lv, Z.; Yang, L.; Liu, D.; Ou, Y.; Xu, C.; Liu, W.; Yuan, D.; Hao, Y.; He, J.; et al. Integrated Aquaculture Contributes to the Transfer of Mcr-1 between Animals and Humans via the Aquaculture Supply Chain. Environ. Int. 2019, 130, 104708. [Google Scholar] [CrossRef]
- Lei, T.; Zhang, J.; Jiang, F.; He, M.; Zeng, H.; Chen, M.; Wu, S.; Wang, J.; Ding, Y.; Wu, Q. First Detection of the Plasmid-Mediated Colistin Resistance Gene Mcr-1 in Virulent Vibrio Parahaemolyticus. Int. J. Food Microbiol. 2019, 308, 108290. [Google Scholar] [CrossRef]
- Hoa, T.T.T.; Nakayama, T.; Huyen, H.M.; Harada, K.; Hinenoya, A.; Phuong, N.T.; Yamamoto, Y. Extended-spectrum Beta-lactamase-producing Escherichia Coli Harbouring Sul and Mcr–1 Genes Isolates from Fish Gut Contents in the Mekong Delta, Vietnam. Lett. Appl. Microbiol. 2020, 71, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Ellis-Iversen, J.; Seyfarth, A.M.; Korsgaard, H.; Bortolaia, V.; Munck, N.; Dalsgaard, A. Antimicrobial Resistant E. Coli and Enterococci in Pangasius Fillets and Prawns in Danish Retail Imported from Asia. Food Control 2020, 114, 106958. [Google Scholar] [CrossRef]
- Slettemeås, J.S.; Urdahl, A.-M.; Mo, S.S.; Johannessen, G.S.; Grave, K.; Norström, M.; Steinbakk, M.; Sunde, M. Imported Food and Feed as Contributors to the Introduction of Plasmid-Mediated Colistin-Resistant Enterobacteriaceae to a ‘Low Prevalence’ Country. J. Antimicrob. Chemother. 2017, 72, 2675–2677. [Google Scholar] [CrossRef]
- Liu, D.; Song, H.; Ke, Y.; Xia, J.; Shen, Y.; Ou, Y.; Hao, Y.; He, J.; Li, X.; Zhou, Y.; et al. Co-Existence of Two Novel Phosphoethanolamine Transferase Gene Variants in Aeromonas Jandaei from Retail Fish. Int. J. Antimicrob. Agents 2020, 55, 105856. [Google Scholar] [CrossRef] [PubMed]
- Eichhorn, I.; Feudi, C.; Wang, Y.; Kaspar, H.; Feßler, A.T.; Lübke-Becker, A.; Michael, G.B.; Shen, J.; Schwarz, S. Identification of Novel Variants of the Colistin Resistance Gene Mcr-3 in Aeromonas Spp. from the National Resistance Monitoring Programme GERM-Vet and from Diagnostic Submissions. J. Antimicrob. Chemother. 2018, 73, 1217–1221. [Google Scholar] [CrossRef]
- Cabello, F.C.; Tomova, A.; Ivanova, L.; Godfrey, H.P. Aquaculture and Mcr Colistin Resistance Determinants. mBio 2017, 8, e01229-17. [Google Scholar] [CrossRef] [Green Version]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A Better, Faster Version of the PHAST Phage Search Tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [Green Version]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [Green Version]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for Predictions of Phenotypes from Genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Hernández, M.; Iglesias, M.R.; Rodríguez-Lázaro, D.; Gallardo, A.; Quijada, N.; Miguela-Villoldo, P.; Campos, M.J.; Píriz, S.; López-Orozco, G.; de Frutos, C.; et al. Co-Occurrence of Colistin-Resistance Genes Mcr-1 and Mcr-3 among Multidrug-Resistant Escherichia Coli Isolated from Cattle, Spain, September 2015. Eurosurveillance 2017, 22, 30586. [Google Scholar] [CrossRef]
- Sun, J.; Li, X.-P.; Fang, L.-X.; Sun, R.-Y.; He, Y.-Z.; Lin, J.; Liao, X.-P.; Feng, Y.; Liu, Y.-H. Co-Occurrence of Mcr-1 in the Chromosome and on an IncHI2 Plasmid: Persistence of Colistin Resistance in Escherichia Coli. Int. J. Antimicrob. Agents 2018, 51, 842–847. [Google Scholar] [CrossRef] [PubMed]
- Gelbíčová, T.; Baráková, A.; Florianová, M.; Jamborová, I.; Zelendová, M.; Pospíšilová, L.; Koláčková, I.; Karpíšková, R. Dissemination and Comparison of Genetic Determinants of Mcr-Mediated Colistin Resistance in Enterobacteriaceae via Retailed Raw Meat Products. Front. Microbiol. 2019, 10, 2824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hala, S.; Antony, C.P.; Momin, A.A.; Alshehri, M.; Ben-Rached, F.; Al-Ahmadi, G.; Zakri, S.; Baadhaim, M.; Alsaedi, A.; Thaqafi, O.A.A.; et al. Co-Occurrence of Mcr-1 and Mcr-8 Genes in Multi-Drug-Resistant Klebsiella Pneumoniae from a 2015 Clinical Isolate. Int. J. Antimicrob. Agents 2021, 57, 106303. [Google Scholar] [CrossRef] [PubMed]
- Lima, W.G.; Alves, M.C.; Cruz, W.S.; Paiva, M.C. Chromosomally Encoded and Plasmid-Mediated Polymyxins Resistance in Acinetobacter Baumannii: A Huge Public Health Threat. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Duan, X.; Peng, Y.; Rui, Y. Molecular Characteristics of Carbapenem-Resistant Acinetobacter Spp. from Clinical Infection Samples and Fecal Survey Samples in Southern China. BMC Infect. Dis. 2019, 19, 900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hameed, F.; Khan, M.A.; Muhammad, H.; Sarwar, T.; Bilal, H.; Rehman, T.U. Plasmid-Mediated Mcr-1 Gene in Acinetobacter Baumannii and Pseudomonas Aeruginosa: First Report from Pakistan. Rev. Soc. Bras. Med. Trop. 2019, 52, e20190237. [Google Scholar] [CrossRef] [Green Version]
- Al-Kadmy, I.M.S.; Ibrahim, S.A.; Al-Saryi, N.; Aziz, S.N.; Besinis, A.; Hetta, H.F. Prevalence of Genes Involved in Colistin Resistance in Acinetobacter Baumannii: First Report from Iraq. Microb. Drug Resist. 2020, 26, 616–622. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, H.; Liu, Y.-H.; Feng, Y. Towards Understanding MCR-like Colistin Resistance. Trends Microbiol. 2018, 26, 794–808. [Google Scholar] [CrossRef] [PubMed]
- Meinersmann, R.J. The Biology of IncI2 Plasmids Shown by Whole-Plasmid Multi-Locus Sequence Typing. Plasmid 2019, 106, 102444. [Google Scholar] [CrossRef]
- Matamoros, S.; van Hattem, J.M.; Arcilla, M.S.; Willemse, N.; Melles, D.C.; Penders, J.; Vinh, T.N.; Thi Hoa, N.; Bootsma, M.C.J.; van Genderen, P.J.; et al. Global Phylogenetic Analysis of Escherichia Coli and Plasmids Carrying the Mcr-1 Gene Indicates Bacterial Diversity but Plasmid Restriction. Sci. Rep. 2017, 7, 15364. [Google Scholar] [CrossRef] [Green Version]
- Zelendova, M.; Papagiannitsis, C.C.; Valcek, A.; Medvecky, M.; Bitar, I.; Hrabak, J.; Gelbicova, T.; Barakova, A.; Kutilova, I.; Karpiskova, R.; et al. Characterization of the Complete Nucleotide Sequences of Mcr-1-Encoding Plasmids From Enterobacterales Isolates in Retailed Raw Meat Products From the Czech Republic. Front. Microbiol. 2021, 11, 604067. [Google Scholar] [CrossRef]
- Madec, J.-Y.; Haenni, M. Antimicrobial Resistance Plasmid Reservoir in Food and Food-Producing Animals. Plasmid 2018, 99, 72–81. [Google Scholar] [CrossRef]
- Ageevets, V.; Lazareva, I.; Mrugova, T.; Gostev, V.; Lobzin, Y.; Sidorenko, S. IncX4 Plasmids Harbouring Mcr-1 Genes: Further Dissemination. J. Global Antimicrob. Resist. 2019, 18, 166–167. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Feng, Y.; Liu, F.; Jiang, H.; Qu, Z.; Lei, M.; Wang, J.; Zhang, B.; Hu, Y.; Ding, J.; et al. A Phage-Like IncY Plasmid Carrying the Mcr-1 Gene in Escherichia Coli from a Pig Farm in China. Antimicrob. Agents Chemother. 2017, 61, e02035-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Xie, M.; Lv, J.; Wai-Chi Chan, E.; Chen, S. Complete Genetic Analysis of Plasmids Carrying Mcr-1 and Other Resistance Genes in an Escherichia Coli Isolate of Animal Origin. J. Antimicrob. Chemother. 2016, 72, 696–699. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Xiong, W.; Liu, P.; Xie, X.; Zeng, J.; Sun, Y.; Zeng, Z. Metagenomic Insights Into the Contribution of Phages to Antibiotic Resistance in Water Samples Related to Swine Feedlot Wastewater Treatment. Front. Microbiol. 2018, 9, 2474. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Xie, X.; Tang, M.; Liu, J.; Tuo, H.; Gu, J.; Tang, Y.; Lei, C.; Wang, H.; Zhang, A. Exploring the Profile of Antimicrobial Resistance Genes Harboring by Bacteriophage in Chicken Feces. Sci. Total Environ. 2020, 700, 134446. [Google Scholar] [CrossRef]
- Shen, Y.; Wu, Z.; Wang, Y.; Zhang, R.; Zhou, H.-W.; Wang, S.; Lei, L.; Li, M.; Cai, J.; Tyrrell, J.; et al. Heterogeneous and Flexible Transmission of Mcr-1 in Hospital-Associated Escherichia Coli. mBio 2018, 9, e00943-18. [Google Scholar] [CrossRef] [Green Version]
- Nordmann, P.; Lienhard, R.; Kieffer, N.; Clerc, O.; Poirel, L. Plasmid-Mediated Colistin-Resistant Escherichia Coli in Bacteremia in Switzerland. Clin. Infect. Dis. 2016, 62, 1322–1323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zając, M.; Sztromwasser, P.; Bortolaia, V.; Leekitcharoenphon, P.; Cavaco, L.M.; Ziȩtek-Barszcz, A.; Hendriksen, R.S.; Wasyl, D. Occurrence and Characterization of Mcr-1-Positive Escherichia Coli Isolated From Food-Producing Animals in Poland, 2011–2016. Front. Microbiol. 2019, 10, 1753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, F.-J.; Lauderdale, T.-L.; Huang, W.-C.; Shiau, Y.-R.; Wang, H.-Y.; Kuo, S.-C. Emergence of Mcr-1, Mcr-3 and Mcr-8 in Clinical Klebsiella Pneumoniae Isolates in Taiwan. Clin. Microbiol. Infect. 2021, 27, 305–307. [Google Scholar] [CrossRef]
- Snesrud, E.; McGann, P.; Chandler, M. The Birth and Demise of the ISApl1-Mcr-1-ISApl1 Composite Transposon: The Vehicle for Transferable Colistin Resistance. mBio 2018, 9, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Yu, H.; Xie, M.; Chen, K.; Dong, N.; Lin, D.; Chan, E.W.-C.; Chen, S. Genetic Basis of Chromosomally-Encoded Mcr-1 Gene. Int. J. Antimicrob. Agents 2018, 51, 578–585. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Kawahara, R.; Harada, K.; Teruya, S.; Nakayama, T.; Motooka, D.; Nakamura, S.; Nguyen, P.D.; Kumeda, Y.; Van Dang, C.; et al. The Presence of Colistin Resistance Gene Mcr-1 and -3 in ESBL Producing Escherichia Coli Isolated from Food in Ho Chi Minh City, Vietnam. FEMS Microbiol. Lett. 2018, 365, fny100. [Google Scholar] [CrossRef]
- Li, R.; Du, P.; Zhang, P.; Li, Y.; Yang, X.; Wang, Z.; Wang, J.; Bai, L. Comprehensive Genomic Investigation of Coevolution of Mcr Genes in Escherichia Coli Strains via Nanopore Sequencing. Global Chall. 2021, 5, 2000014. [Google Scholar] [CrossRef]
- Shen, C.; Zhong, L.-L.; Ma, F.; El-Sayed Ahmed, M.A.E.-G.; Doi, Y.; Zhang, G.; Liu, Y.; Huang, S.; Li, H.-Y.; Zhang, L.; et al. Genomic Patterns and Characterizations of Chromosomally-Encoded Mcr-1 in Escherichia Coli Populations. Gut. Pathog. 2020, 12, 55. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Kawahara, R.; Hamamoto, K.; Hirai, I.; Khong, D.T.; Nguyen, T.N.; Tran, H.T.; Motooka, D.; Nakamura, S.; Yamamoto, Y. High Prevalence of Colistin-Resistant Escherichia Coli with Chromosomally Carried Mcr-1 in Healthy Residents in Vietnam. mSphere 2020, 5, e00117-20. [Google Scholar] [CrossRef] [Green Version]
- Tada, T.; Nhung, P.H.; Shimada, K.; Tsuchiya, M.; Phuong, D.M.; Anh, N.Q.; Ohmagari, N.; Kirikae, T. Emergence of Colistin-Resistant Escherichia Coli Clinical Isolates Harboring Mcr-1 in Vietnam. Int. J. Infect. Dis. 2017, 63, 72–73. [Google Scholar] [CrossRef] [Green Version]
- Krutova, M.; Kalova, A.; Nycova, E.; Gelbicova, T.; Karpiskova, R.; Smelikova, E.; Nyc, O.; Drevinek, P.; Tkadlec, J. The Colonisation of Czech Travellers and Expatriates Living in the Czech Republic by Colistin-Resistant Enterobacteriaceae and Whole Genome Characterisation of E. Coli Isolates Harbouring the Mcr-1 Genes on a Plasmid or Chromosome: A Cross-Sectional Study. Travel Med. Infect. Dis. 2021, 39, 101914. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, 22. [Google Scholar] [CrossRef] [Green Version]
- Zankari, E.; Allesøe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A Novel Web Tool for WGS-Based Detection of Antimicrobial Resistance Associated with Chromosomal Point Mutations in Bacterial Pathogens. J. Antimicrob. Chemother. 2017, 72, 2764–2768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2019, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alikhan, N.-F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple Prokaryote Genome Comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [Green Version]

| Strain ID | Source | Species | Colistin MIC (mg/L) | MLST | mcr Gene | mcr Gene Localisation (Plasmid Type/ Chromosome) |
|---|---|---|---|---|---|---|
| CT225 | pangasius | E. coli | 4 | ST155 | mcr-1.1 | IncFIB(AP001918)-FIC(FII) |
| CT226 | pangasius | E. coli | >16 | ST2253 | mcr-1.1 | IncX4, IncI2 |
| CT227 | pangasius | E. coli | 4 | ST206 | mcr-1.1 | chromosome |
| CT228 | pangasius | E. coli | 8 | ST156 | mcr-1.1 | IncI2 |
| CT229 | crab | E. coli | 4 | ST1011 | mcr-1.1 | p0111 |
| CT230 | crab | E. coli | 8 | ST6745 | mcr-1.1 | chromosome |
| CT248 | frog legs | E. coli | 4 | ST4481 | mcr-1.1 | IncHI2 |
| CT249 | frog legs | E. coli | 4 | ST48 | mcr-1.1 | IncHI2-N |
| CT250 | frog legs | E. coli | 4 | ST2179 | mcr-1.1 | IncFIB(K) |
| CT258 | frog legs | E. coli | 4 | ST48 | mcr-1.1 | IncHI2 |
| CT259 | frog legs | E. coli | 8 | ST8680 | mcr-1.1 | IncHI2-N |
| CT260 | frog legs | E. coli | 4 | ST48 | mcr-1.1 | IncHI2 |
| CT262 | frog legs | E. coli | 4 | ST609 | mcr-1.1, mcr-3.5 | mcr-1/IncHI2, mcr-3/IncFII(pCoo) |
| CT267 | frog legs | E. coli | 4 | ST48 | mcr-1.1 | IncHI2 |
| CT251 | frog legs | K. pneumoniae | >16 | ST11 | mcr-1.1 + mcr-8.2 | IncFIA(HI1) |
| CT237 | pangasius | A. nosocomialis | >16 | ST279 | mcr-4.3 | untypeable plasmid |
| CT263 | frog legs | A. baumannii | >16 | ST490 | mcr-4.3 | untypeable plasmid |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalová, A.; Gelbíčová, T.; Overballe-Petersen, S.; Litrup, E.; Karpíšková, R. Characterisation of Colistin -Resistant Enterobacterales and Acinetobacter Strains Carrying mcr Genes from Asian Aquaculture Products. Antibiotics 2021, 10, 838. https://doi.org/10.3390/antibiotics10070838
Kalová A, Gelbíčová T, Overballe-Petersen S, Litrup E, Karpíšková R. Characterisation of Colistin -Resistant Enterobacterales and Acinetobacter Strains Carrying mcr Genes from Asian Aquaculture Products. Antibiotics. 2021; 10(7):838. https://doi.org/10.3390/antibiotics10070838
Chicago/Turabian StyleKalová, Alžběta, Tereza Gelbíčová, Søren Overballe-Petersen, Eva Litrup, and Renáta Karpíšková. 2021. "Characterisation of Colistin -Resistant Enterobacterales and Acinetobacter Strains Carrying mcr Genes from Asian Aquaculture Products" Antibiotics 10, no. 7: 838. https://doi.org/10.3390/antibiotics10070838
APA StyleKalová, A., Gelbíčová, T., Overballe-Petersen, S., Litrup, E., & Karpíšková, R. (2021). Characterisation of Colistin -Resistant Enterobacterales and Acinetobacter Strains Carrying mcr Genes from Asian Aquaculture Products. Antibiotics, 10(7), 838. https://doi.org/10.3390/antibiotics10070838





