Polymicrobial Interactions in the Cystic Fibrosis Airway Microbiome Impact the Antimicrobial Susceptibility of Pseudomonas aeruginosa
Abstract
1. Introduction
2. P. aeruginosa Infection in CF
3. The CF Airway Microbiome
Antibiotics and the CF Airway Microbiome
4. Polymicrobial Interactions with P. aeruginosa in the CF Airway Microbiome
4.1. P. aeruginosa and S. aureus Interactions
Impacts on Antibacterial Resistance
4.2. P. aeruginosa and S. Maltophilia Interactions
Impacts on Antibacterial Resistance
4.3. Anaerobes Impact P. aeruginosa Antibiotic Resistance
4.4. P. aeruginosa and Fungal Interactions
4.4.1. P. aeruginosa Interacts with A. fumigatus
4.4.2. Impact on Antimicrobial Resistance
4.5. P. aeruginosa and C. albicans Interactions
Impact on Antimicrobial Resistance
4.6. Other Fungi
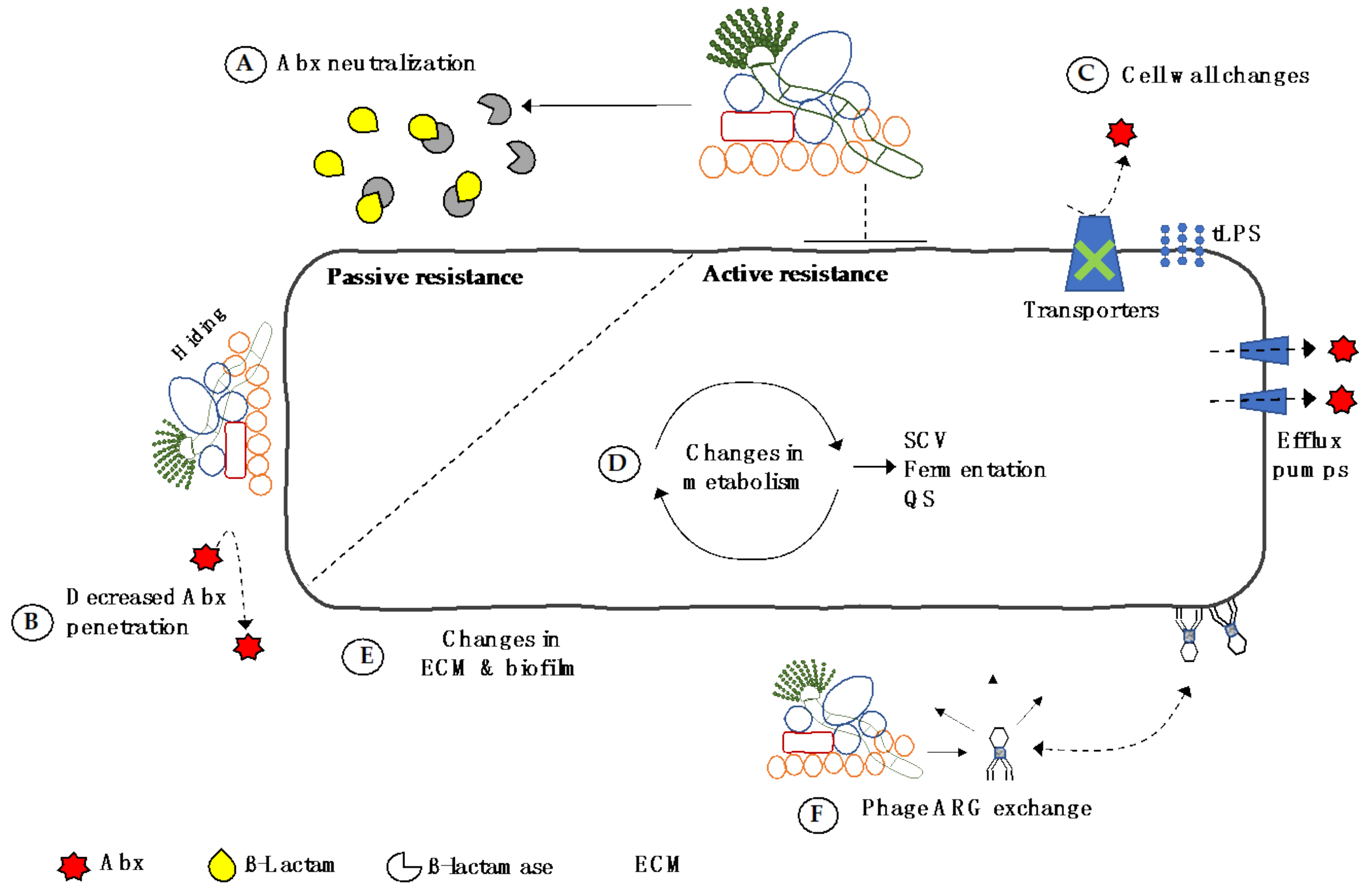
5. Direct Transfer of Resistance in the CF Airway Microbiome
6. Summary
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- CFF. Cystic Fibrosis Foundation 2018 Report; CFF: Bethesda, MD, USA, 2018. [Google Scholar]
- Rao, S.; Grigg, J. New insights into pulmonary inflammation in cystic fibrosis. Arch. Dis. Child. 2006, 91, 786–788. [Google Scholar] [CrossRef] [PubMed]
- Elborn, J.S.; Bell, S.C.; Madge, S.L.; Burgel, P.R.; Castellani, C.; Conway, S.; De Rijcke, K.; Dembski, B.; Drevinek, P.; Heijerman, H.G.; et al. Report of the European Respiratory Society/European Cystic Fibrosis Society task force on the care of adults with cystic fibrosis. Eur. Respir. J. 2019, 47, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Saint-Criq, V.; Gray, M.A. Role of CFTR in epithelial physiology. Cell Mol. Life Sci. 2017, 74, 93–115. [Google Scholar] [CrossRef] [PubMed]
- Bhagirath, A.Y.; Li, Y.; Somayajula, D.; Dadashi, M.; Badr, S.; Duan, K. Cystic fibrosis lung environment and Pseudomonas aeruginosa infection. BMC Pulm. Med. 2016, 16, 174. [Google Scholar] [CrossRef] [PubMed]
- Granchelli, A.M.; Adler, F.R.; Keogh, R.H.; Kartsonaki, C.; Cox, D.R.; Liou, T.G. Microbial interactions in the cystic fibrosis airway. J. Clin. Microbiol. 2018, 56, e00354-18. [Google Scholar] [CrossRef]
- Nixon, G.M.; Armstrong, D.S.; Carzino, R.; Carlin, J.B.; Olinsky, A.; Robertson, C.F.; Grimwood, K. Clinical outcome after early Pseudomonas aeruginosa infection in cystic fibrosis. J. Pediatrics 2001, 138, 699–704. [Google Scholar] [CrossRef]
- Emerson, J.; Rosenfeld, M.; McNamara, S.; Ramsey, B.; Gibson, R.L. Pseudomonas aeruginosa and other predictors of mortality and morbidity in young children with cystic fibrosis. Pediatric Pulmonol. 2002, 34, 91–100. [Google Scholar] [CrossRef]
- Kosorok, M.R.; Zeng, L.; West, S.E.; Rock, M.J.; Splaingard, M.L.; Laxova, A.; Green, C.G.; Collins, J.; Farrell, P.M. Acceleration of lung disease in children with cystic fibrosis after Pseudomonas aeruginosa acquisition. Pediatr. Pulmonol. 2001, 32, 277–287. [Google Scholar] [CrossRef]
- Li, Z.; Kosorok, M.R.; Farrell, P.M.; Laxova, A.; West, S.E.; Green, C.G.; Collins, J.; Rock, M.J.; Splaingard, M.L. Longitudinal development of mucoid Pseudomonas aeruginosa infection and lung disease progression in children with cystic fibrosis. JAMA 2005, 293, 581–588. [Google Scholar] [CrossRef]
- Smith, D.J.; Ramsay, K.A.; Yerkovich, S.T.; Reid, D.W.; Wainwright, C.E.; Grimwood, K.; Bell, S.C.; Kidd, T.J. Pseudomonas aeruginosa antibiotic resistance in Australian cystic fibrosis centres. Respirology 2016, 21, 329–337. [Google Scholar] [CrossRef]
- Lucca, F.; Guarnieri, M.; Ros, M.; Muffato, G.; Rigoli, R.; Da Dalt, L. Antibiotic resistance evolution of Pseudomonas aeruginosa in cystic fibrosis patients (2010–2013). Clin. Respir. J. 2018, 12, 2189–2196. [Google Scholar] [CrossRef] [PubMed]
- Lechtzin, N.; John, M.; Irizarry, R.; Merlo, C.; Diette, G.B.; Boyle, M.P. Outcomes of adults with cystic fibrosis infected with antibiotic-resistant Pseudomonas aeruginosa. Respiration 2006, 73, 27–33. [Google Scholar] [CrossRef]
- Stanojevic, S.; McDonald, A.; Waters, V.; MacDonald, S.; Horton, E.; Tullis, E.; Ratjen, F. Effect of pulmonary exacerbations treated with oral antibiotics on clinical outcomes in cystic fibrosis. Thorax 2017, 72, 327. [Google Scholar] [CrossRef]
- Hahn, A.; Burrell, A.; Fanous, H.; Chaney, H.; Sami, I.; Perez, G.F.; Koumbourlis, A.C.; Freishtat, R.J.; Crandall, K.A. Antibiotic multidrug resistance in the cystic fibrosis airway microbiome is associated with decreased diversity. Heliyon 2018, 4, e00795. [Google Scholar] [CrossRef]
- Reece, E.; Segurado, R.; Jackson, A.; McClean, S.; Renwick, J.; Greally, P. Co-colonisation with Aspergillus fumigatus and Pseudomonas aeruginosa is associated with poorer health in cystic fibrosis patients: An Irish registry analysis. BMC Pulm. Med. 2017, 17, 70. [Google Scholar] [CrossRef]
- Pallett, R.; Leslie, L.J.; Lambert, P.A.; Milic, I.; Devitt, A.; Marshall, L.J. Anaerobiosis influences virulence properties of Pseudomonas aeruginosa cystic fibrosis isolates and the interaction with Staphylococcus aureus. Sci. Rep. 2019, 9, 6748. [Google Scholar] [CrossRef]
- Pompilio, A.; Crocetta, V.; De Nicola, S.; Verginelli, F.; Fiscarelli, E.; Di Bonaventura, G. Cooperative pathogenicity in cystic fibrosis: Stenotrophomonas maltophilia modulates Pseudomonas aeruginosa virulence in mixed biofilm. Front. Microbiol. 2015, 6, 951. [Google Scholar] [CrossRef]
- Bragonzi, A.; Farulla, I.; Paroni, M.; Twomey, K.B.; Pirone, L.; Lorè, N.I.; Bianconi, I.; Dalmastri, C.; Ryan, R.P.; Bevivino, A. Modelling co-infection of the cystic fibrosis lung by Pseudomonas aeruginosa and Burkholderia cenocepacia reveals influences on biofilm formation and host response. PLoS ONE 2012, 7, e52330. [Google Scholar] [CrossRef]
- Hoffman, L.R.; Deziel, E.; D’Argenio, D.A.; Lepine, F.; Emerson, J.; McNamara, S.; Gibson, R.L.; Ramsey, B.W.; Miller, S.I. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2006, 103, 19890–19895. [Google Scholar] [CrossRef] [PubMed]
- Manavathu, E.K.; Vager, D.L.; Vazquez, J.A. Development and antimicrobial susceptibility studies of in vitro monomicrobial and polymicrobial biofilm models with Aspergillus fumigatus and Pseudomonas aeruginosa. BMC Microbiol. 2014, 14, 53. [Google Scholar] [CrossRef] [PubMed]
- Fourine, R.; Pohl, C.H. Beyond Antagonism: The Interaction Between Candida Species and Pseudomonas aeruginosa. J. Fungy 2019, 5, 34. [Google Scholar] [CrossRef]
- Briard, B.; Mislin, G.L.A.; Latge, J.P.; Beauvais, A. Interactions between Aspergillus fumigatus and Pulmonary Bacteria: Current State of the Field, New Data, and Future Perspective. J. Fungi 2019, 5. [Google Scholar] [CrossRef]
- Folkesson, A.; Jelsbak, L.; Yang, L.; Johansen, H.K.; Ciofu, O.; Høiby, N.; Molin, S. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: An evolutionary perspective. Nat. Rev. 2012, 10, 841–851. [Google Scholar] [CrossRef] [PubMed]
- Tariq, M.A.; Everest, F.L.C.; Cowley, L.A.; Wright, R.; Holt, G.S.; Ingram, H.; Duignan, L.A.M.; Nelson, A.; Lanyon, C.V.; Perry, A.; et al. Temperate Bacteriophages from Chronic Pseudomonas aeruginosa Lung Infections Show Disease-Specific Changes in Host Range and Modulate Antimicrobial Susceptibility. mSystems 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Vandeplassche, E.; Tavernier, S.; Coenye, T.; Crabbe, A. Influence of the lung microbiome on antibiotic susceptibility of cystic fibrosis pathogens. Eur. Respir. Rev. 2019, 28. [Google Scholar] [CrossRef]
- Briaud, P.; Camus, L.; Bastien, S.; Doleans-Jordheim, A.; Vandenesch, F.; Moreau, K. Coexistence with Pseudomonas aeruginosa alters Staphylococcus aureus transcriptome, antibiotic resistance and internalization into epithelial cells. Sci. Rep. 2019, 9, 16564. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin, T.; Yau, Y.C.W.; Stapleton, P.J.; Gong, Y.; Wang, P.W.; Guttman, D.S.; Waters, V. Staphylococcus aureus interaction with Pseudomonas aeruginosa biofilm enhances tobramycin resistance. NPJ Biofilms Microbiomes 2017, 3, 25. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.L.; Fiel, S.B.; Mayer-Hamblett, N.; Ramsey, B.; Burns, J.L. Susceptibility Testing of Pseudomonas aeruginosa Isolates and Clinical Response to Parenteral Antibiotic Administration: Lack of Association in Cystic Fibrosis. Chest 2003, 123, 1495–1502. [Google Scholar] [CrossRef]
- Hurley, M.N.; Ariff, A.H.A.; Bertenshaw, C.; Bhatt, J.; Smyth, A.R. Results of antibiotic susceptibility testing do not influence clinical outcome in children with cystic fibrosis. J. Cyst. Fibros. 2012, 11, 288–292. [Google Scholar] [CrossRef] [PubMed]
- Koch, C. Early infection and progression of cystic fibrosis lung disease. Pediatr. Pulmonol. 2002, 34, 232–236. [Google Scholar] [CrossRef]
- Tingpej, P.; Smith, L.; Rose, B.; Zhu, H.; Conibear, T.; Al Nassafi, K.; Manos, J.; Elkins, M.; Bye, P.; Willcox, M.; et al. Phenotypic characterization of clonal and nonclonal Pseudomonas aeruginosa strains isolated from lungs of adults with cystic fibrosis. J. Clin. Microbiol. 2007, 45, 1697–1704. [Google Scholar] [CrossRef]
- Pamukcu, A.; Bush, A.; Buchdahl, R. Effects of pseudomonas aeruginosa colonization on lung function and anthropometric variables in children with cystic fibrosis. Pediatr. Pulmonol. 1995, 19, 10–15. [Google Scholar] [CrossRef]
- Anantharajah, A.; Mingeot-Leclercq, M.P.; Van Bambeke, F. Targeting the Type Three Secretion System in Pseudomonas aeruginosa. Trends Pharm. Sci. 2016, 37, 734–749. [Google Scholar] [CrossRef]
- King, J.D.; Kocincova, D.; Westman, E.L.; Lam, J.S. Review: Lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate Immun. 2009, 15, 261–312. [Google Scholar] [CrossRef]
- Leid, J.G.; Willson, C.J.; Shirtliff, M.E.; Hassett, D.J.; Parsek, M.R.; Jeffers, A.K. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J. Immunol. 2005, 175, 7512–7518. [Google Scholar] [CrossRef] [PubMed]
- Bleves, S.; Viarre, V.; Salacha, R.; Michel, G.P.; Filloux, A.; Voulhoux, R. Protein secretion systems in Pseudomonas aeruginosa: A wealth of pathogenic weapons. Int. J. Med. Microbiol. 2010, 300, 534–543. [Google Scholar] [CrossRef]
- Zhao, K.; Li, W.; Li, J.; Ma, T.; Wang, K.; Yuan, Y.; Li, J.S.; Xie, R.; Huang, T.; Zhang, Y.; et al. TesG is a type I secretion effector of Pseudomonas aeruginosa that suppresses the host immune response during chronic infection. Nat. Microbiol. 2019, 4, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Hall, S.; McDermott, C.; Anoopkumar-Dukie, S.; McFarland, A.J.; Forbes, A.; Perkins, A.V.; Davey, A.K.; Chess-Williams, R.; Kiefel, M.J.; Arora, D.; et al. Cellular Effects of Pyocyanin, a Secreted Virulence Factor of Pseudomonas aeruginosa. Toxins 2016, 8, 236. [Google Scholar] [CrossRef]
- Cornelis, P.; Dingemans, J. Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front. Cell Infect. Microbiol. 2013, 3, 75. [Google Scholar] [CrossRef] [PubMed]
- Faure, E.; Mear, J.B.; Faure, K.; Normand, S.; Couturier-Maillard, A.; Grandjean, T.; Balloy, V.; Ryffel, B.; Dessein, R.; Chignard, M.; et al. Pseudomonas aeruginosa type-3 secretion system dampens host defense by exploiting the NLRC4-coupled inflammasome. Am. J. Respir. Crit. Care Med. 2014, 189, 799–811. [Google Scholar] [CrossRef]
- Rangel, S.M.; Logan, L.K.; Hauser, A.R. The ADP-ribosyltransferase domain of the effector protein ExoS inhibits phagocytosis of Pseudomonas aeruginosa during pneumonia. mBio 2014, 5. [Google Scholar] [CrossRef]
- Zhao, K.; Tseng, B.S.; Beckerman, B.; Jin, F.; Gibiansky, M.L.; Harrison, J.J.; Luijten, E.; Parsek, M.R.; Wong, G.C. Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature 2013, 497, 388–391. [Google Scholar] [CrossRef]
- Favre-Bonté, S.; Köhler, T.; Van Delden, C. Biofilm formation by Pseudomonas aeruginosa: Role of the C4-HSL cell-to-cell signal and inhibition by azithromycin. J. Antimicrob. Chemother. 2003, 52, 598–604. [Google Scholar] [CrossRef]
- Ryder, C.; Byrd, M.; Wozniak, D.J. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 2007, 10, 644–648. [Google Scholar] [CrossRef]
- Abdulhaq, N.; Nawaz, Z.; Zahoor, M.A.; Siddique, A.B. Association of biofilm formation with multi drug resistance in clinical isolates of Pseudomonas aeruginosa. EXCLI J. 2020, 19, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Orazi, G.; O’Toole, G.A. Pseudomonas aeruginosa Alters Staphylococcus aureus Sensitivity to Vancomycin in a Biofilm Model of Cystic Fibrosis Infection. mBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Karami, P.; Khaledi, A.; Mashoof, R.Y.; Yaghoobi, M.H.; Karami, M.; Dastan, D.; Alikhani, M.Y. The correlation between biofilm formation capability and antibiotic resistance pattern in Pseudomonas aeruginosa. Gene Rep. 2020, 18, 100561. [Google Scholar] [CrossRef]
- Schurr, M.J. Pseudomonas aeruginosa Alginate Benefits Staphylococcus aureus? J. Bacteriol. 2020. [Google Scholar] [CrossRef]
- McDaniel, M.S.; Schoeb, T.; Swords, W.E. Cooperativity between Stenotrophomonas maltophilia and Pseudomonas aeruginosa during polymicrobial airway infections. Infect. Immun. 2020. [Google Scholar] [CrossRef]
- Sousa, A.M.; Pereira, M.O. Pseudomonas aeruginosa Diversification during Infection Development in Cystic Fibrosis Lungs-A Review. Pathogens 2014, 3, 680–703. [Google Scholar] [CrossRef]
- Rasamiravaka, T.; Labtani, Q.; Duez, P.; El Jaziri, M. The formation of biofilms by Pseudomonas aeruginosa: A review of the natural and synthetic compounds interfering with control mechanisms. Biomed. Res. Int. 2015, 2015, 759348. [Google Scholar] [CrossRef] [PubMed]
- De Kievit, T.R.; Gillis, R.; Marx, S.; Brown, C.; Iglewski, B.H. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: Their role and expression patterns. Appl. Environ. Microbiol 2001, 67, 1865–1873. [Google Scholar] [CrossRef] [PubMed]
- Carty, N.L.; Layland, N.; Colmer-Hamood, J.A.; Calfee, M.W.; Pesci, E.C.; Hamood, A.N. PtxR modulates the expression of QS-controlled virulence factors in the Pseudomonas aeruginosa strain PAO1. Mol. Microbiol. 2006, 61, 782–794. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Li, X.H.; Kim, S.K.; Lee, J.H. Post-secretional activation of Protease IV by quorum sensing in Pseudomonas aeruginosa. Sci. Rep. 2017, 7, 4416. [Google Scholar] [CrossRef]
- Zulianello, L.; Canard, C.; Kohler, T.; Caille, D.; Lacroix, J.S.; Meda, P. Rhamnolipids are virulence factors that promote early infiltration of primary human airway epithelia by Pseudomonas aeruginosa. Infect. Immun. 2006, 74, 3134–3147. [Google Scholar] [CrossRef]
- Govan, J.R.; Deretic, V. Microbial pathogenesis in cystic fibrosis: Mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 1996, 60, 539–574. [Google Scholar] [CrossRef] [PubMed]
- Lujan, A.M.; Macia, M.D.; Yang, L.; Molin, S.; Oliver, A.; Smania, A.M. Evolution and adaptation in Pseudomonas aeruginosa biofilms driven by mismatch repair system-deficient mutators. PLoS ONE 2011, 6, e27842. [Google Scholar] [CrossRef]
- Palmer, K.L.; Aye, L.M.; Whiteley, M. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 2007, 189, 8079–8087. [Google Scholar] [CrossRef]
- Winstanley, C.; O’Brien, S.; Brockhurst, M.A. Pseudomonas aeruginosa Evolutionary Adaptation and Diversification in Cystic Fibrosis Chronic Lung Infections. Trends Microbiol. 2016, 24, 327–337. [Google Scholar] [CrossRef]
- Boles, B.R.; Singh, P.K. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc. Natl. Acad. Sci. USA 2008, 105, 12503–12508. [Google Scholar] [CrossRef]
- Marvig, R.L.; Sommer, L.M.; Jelsbak, L.; Molin, S.; Johansen, H.K. Evolutionary insight from whole-genome sequencing of Pseudomonas aeruginosa from cystic fibrosis patients. Future Microbiol. 2015, 10, 599–611. [Google Scholar] [CrossRef]
- D’Argenio, D.A.; Wu, M.; Hoffman, L.R.; Kulasekara, H.D.; Deziel, E.; Smith, E.E.; Nguyen, H.; Ernst, R.K.; Larson Freeman, T.J.; Spencer, D.H.; et al. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol. Microbiol. 2007, 64, 512–533. [Google Scholar] [CrossRef]
- Jorgensen, K.M.; Wassermann, T.; Johansen, H.K.; Christiansen, L.E.; Molin, S.; Hoiby, N.; Ciofu, O. Diversity of metabolic profiles of cystic fibrosis Pseudomonas aeruginosa during the early stages of lung infection. Microbiology 2015, 161, 1447–1462. [Google Scholar] [CrossRef]
- Mahenthiralingam, E.; Campbell, M.E.; Speert, D.P. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect. Immun. 1994, 62, 596–605. [Google Scholar] [CrossRef] [PubMed]
- Hentzer, M.; Teitzel, G.M.; Balzer, G.J.; Heydorn, A.; Molin, S.; Givskov, M.; Parsek, M.R. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol 2001, 183, 5395–5401. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef]
- Santi, I.; Manfredi, P.; Maffei, E.; Egli, A.; Jenal, U. Evolution of Antibiotic Tolerance Shapes Resistance Development in Chronic Pseudomonas aeruginosa Infections. mBio 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, S.; Fothergill, J.L. The role of multispecies social interactions in shaping Pseudomonas aeruginosa pathogenicity in the cystic fibrosis lung. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef]
- Armbruster, C.R.; Coenye, T.; Touqui, L.; Bomberger, J.M. Interplay between host-microbe and microbe-microbe interactions in cystic fibrosis. J. Cyst. Fibros. 2019. [Google Scholar] [CrossRef]
- Bittar, F.; Richet, H.; Dubus, J.C.; Reynaud-Gaubert, M.; Stremler, N.; Sarles, J.; Raoult, D.; Rolain, J.M. Molecular detection of multiple emerging pathogens in sputa from cystic fibrosis patients. PLoS ONE 2008, 3, e2908. [Google Scholar] [CrossRef]
- Rogers, G.B.; Hart, C.A.; Mason, J.R.; Hughes, M.; Walshaw, M.J.; Bruce, K.D. Bacterial diversity in cases of lung infection in cystic fibrosis patients: 16S ribosomal DNA (rDNA) length heterogeneity PCR and 16S rDNA terminal restriction fragment length polymorphism profiling. J. Clin. Microbiol. 2003, 41, 3548–3558. [Google Scholar] [CrossRef]
- Zemanick, E.T.; Wagner, B.D.; Robertson, C.E.; Ahrens, R.C.; Chmiel, J.F.; Clancy, J.P.; Gibson, R.L.; Harris, W.T.; Kurland, G.; Laguna, T.A.; et al. Airway microbiota across age and disease spectrum in cystic fibrosis. Eur. Respir. J. 2017, 50. [Google Scholar] [CrossRef]
- Gangell, C.; Gard, S.; Douglas, T.; Park, J.; de Klerk, N.; Keil, T.; Brennan, S.; Ranganathan, S.; Robins-Browne, R.; Sly, P.D.; et al. Inflammatory Responses to Individual Microorganisms in the Lungs of Children With Cystic Fibrosis. Clin. Infect. Dis. 2011, 53, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.S.; Pope, C.E.; Marsh, R.L.; Qin, X.; McNamara, S.; Gibson, R.; Burns, J.L.; Deutsch, G.; Hoffman, L.R. Directly sampling the lung of a young child with cystic fibrosis reveals diverse microbiota. Ann. Am. Thorac. Soc. 2014, 11, 1049–1055. [Google Scholar] [CrossRef] [PubMed]
- Coburn, B.; Wang, P.W.; Diaz Caballero, J.; Clark, S.T.; Brahma, V.; Donaldson, S.; Zhang, Y.; Surendra, A.; Gong, Y.; Elizabeth Tullis, D.; et al. Lung microbiota across age and disease stage in cystic fibrosis. Sci. Rep. 2015. [Google Scholar] [CrossRef]
- Cox, M.J.; Allgaier, M.; Taylor, B.; Baek, M.S.; Huang, Y.J.; Daly, R.A.; Karaoz, U.; Andersen, G.L.; Brown, R.; Fujimura, K.E.; et al. Airway Microbiota and Pathogen Abundance in Age-Stratified Cystic Fibrosis Patients. PLoS ONE 2010, 5, e11044. [Google Scholar] [CrossRef]
- Carmody, L.A.; Zhao, J.; Schloss, P.D.; Petrosino, J.F.; Murray, S.; Young, V.B.; Li, J.Z.; LiPuma, J.J. Changes in cystic fibrosis airway microbiota at pulmonary exacerbation. Ann. Am. Thorac. Soc. 2013, 10, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Flight, W.G.; Smith, A.; Paisey, C.; Marchesi, J.R.; Bull, M.J.; Norville, P.J.; Mutton, K.J.; Webb, A.K.; Bright-Thomas, R.J.; Jones, A.M.; et al. Rapid Detection of Emerging Pathogens and Loss of Microbial Diversity Associated with Severe Lung Disease in Cystic Fibrosis. J. Clin. Microbiol. 2015, 53, 2022–2029. [Google Scholar] [CrossRef] [PubMed]
- Tunney, M.M.; Field, T.R.; Moriarty, T.F.; Patrick, S.; Doering, G.; Muhlebach, M.S.; Wolfgang, M.C.; Boucher, R.; Gilpin, D.F.; McDowell, A.; et al. Detection of Anaerobic Bacteria in High Numbers in Sputum from Patients with Cystic Fibrosis. Am. J. Respir. Crit. Care Med. 2008, 177, 995–1001. [Google Scholar] [CrossRef] [PubMed]
- Muhlebach, M.S.; Hatch, J.E.; Einarsson, G.G.; McGrath, S.J.; Gilipin, D.F.; Lavelle, G.; Mirkovic, B.; Murray, M.A.; McNally, P.; Gotman, N.; et al. Anaerobic bacteria cultured from cystic fibrosis airways correlate to milder disease: A multisite study. Eur. Respir. J. 2018, 52, 1800242. [Google Scholar] [CrossRef]
- Chmiel, J.F.; Aksamit, T.R.; Chotirmall, S.H.; Dasenbrook, E.C.; Elborn, J.S.; LiPuma, J.J.; Ranganathan, S.C.; Waters, V.J.; Ratjen, F.A. Antibiotic management of lung infections in cystic fibrosis. II. Nontuberculous mycobacteria, anaerobic bacteria, and fungi. Ann. Am. Thorac. Soc. 2014, 11, 1298–1306. [Google Scholar] [CrossRef]
- Lamoureux, C.; Guilloux, C.A.; Beauruelle, C.; Jolivet-Gougeon, A.; Héry-Arnaud, G. Anaerobes in cystic fibrosis patients’ airways. Crit. Rev. Microbiol. 2019, 45, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Guss, A.M.; Roeselers, G.; Newton, I.L.; Young, C.R.; Klepac-Ceraj, V.; Lory, S.; Cavanaugh, C.M. Phylogenetic and metabolic diversity of bacteria associated with cystic fibrosis. ISME J. 2011, 5, 20–29. [Google Scholar] [CrossRef]
- Linnane, B.; Walsh, A.M.; Walsh, C.J.; Crispie, F.; O’Sullivan, O.; Cotter, P.D.; McDermott, M.; Renwick, J.; McNally, P. The Lung Microbiome in Young Children with Cystic Fibrosis: A Prospective Cohort Study. Microorganisms 2021, 9. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.M.; Niccum, D.; Dunitz, J.M.; Hunter, R.C. Evidence and Role for Bacterial Mucin Degradation in Cystic Fibrosis Airway Disease. PLoS Pathog. 2016, 12, e1005846. [Google Scholar] [CrossRef]
- Ghorbani, P.; Santhakumar, P.; Hu, Q.; Djiadeu, P.; Wolever, T.M.; Palaniyar, N.; Grasemann, H. Short-chain fatty acids affect cystic fibrosis airway inflammation and bacterial growth. Eur. Respir. J. 2015, 46, 1033–1045. [Google Scholar] [CrossRef] [PubMed]
- Mirkovic, B.; Murray, M.A.; Lavelle, G.M.; Molloy, K.; Azim, A.A.; Gunaratnam, C.; Healy, F.; Slattery, D.; McNally, P.; Hatch, J.; et al. The Role of Short-Chain Fatty Acids, Produced by Anaerobic Bacteria, in the Cystic Fibrosis Airway. Am. J. Respir. Crit. Care Med. 2015, 192, 1314–1324. [Google Scholar] [CrossRef] [PubMed]
- Carmody, L.A.; Caverly, L.J.; Foster, B.K.; Rogers, M.A.M.; Kalikin, L.M.; Simon, R.H.; VanDevanter, D.R.; LiPuma, J.J. Fluctuations in airway bacterial communities associated with clinical states and disease stages in cystic fibrosis. PLoS ONE 2018, 13, e0194060. [Google Scholar] [CrossRef]
- Fodor, A.A.; Klem, E.R.; Gilpin, D.F.; Elborn, J.S.; Boucher, R.C.; Tunney, M.M.; Wolfgang, M.C. The adult cystic fibrosis airway microbiota is stable over time and infection type, and highly resilient to antibiotic treatment of exacerbations. PLoS ONE 2012, 7, e45001. [Google Scholar] [CrossRef]
- Cuthbertson, L.; Rogers, G.B.; Walker, A.W.; Oliver, A.; Green, L.E.; Daniels, T.W.; Carroll, M.P.; Parkhill, J.; Bruce, K.D.; van der Gast, C.J. Respiratory microbiota resistance and resilience to pulmonary exacerbation and subsequent antimicrobial intervention. ISME J. 2016, 10, 1081–1091. [Google Scholar] [CrossRef]
- Acosta, N.; Whelan, F.J.; Somayaji, R.; Poonja, A.; Surette, M.G.; Rabin, H.R.; Parkins, M.D. The Evolving Cystic Fibrosis Microbiome: A Comparative Cohort Study Spanning 16 Years. Ann. Am. Thorac. Soc. 2017, 14, 1288–1297. [Google Scholar] [CrossRef]
- Delhaes, L.; Monchy, S.; Frealle, E.; Hubans, C.; Salleron, J.; Leroy, S.; Prevotat, A.; Wallet, F.; Wallaert, B.; Dei-Cas, E.; et al. The airway microbiota in cystic fibrosis: A complex fungal and bacterial community--implications for therapeutic management. PLoS ONE 2012, 7, e36313. [Google Scholar] [CrossRef]
- Stressmann, F.A.; Rogers, G.B.; van der Gast, C.J.; Marsh, P.; Vermeer, L.S.; Carroll, M.P.; Hoffman, L.; Daniels, T.W.; Patel, N.; Forbes, B.; et al. Long-term cultivation-independent microbial diversity analysis demonstrates that bacterial communities infecting the adult cystic fibrosis lung show stability and resilience. Thorax 2012, 67, 867–873. [Google Scholar] [CrossRef] [PubMed]
- Cuthbertson, L.; Walker, A.W.; Oliver, A.E.; Rogers, G.B.; Rivett, D.W.; Hampton, T.H.; Ashare, A.; Elborn, J.S.; De Soyza, A.; Carroll, M.P.; et al. Lung function and microbiota diversity in cystic fibrosis. Microbiome 2020, 8, 45. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Schloss, P.D.; Kalikin, L.M.; Carmody, L.A.; Foster, B.K.; Petrosino, J.F.; Cavalcoli, J.D.; VanDevanter, D.R.; Murray, S.; Li, J.Z.; et al. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc. Natl. Acad. Sci. USA 2012, 109, 5809–5814. [Google Scholar] [CrossRef]
- Bacci, G.; Taccetti, G.; Dolce, D.; Armanini, F.; Segata, N.; Di Cesare, F.; Lucidi, V.; Fiscarelli, E.; Morelli, P.; Casciaro, R.; et al. Untargeted Metagenomic Investigation of the Airway Microbiome of Cystic Fibrosis Patients with Moderate-Severe Lung Disease. Microorganisms 2020, 8. [Google Scholar] [CrossRef]
- Cystic Fibrosis Trust UK. Antibiotic Treatment for Cystic Fibrosis—Recommendations; Cystic Fibrosis Trust: London, UK, 2009. [Google Scholar]
- Lahiri, T.; Hempstead, S.E.; Brady, C.; Cannon, C.L.; Clark, K.; Condren, M.E.; Guill, M.F.; Guillerman, R.P.; Leone, C.G.; Maguiness, K.; et al. Clinical Practice Guidelines From the Cystic Fibrosis Foundation for Preschoolers With Cystic Fibrosis. Pediatrics 2016, 137. [Google Scholar] [CrossRef]
- Pittman, J.E.; Wylie, K.M.; Akers, K.; Storch, G.A.; Hatch, J.; Quante, J.; Frayman, K.B.; Clarke, N.; Davis, M.; Stick, S.M.; et al. Association of Antibiotics, Airway Microbiome, and Inflammation in Infants with Cystic Fibrosis. Ann. Am. Thorac. Soc. 2017, 14, 1548–1555. [Google Scholar] [CrossRef]
- Stutman, H.R.; Lieberman, J.M.; Nussbaum, E.; Marks, M.I. Antibiotic prophylaxis in infants and young children with cystic fibrosis: A randomized controlled trial. J. Pediatr. 2002, 140, 299–305. [Google Scholar] [CrossRef]
- Smyth, A.; Walters, S. Prophylactic antibiotics for cystic fibrosis. Cochrane Database Syst. Rev. 2003, CD001912. [Google Scholar] [CrossRef]
- McCaffery, K.; Olver, R.E.; Franklin, M.; Mukhopadhyay, S. Systematic review of antistaphylococcal antibiotic therapy in cystic fibrosis. Thorax 1999, 54, 380–383. [Google Scholar] [CrossRef] [PubMed]
- Daniels, T.W.; Rogers, G.B.; Stressmann, F.A.; van der Gast, C.J.; Bruce, K.D.; Jones, G.R.; Connett, G.J.; Legg, J.P.; Carroll, M.P. Impact of antibiotic treatment for pulmonary exacerbations on bacterial diversity in cystic fibrosis. J. Cyst. Fibros. 2013, 12, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Heirali, A.A.; Workentine, M.L.; Acosta, N.; Poonja, A.; Storey, D.G.; Somayaji, R.; Rabin, H.R.; Whelan, F.J.; Surette, M.G.; Parkins, M.D. The effects of inhaled aztreonam on the cystic fibrosis lung microbiome. Microbiome 2017, 5, 51. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.J.; Badrick, A.C.; Zakrzewski, M.; Krause, L.; Bell, S.C.; Anderson, G.J.; Reid, D.W. Pyrosequencing reveals transient cystic fibrosis lung microbiome changes with intravenous antibiotics. Eur. Respir. J. 2014, 44, 922–930. [Google Scholar] [CrossRef] [PubMed]
- Medina, G.; Juárez, K.; Díaz, R.; Soberón-Chávez, G. Transcriptional regulation of Pseudomonas aeruginosa rhlR, encoding a quorum-sensing regulatory protein. Microbiology 2003, 149, 3073–3081. [Google Scholar] [CrossRef] [PubMed]
- Dekimpe, V.; Déziel, E. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: The transcriptional regulator RhlR regulates LasR-specific factors. Microbiology 2009, 155, 712–723. [Google Scholar] [CrossRef]
- Zhao, J.; Cheng, W.; He, X.; Liu, Y. The co-colonization prevalence of Pseudomonas aeruginosa and Aspergillus fumigatus in cystic fibrosis: A systematic review and meta-analysis. Microb. Pathog. 2018, 125, 122–128. [Google Scholar] [CrossRef]
- Fischer, A.J.; Singh, S.B.; LaMarche, M.M.; Maakestad, L.J.; Kienenberger, Z.E.; Peña, T.A.; Stoltz, D.A.; Limoli, D.H. Sustained Coinfections with Staphylococcus aureus and Pseudomonas aeruginosa in Cystic Fibrosis. Am. J. Respir. Crit. Care Med. 2020. [Google Scholar] [CrossRef]
- Ryan, R.P.; Fouhy, Y.; Garcia, B.F.; Watt, S.A.; Niehaus, K.; Yang, L.; Tolker-Nielsen, T.; Dow, J.M. Interspecies signalling via the Stenotrophomonas maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa. Mol. Microbiol. 2008, 68, 75–86. [Google Scholar] [CrossRef]
- Tognon, M.; Kohler, T.; Gdaniec, B.G.; Hao, Y.; Lam, J.S.; Beaume, M.; Luscher, A.; Buckling, A.; van Delden, C. Co-evolution with Staphylococcus aureus leads to lipopolysaccharide alterations in Pseudomonas aeruginosa. ISME J. 2017, 11, 2233–2243. [Google Scholar] [CrossRef]
- Magalhães, A.P.; Azevedo, N.F.; Pereira, M.O.; Lopes, S.P. The cystic fibrosis microbiome in an ecological perspective and its impact in antibiotic therapy. Appl. Microbiol. Biotechnol. 2016, 100, 1163–1181. [Google Scholar] [CrossRef]
- Korgaonkar, A.; Trivedi, U.; Rumbaugh, K.P.; Whiteley, M. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc. Natl. Acad. Sci. USA 2013, 110, 1059–1064. [Google Scholar] [CrossRef] [PubMed]
- Korgaonkar, A.K.; Whiteley, M. Pseudomonas aeruginosa enhances production of an antimicrobial in response to N-acetylglucosamine and peptidoglycan. J. Bacteriol. 2011, 193, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Amin, R.; Dupuis, A.; Aaron, S.D.; Ratjen, F. The effect of chronic infection with Aspergillus fumigatus on lung function and hospitalization in patients with cystic fibrosis. Chest 2010, 137, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Pernet, E.; Guillemot, L.; Burgel, P.R.; Martin, C.; Lambeau, G.; Sermet-Gaudelus, I.; Sands, D.; Leduc, D.; Morand, P.C.; Jeammet, L.; et al. Pseudomonas aeruginosa eradicates Staphylococcus aureus by manipulating the host immunity. Nat. Commun. 2014, 5, 5105. [Google Scholar] [CrossRef] [PubMed]
- Chew, S.C.; Kundukad, B.; Seviour, T.; van der Maarel, J.R.; Yang, L.; Rice, S.A.; Doyle, P.; Kjelleberg, S. Dynamic remodeling of microbial biofilms by functionally distinct exopolysaccharides. mBio 2014, 5, e01536-14. [Google Scholar] [CrossRef]
- Kessler, E.; Safrin, M.; Olson, J.C.; Ohman, D.E. Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J. Biol. Chem. 1993, 268, 7503–7508. [Google Scholar] [CrossRef]
- Mashburn, L.M.; Jett, A.M.; Akins, D.R.; Whiteley, M. Staphylococcus aureus Serves as an Iron Source for Pseudomonas aeruginosa during In Vivo Coculture. J. Bacteriol. 2004, 187, 554–556. [Google Scholar] [CrossRef]
- Wainwright, C.E.; France, M.W.; O’Rourke, P.; Anuj, S.; Kidd, T.J.; Nissen, M.D.; Sloots, T.P.; Coulter, C.; Ristovski, Z.; Hargreaves, M.; et al. Cough-generated aerosols of Pseudomonas aeruginosa and other Gram-negative bacteria from patients with cystic fibrosis. Thorax 2009, 64, 926–931. [Google Scholar] [CrossRef]
- Scott, J.E.; Li, K.; Filkins, L.M.; Zhu, B.; Kuchma, S.L.; Schwartzman, J.D.; O’Toole, G.A. Pseudomonas aeruginosa Can Inhibit Growth of Streptococcal Species via Siderophore Production. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef]
- Tavernier, S.; Crabbe, A.; Hacioglu, M.; Stuer, L.; Henry, S.; Rigole, P.; Dhondt, I.; Coenye, T. Community Composition Determines Activity of Antibiotics against Multispecies Biofilms. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Scoffield, J.A.; Duan, D.; Zhu, F.; Wu, H. A commensal streptococcus hijacks a Pseudomonas aeruginosa exopolysaccharide to promote biofilm formation. PLoS Pathog. 2017, 13, e1006300. [Google Scholar] [CrossRef] [PubMed]
- Price, K.E.; Naimie, A.A.; Griffin, E.F.; Bay, C.; O’Toole, G.A. Tobramycin-Treated Pseudomonas aeruginosa PA14 Enhances Streptococcus constellatus 7155 Biofilm Formation in a Cystic Fibrosis Model System. J. Bacteriol. 2016, 198, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Scoffield, J.A.; Wu, H. Nitrite reductase is critical for Pseudomonas aeruginosa survival during co-infection with the oral commensal Streptococcus parasanguinis. Microbiology 2016, 162, 376–383. [Google Scholar] [CrossRef]
- Malesevic, M.; Stanisavljevic, N.; Novovic, K.; Polovic, N.; Vasiljevic, Z.; Kojic, M.; Jovcic, B. Burkholderia cepacia YtnP and Y2-aiiA lactonases inhibit virulence of Pseudomonas aeruginosa via quorum quenching activity. Microb. Pathog. 2020, 149, 104561. [Google Scholar] [CrossRef] [PubMed]
- McKenney, D.; Brown, K.E.; Allison, D.G. Influence of Pseudomonas aeruginosa exoproducts on virulence factor production in Burkholderia cepacia: Evidence of interspecies communication. J. Bacteriol. 1995, 177, 6989–6992. [Google Scholar] [CrossRef]
- Schwab, U.; Abdullah, L.H.; Perlmutt, O.S.; Albert, D.; Davis, C.W.; Arnold, R.R.; Yankaskas, J.R.; Gilligan, P.; Neubauer, H.; Randell, S.H.; et al. Localization of Burkholderia cepacia complex bacteria in cystic fibrosis lungs and interactions with Pseudomonas aeruginosa in hypoxic mucus. Infect. Immun. 2014, 82, 4729–4745. [Google Scholar] [CrossRef]
- Bakkal, S.; Robinson, S.M.; Ordonez, C.L.; Waltz, D.A.; Riley, M.A. Role of bacteriocins in mediating interactions of bacterial isolates taken from cystic fibrosis patients. Microbiology 2010, 156, 2058–2067. [Google Scholar] [CrossRef][Green Version]
- Briard, B.; Heddergott, C.; Latge, J.P. Volatile Compounds Emitted by Pseudomonas aeruginosa Stimulate Growth of the Fungal Pathogen Aspergillus fumigatus. mBio 2016, 7, e00219. [Google Scholar] [CrossRef]
- Shirazi, F.; Ferreira, J.A.; Stevens, D.A.; Clemons, K.V.; Kontoyiannis, D.P. Biofilm Filtrates of Pseudomonas aeruginosa Strains Isolated from Cystic Fibrosis Patients Inhibit Preformed Aspergillus fumigatus Biofilms via Apoptosis. PLoS ONE 2016, 11, e0150155. [Google Scholar] [CrossRef]
- Smith, K.; Rajendran, R.; Kerr, S.; Lappin, D.F.; Mackay, W.G.; Williams, C.; Ramage, G. Aspergillus fumigatus enhances elastase production in Pseudomonas aeruginosa co-cultures. Med. Mycol. 2015, 53, 645–655. [Google Scholar] [CrossRef]
- Reece, E.; Doyle, S.; Greally, P.; Renwick, J.; McClean, S. Aspergillus fumigatus Inhibits Pseudomonas aeruginosa in Co-culture: Implications of a Mutually Antagonistic Relationship on Virulence and Inflammation in the CF Airway. Front. Microbiol. 2018, 9, 1205. [Google Scholar] [CrossRef]
- Briard, B.; Rasoldier, V.; Bomme, P.; ElAouad, N.; Guerreiro, C.; Chassagne, P.; Muszkieta, L.; Latge, J.P.; Mulard, L.; Beauvais, A. Dirhamnolipids secreted from Pseudomonas aeruginosa modify anjpegungal susceptibility of Aspergillus fumigatus by inhibiting beta1,3 glucan synthase activity. ISME J. 2017, 11, 1578–1591. [Google Scholar] [CrossRef]
- Briard, B.; Bomme, P.; Lechner, B.E.; Mislin, G.L.A.; Lair, V.; Prévost, M.-C.; Latgé, J.-P.; Haas, H.; Beauvais, A. Pseudomonas aeruginosa manipulates redox and iron homeostasis of its microbiota partner Aspergillus fumigatus via phenazines. Sci. Rep. 2015, 5, 8220. [Google Scholar] [CrossRef]
- Phelan, V.V.; Moree, W.J.; Aguilar, J.; Cornett, D.S.; Koumoutsi, A.; Noble, S.M.; Pogliano, K.; Guerrero, C.A.; Dorrestein, P.C. Impact of a transposon insertion in phzF2 on the specialized metabolite production and interkingdom interactions of Pseudomonas aeruginosa. J. Bacteriol. 2014, 196, 1683–1693. [Google Scholar] [CrossRef] [PubMed]
- Sass, G.; Nazik, H.; Penner, J.; Shah, H.; Ansari, S.R.; Clemons, K.V.; Groleau, M.C.; Dietl, A.M.; Visca, P.; Haas, H.; et al. Studies of Pseudomonas aeruginosa Mutants Indicate Pyoverdine as the Central Factor in Inhibition of Aspergillus fumigatus Biofilm. J. Bacteriol. 2018, 200. [Google Scholar] [CrossRef]
- Anand, R.; Moss, R.B.; Sass, G.; Banaei, N.; Clemons, K.V.; Martinez, M.; Stevens, D.A. Small Colony Variants of Pseudomonas aeruginosa Display Heterogeneity in Inhibiting Aspergillus fumigatus Biofilm. Mycopathologia 2018, 183, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Moree, W.J.; Phelan, V.V.; Wu, C.H.; Bandeira, N.; Cornett, D.S.; Duggan, B.M.; Dorrestein, P.C. Interkingdom metabolic transformations captured by microbial imaging mass spectrometry. Proc. Natl. Acad. Sci. USA 2012, 109, 13811–13816. [Google Scholar] [CrossRef] [PubMed]
- Mowat, E.; Rajendran, R.; Williams, C.; McCulloch, E.; Jones, B.; Lang, S.; Ramage, G. Pseudomonas aeruginosa and their small diffusible extracellular molecules inhibit Aspergillus fumigatus biofilm formation. FEMS Microbiol. Lett 2010, 313, 96–102. [Google Scholar] [CrossRef]
- Reen, F.J.; Phelan, J.P.; Woods, D.F.; Shanahan, R.; Cano, R.; Clarke, S.; McGlacken, G.P.; O’Gara, F. Harnessing Bacterial Signals for Suppression of Biofilm Formation in the Nosocomial Fungal Pathogen Aspergillus fumigatus. Front. Microbiol. 2016, 7, 2074. [Google Scholar] [CrossRef]
- Bandara, H.M.; Cheung, B.P.; Watt, R.M.; Jin, L.J.; Samaranayake, L.P. Pseudomonas aeruginosa lipopolysaccharide inhibits Candida albicans hyphae formation and alters gene expression during biofilm development. Mol. Oral Microbiol. 2013, 28, 54–69. [Google Scholar] [CrossRef]
- McAlester, G.; O’Gara, F.; Morrissey, J.P. Signal-mediated interactions between Pseudomonas aeruginosa and Candida albicans. J. Med. Microbiol. 2008, 57, 563–569. [Google Scholar] [CrossRef]
- Reen, F.J.; Mooij, M.J.; Holcombe, L.J.; McSweeney, C.M.; McGlacken, G.P.; Morrissey, J.P.; O’Gara, F. The Pseudomonas quinolone signal (PQS), and its precursor HHQ, modulate interspecies and interkingdom behaviour. FEMS Microbiol. Ecol. 2011, 77, 413–428. [Google Scholar] [CrossRef] [PubMed]
- Cugini, C.; Calfee, M.W.; Farrow, J.M., 3rd; Morales, D.K.; Pesci, E.C.; Hogan, D.A. Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Mol. Microbiol. 2007, 65, 896–906. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Rhman, S.H.; El-Mahdy, A.M.; El-Mowafy, M. Effect of Tyrosol and Farnesol on Virulence and Antibiotic Resistance of Clinical Isolates of Pseudomonas aeruginosa. Biomed. Res. Int. 2015, 2015, 456463. [Google Scholar] [CrossRef] [PubMed]
- Goss, C.H.; Muhlebach, M.S. Review: Staphylococcus aureus and MRSA in cystic fibrosis. J. Cyst. Fibros. 2011, 10, 298–306. [Google Scholar] [CrossRef] [PubMed]
- CFF. Cystic Fibrosis Foundation Patient Registry; CFRI: Bethesda, MD, USA, 2016. [Google Scholar]
- Cystic Fibrosis Registry Ireland. CFRI Annual Data Report 2016; CFI Cystic Fibrosis Registry of Ireland: Dublin, Irleand, 2016. [Google Scholar]
- Limoli, D.; Yang, J.; Khansaheb, M.; Helfman, B.; Peng, L.; Stecenko, A.; Goldberg, J. Staphylococcus aureus and Pseudomonas aeruginosa co-infection is associated with cystic fibrosis-related diabetes and poor clinical outcomes. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35. [Google Scholar] [CrossRef]
- Limoli, D.H.; Hoffman, L.R. hinder, hide and harm: What can we learn from the interactions between Pseudomonas aeruginosa and Staphylococcus aureus during respiratory infections? Thorax 2019, 74, 684–692. [Google Scholar] [CrossRef]
- Diaz De Rienzo, M.A.; Stevenson, P.S.; Marchant, R.; Banat, I.M. Effect of biosurfactants on Pseudomonas aeruginosa and Staphylococcus aureus biofilms in a BioFlux channel. Appl. Microbiol. Biotechnol. 2016, 100, 5773–5779. [Google Scholar] [CrossRef] [PubMed]
- Radlinski, L.; Rowe, S.E.; Kartchner, L.B.; Maile, R.; Cairns, B.A.; Vitko, N.P.; Gode, C.J.; Lachiewicz, A.M.; Wolfgang, M.C.; Conlon, B.P. Pseudomonas aeruginosa exoproducts determine antibiotic efficacy against Staphylococcus aureus. PLoS Biol. 2017, 15, e2003981. [Google Scholar] [CrossRef]
- DeLeon, S.; Clinton, A.; Fowler, H.; Everett, J.; Horswill, A.R.; Rumbaugh, K.P. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect. Immun. 2014, 82, 4718–4728. [Google Scholar] [CrossRef]
- Bernardy, E.E.; Petit, R.A., 3rd; Raghuram, V.; Alexander, A.M.; Read, T.D.; Goldberg, J.B. Genotypic and Phenotypic Diversity of Staphylococcus aureus Isolates from Cystic Fibrosis Patient Lung Infections and Their Interactions with Pseudomonas aeruginosa. mBio 2020, 11. [Google Scholar] [CrossRef]
- Michelsen, C.F.; Christensen, A.M.; Bojer, M.S.; Hoiby, N.; Ingmer, H.; Jelsbak, L. Staphylococcus aureus alters growth activity, autolysis, and antibiotic tolerance in a human host-adapted Pseudomonas aeruginosa lineage. J. Bacteriol. 2014, 196, 3903–3911. [Google Scholar] [CrossRef] [PubMed]
- Frydenlund Michelsen, C.; Hossein Khademi, S.M.; Krogh Johansen, H.; Ingmer, H.; Dorrestein, P.C.; Jelsbak, L. Evolution of metabolic divergence in Pseudomonas aeruginosa during long-term infection facilitates a proto-cooperative interspecies interaction. ISME J. 2016, 10, 1323–1336. [Google Scholar] [CrossRef]
- Filkins, L.M.; Graber, J.A.; Olson, D.G.; Dolben, E.L.; Lynd, L.R.; Bhuju, S.; O’Toole, G.A. Coculture of Staphylococcus aureus with Pseudomonas aeruginosa Drives S. aureus towards Fermentative Metabolism and Reduced Viability in a Cystic Fibrosis Model. J. Bacteriol. 2015, 197, 2252–2264. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, G.; Seguin, D.L.; Asselin, A.E.; Deziel, E.; Cantin, A.M.; Frost, E.H.; Michaud, S.; Malouin, F. Staphylococcus aureus sigma B-dependent emergence of small-colony variants and biofilm production following exposure to Pseudomonas aeruginosa 4-hydroxy-2-heptylquinoline-N-oxide. BMC Microbiol. 2010, 10, 33. [Google Scholar] [CrossRef] [PubMed]
- Fugere, A.; Lalonde Seguin, D.; Mitchell, G.; Deziel, E.; Dekimpe, V.; Cantin, A.M.; Frost, E.; Malouin, F. Interspecific small molecule interactions between clinical isolates of Pseudomonas aeruginosa and Staphylococcus aureus from adult cystic fibrosis patients. PLoS ONE 2014, 9, e86705. [Google Scholar] [CrossRef] [PubMed]
- Trizna, E.Y.; Yarullina, M.N.; Baidamshina, D.R.; Mironova, A.V.; Akhatova, F.S.; Rozhina, E.V.; Fakhrullin, R.F.; Khabibrakhmanova, A.M.; Kurbangalieva, A.R.; Bogachev, M.I.; et al. Bidirectional alterations in antibiotics susceptibility in Staphylococcus aureus-Pseudomonas aeruginosa dual-species biofilm. Sci. Rep. 2020, 10, 14849. [Google Scholar] [CrossRef]
- Orazi, G.; Ruoff, K.L.; O’Toole, G.A. Pseudomonas aeruginosa Increases the Sensitivity of Biofilm-Grown Staphylococcus aureus to Membrane-Targeting Antiseptics and Antibiotics. mBio 2019, 10. [Google Scholar] [CrossRef]
- Proctor, R.A.; von Eiff, C.; Kahl, B.C.; Becker, K.; McNamara, P.; Herrmann, M.; Peters, G. Small colony variants: A pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 2006, 4, 295–305. [Google Scholar] [CrossRef]
- Biswas, L.; Biswas, R.; Schlag, M.; Bertram, R.; Gotz, F. Small-colony variant selection as a survival strategy for Staphylococcus aureus in the presence of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2009, 75, 6910–6912. [Google Scholar] [CrossRef]
- Sadeqi Nezhad, M.; Pordeli, H.; Ghasemi, N.; Ahani, A. Evaluation of multidrug resistance patterns in siderophore-producing Pseudomonas aeruginosa from clinical and environmental samples in Gorgan, Iran. New Microbes New Infect. 2018, 24, 38–41. [Google Scholar] [CrossRef]
- Soberon-Chavez, G.; Lepine, F.; Deziel, E. Production of rhamnolipids by Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2005, 68, 718–725. [Google Scholar] [CrossRef]
- Kataoka, D.; Fujiwara, H.; Kawakami, T.; Tanaka, Y.; Tanimoto, A.; Ikawa, S.; Tanaka, Y. The indirect pathogenicity of Stenotrophomonas maltophilia. Int. J. Antimicrob. Agents 2003, 22, 601–606. [Google Scholar] [CrossRef]
- Sherrard, L.J.; McGrath, S.J.; McIlreavey, L.; Hatch, J.; Wolfgang, M.C.; Muhlebach, M.S.; Gilpin, D.F.; Elborn, J.S.; Tunney, M.M. Production of extended-spectrum beta-lactamases and the potential indirect pathogenic role of Prevotella isolates from the cystic fibrosis respiratory microbiota. Int. J. Antimicrob. Agents 2016, 47, 140–145. [Google Scholar] [CrossRef]
- Margalit, A.; Carolan, J.C.; Sheehan, D.; Kavanagh, K. The Aspergillus fumigatus Secretome Alters the Proteome of Pseudomonas aeruginosa to Stimulate Bacterial Growth: Implications for Co-infection. Mol. Cell Proteom. 2020, 19, 1346–1359. [Google Scholar] [CrossRef]
- Phuengmaung, P.; Somparn, P.; Panpetch, W.; Singkham-In, U.; Wannigama, D.L.; Chatsuwan, T.; Leelahavanichkul, A. Coexistence of Pseudomonas aeruginosa With Candida albicans Enhances Biofilm Thickness Through Alginate-Related Extracellular Matrix but Is Attenuated by N-acetyl-l-cysteine. Front. Cell Infect. Microbiol. 2020, 10, 594336. [Google Scholar] [CrossRef] [PubMed]
- Alam, F.; Catlow, D.; Di Maio, A.; Blair, J.M.A.; Hall, R.A. Candida albicans enhances meropenem tolerance of Pseudomonas aeruginosa in a dual-species biofilm. J. Antimicrob. Chemother. 2020, 75, 925–935. [Google Scholar] [CrossRef] [PubMed]
- Waters, V.; Atenafu, E.G.; Lu, A.; Yau, Y.; Tullis, E.; Ratjen, F. Chronic Stenotrophomonas maltophilia infection and mortality or lung transplantation in cystic fibrosis patients. J. Cyst. Fibros. 2013, 12, 482–486. [Google Scholar] [CrossRef] [PubMed]
- ECFS Patient Registry. ECFS Patient Registry Annual Data Report. 2018. [Google Scholar]
- Spicuzza, L.; Sciuto, C.; Vitaliti, G.; Di Dio, G.; Leonardi, S.; La Rosa, M. Emerging pathogens in cystic fibrosis: Ten years of follow-up in a cohort of patients. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 191–195. [Google Scholar] [CrossRef]
- Blau, H.; Linnane, B.; Carzino, R.; Tannenbaum, E.L.; Skoric, B.; Robinson, P.J.; Robertson, C.; Ranganathan, S.C. Induced sputum compared to bronchoalveolar lavage in young, non-expectorating cystic fibrosis children. J. Cyst. Fibros. 2014, 13, 106–110. [Google Scholar] [CrossRef]
- Zemanick, E.T.; Wagner, B.D.; Robertson, C.E.; Stevens, M.J.; Szefler, S.J.; Accurso, F.J.; Sagel, S.D.; Harris, J.K. Assessment of airway microbiota and inflammation in cystic fibrosis using multiple sampling methods. Ann. Am. Thorac. Soc. 2015, 12, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Nas, M.Y.; White, R.C.; DuMont, A.L.; Lopez, A.E.; Cianciotto, N.P. Stenotrophomonas maltophilia Encodes a VirB/VirD4 Type IV Secretion System That Modulates Apoptosis in Human Cells and Promotes Competition against Heterologous Bacteria, Including Pseudomonas aeruginosa. Infect. Immun. 2019, 87. [Google Scholar] [CrossRef] [PubMed]
- Lamoureux, C.; Guilloux, C.A.; Beauruelle, C.; Gouriou, S.; Ramel, S.; Dirou, A.; Le Bihan, J.; Revert, K.; Ropars, T.; Lagrafeuille, R.; et al. An observational study of anaerobic bacteria in cystic fibrosis lung using culture dependant and independent approaches. Sci. Rep. 2021, 11, 6845. [Google Scholar] [CrossRef]
- Field, T.R.; Sibley, C.D.; Parkins, M.D.; Rabin, H.R.; Surette, M.G. The genus Prevotella in cystic fibrosis airways. Anaerobe 2010, 16, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Sherrard, L.J.; Graham, K.A.; McGrath, S.J.; McIlreavey, L.; Hatch, J.; Muhlebach, M.S.; Wolfgang, M.C.; Gilpin, D.F.; Elborn, J.S.; Schneiders, T.; et al. Antibiotic resistance in Prevotella species isolated from patients with cystic fibrosis. J. Antimicrob. Chemother. 2013, 68, 2369–2374. [Google Scholar] [CrossRef] [PubMed]
- Scoffield, J.A.; Wu, H. Oral streptococci and nitrite-mediated interference of Pseudomonas aeruginosa. Infect. Immun. 2015, 83, 101–107. [Google Scholar] [CrossRef]
- Vandeplassche, E.; Sass, A.; Ostyn, L.; Burmolle, M.; Kragh, K.N.; Bjarnsholt, T.; Coenye, T.; Crabbe, A. Antibiotic susceptibility of cystic fibrosis lung microbiome members in a multispecies biofilm. Biofilm 2020, 2, 100031. [Google Scholar] [CrossRef]
- Bakare, N.; Rickerts, V.; Bargon, J.; Just-Nübling, G. Prevalence of Aspergillus fumigatus and other fungal species in the sputum of adult patients with cystic fibrosis. Mycoses 2003, 46, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Valenza, G.; Tappe, D.; Turnwald, D.; Frosch, M.; Konig, C.; Hebestreit, H.; Abele-Horn, M. Prevalence and antimicrobial susceptibility of microorganisms isolated from sputa of patients with cystic fibrosis. J. Cyst. Fibros. 2008, 7, 123–127. [Google Scholar] [CrossRef] [PubMed]
- Ziesing, S.; Suerbaum, S.; Sedlacek, L. Fungal epidemiology and diversity in cystic fibrosis patients over a 5-year period in a national reference center. Med. Mycol. 2016, 54, 781–786. [Google Scholar] [CrossRef]
- Coughlan, C.A.; Chotirmall, S.H.; Renwick, J.; Hassan, T.; Low, T.B.; Bergsson, G.; Eshwika, A.; Bennett, K.; Dunne, K.; Greene, C.M.; et al. The effect of Aspergillus fumigatus infection on vitamin D receptor expression in cystic fibrosis. Am. J. Respir. Crit. Care Med. 2012, 186, 999–1007. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Meng, J.; Jia, M.; Ma, X.; He, G.; Yu, J.; Wang, R.; Bai, H.; Hou, Z.; Luo, X. oprM as a new target for reversion of multidrug resistance in Pseudomonas aeruginosa by antisense phosphorothioate oligodeoxynucleotides. FEMS Immunol. Med. Microbiol. 2010, 60, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Horna, G.; Lopez, M.; Guerra, H.; Saenz, Y.; Ruiz, J. Interplay between MexAB-OprM and MexEF-OprN in clinical isolates of Pseudomonas aeruginosa. Sci. Rep. 2018, 8, 16463. [Google Scholar] [CrossRef]
- Gupta, N.; Haque, A.; Mukhopadhyay, G.; Narayan, R.P.; Prasad, R. Interactions between bacteria and Candida in the burn wound. Burns 2005, 31, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Kaleli, I.; Cevahir, N.; Demir, M.; Yildirim, U.; Sahin, R. Anticandidal activity of Pseudomonas aeruginosa strains isolated from clinical specimens. Mycoses 2007, 50, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Hogan, D.A.; Vik, A.; Kolter, R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol. Microbiol. 2004, 54, 1212–1223. [Google Scholar] [CrossRef]
- Hogan, D.A.; Kolter, R. Pseudomonas-Candida interactions: An ecological role for virulence factors. Science 2002, 296, 2229–2232. [Google Scholar] [CrossRef]
- Homa, M.; Sandor, A.; Toth, E.; Szebenyi, C.; Nagy, G.; Vagvolgyi, C.; Papp, T. In vitro Interactions of Pseudomonas aeruginosa With Scedosporium Species Frequently Associated With Cystic Fibrosis. Front. Microbiol. 2019, 10, 441. [Google Scholar] [CrossRef]
- Chen, S.C.; Patel, S.; Meyer, W.; Chapman, B.; Yu, H.; Byth, K.; Middleton, P.G.; Nevalainen, H.; Sorrell, T.C. Pseudomonas aeruginosa Inhibits the Growth of Scedosporium and Lomentospora In Vitro. Mycopathologia 2018, 183, 251–261. [Google Scholar] [CrossRef]
- Kaur, J.; Pethani, B.P.; Kumar, S.; Kim, M.; Sunna, A.; Kautto, L.; Penesyan, A.; Paulsen, I.T.; Nevalainen, H. Pseudomonas aeruginosa inhibits the growth of Scedosporium aurantiacum, an opportunistic fungal pathogen isolated from the lungs of cystic fibrosis patients. Front. Microbiol. 2015, 6, 866. [Google Scholar] [CrossRef]
- Mac Aogain, M.; Lau, K.J.X.; Cai, Z.; Kumar Narayana, J.; Purbojati, R.W.; Drautz-Moses, D.I.; Gaultier, N.E.; Jaggi, T.K.; Tiew, P.Y.; Ong, T.H.; et al. Metagenomics Reveals a Core Macrolide Resistome Related to Microbiota in Chronic Respiratory Disease. Am. J. Respir. Crit. Care Med. 2020, 202, 433–447. [Google Scholar] [CrossRef] [PubMed]
- Bacci, G.; Mengoni, A.; Fiscarelli, E.; Segata, N.; Taccetti, G.; Dolce, D.; Paganin, P.; Morelli, P.; Tuccio, V.; De Alessandri, A.; et al. A Different Microbiome Gene Repertoire in the Airways of Cystic Fibrosis Patients with Severe Lung Disease. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Willner, D.; Furlan, M.; Haynes, M.; Schmieder, R.; Angly, F.E.; Silva, J.; Tammadoni, S.; Nosrat, B.; Conrad, D.; Rohwer, F. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS ONE 2009, 4, e7370. [Google Scholar] [CrossRef]
- Fothergill, J.L.; Mowat, E.; Ledson, M.J.; Walshaw, M.J.; Winstanley, C. Fluctuations in phenotypes and genotypes within populations of Pseudomonas aeruginosa in the cystic fibrosis lung during pulmonary exacerbations. J. Med. Microbiol. 2010, 59, 472–481. [Google Scholar] [CrossRef]
- Winstanley, C.; Langille, M.G.; Fothergill, J.L.; Kukavica-Ibrulj, I.; Paradis-Bleau, C.; Sanschagrin, F.; Thomson, N.R.; Winsor, G.L.; Quail, M.A.; Lennard, N.; et al. Newly introduced genomic prophage islands are critical determinants of in vivo competitiveness in the Liverpool Epidemic Strain of Pseudomonas aeruginosa. Genome Res. 2009, 19, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Rolain, J.M.; Francois, P.; Hernandez, D.; Bittar, F.; Richet, H.; Fournous, G.; Mattenberger, Y.; Bosdure, E.; Stremler, N.; Dubus, J.C.; et al. Genomic analysis of an emerging multiresistant Staphylococcus aureus strain rapidly spreading in cystic fibrosis patients revealed the presence of an antibiotic inducible bacteriophage. Biol. Direct 2009, 4, 1. [Google Scholar] [CrossRef]
- Allemann, A.; Kraemer, J.G.; Korten, I.; Ramsey, K.; Casaulta, C.; Group, S.S.; Wuthrich, D.; Ramette, A.; Endimiani, A.; Latzin, P.; et al. Nasal Resistome Development in Infants With Cystic Fibrosis in the First Year of Life. Front. Microbiol. 2019, 10, 212. [Google Scholar] [CrossRef]
- Elphick, H.E.; Scott, A. Single versus combination intravenous anti-pseudomonal antibiotic therapy for people with cystic fibrosis. Cochrane Database Syst. Rev. 2016, 12, CD002007. [Google Scholar] [CrossRef]
- Boulanger, S.; Mitchell, G.; Bouarab, K.; Marsault, E.; Cantin, A.; Frost, E.H.; Deziel, E.; Malouin, F. Bactericidal Effect of Tomatidine-Tobramycin Combination against Methicillin-Resistant Staphylococcus aureus and Pseudomonas aeruginosa Is Enhanced by Interspecific Small-Molecule Interactions. Antimicrob. Agents Chemother. 2015, 59, 7458–7464. [Google Scholar] [CrossRef]
- Aoki, T.; Yoshizawa, H.; Yamawaki, K.; Yokoo, K.; Sato, J.; Hisakawa, S.; Hasegawa, Y.; Kusano, H.; Sano, M.; Sugimoto, H.; et al. Cefiderocol (S-649266), A new siderophore cephalosporin exhibiting potent activities against Pseudomonas aeruginosa and other gram-negative pathogens including multi-drug resistant bacteria: Structure activity relationship. Eur. J. Med. Chem. 2018, 155, 847–868. [Google Scholar] [CrossRef] [PubMed]
- Saisho, Y.; Katsube, T.; White, S.; Fukase, H.; Shimada, J. Pharmacokinetics, Safety, and Tolerability of Cefiderocol, a Novel Siderophore Cephalosporin for Gram-Negative Bacteria, in Healthy Subjects. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [PubMed]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; Gonzalez, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef] [PubMed]
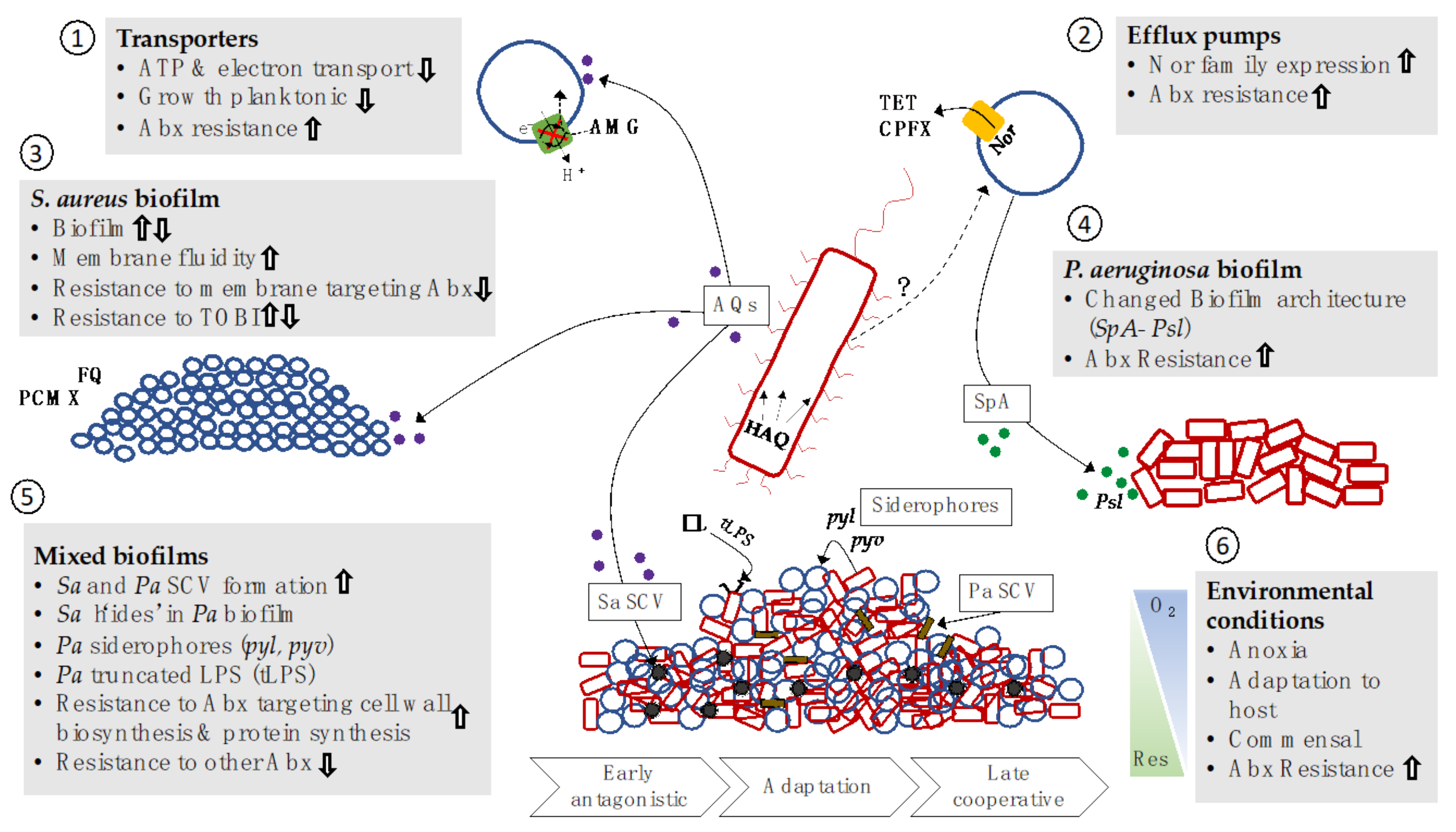
| Sp Interacting with PA | Microbial/Host Response | Potential Implications on Disease | Ref(s). |
|---|---|---|---|
| Gram-positives | ↑ lytic activity by PA ↓ Gram+ in vivo models | PA more toxic in co-infections with Gram+ | [114] |
| ↑ pyocyanin production by PA | PA mechanisms of dominance | [115] | |
| S. aureus | Co-infection strains less competitive than mono-infection strains | Adaptation to coexistence in the lung | [116] |
| PA induces bronchial epithelial cells to produce phospholipase, sPLA2-IIA | Manipulation of host immune response, enhanced survival of PA, and killing of SA and other Gram+ | [117] | |
| PA EPS can affect mixed species biofilm architecture | Proximity of SA and PA in mixed biofilms | [118] | |
| ↑ PA siderophore production Lysis of SA | Iron competition LasA protease | [112] [119,120] | |
| PA LPS inactivation mutations ↓ production of PA LPS in anoxia | Reduced recognition by immune system, persistence Immune evasion | [112] [112] | |
| ↑ PA swimming motility in anoxia | Reseeding of infection in lung | [17] | |
| S. maltophilia | Co-colonise the CF airway | Opportunity to interact | [121] |
| ↓ SM growth ↑ PA biofilm | Altered virulence and persistence | [18] [111] | |
| ↓ Adhesion of PA to CFBE | Evasion of immune system and persistence | [18] | |
| Streptococci spp. | ↓ SMG growth—PA competition for iron ↓ S. anginosus growth and biofilm formation ↑ Strep spp. biofilm formation—hijacks PA EPS ↓ PA viability—Strep H2O2 production | Pathogen dominance and persistence Altered virulence and persistence Persistence Strep beneficial to host | [122] [123] [124,125] [126] |
| Burkholderia cepacia complex | ↓ PA virulence factors | Altered virulence | [127] |
| PA enhances Bcc virulence | Altered virulence | [128] | |
| ↓ PA growth in vivo | Altered persistence | [129] | |
| Reduced growth of Bcc and PA | Competition beneficial to host? | [130] | |
| Co-infection ↑ inflammatory markers | Increased host inflammation | [19] | |
| A. fumigatus | Co-colonise the CF airway | Opportunity to interact in airway | [131] |
| Co-colonised patients—↓ lung function, ↑ hospitalisations, exacerbations and Abx usage | Poorer disease outcome | [16] | |
| PA SNs stimulate AF growth | Increased AF abundance in co-infections * | [131] | |
| Metacaspases from Pa SNs inhibit and damage AF biofilms | Reduced AF abundance in co-infections | [132] | |
| ↑ elastase production by PA in presence of AF | More damaging pathology | [133] | |
| SNs from co-cultures more toxic to epithelial cells lines | More damaging pathology | [133] | |
| Mutually antagonistic | Competition beneficial to host? | [134] | |
| Gliotoxin produced by AF reduces PA biofilm | Competition beneficial to host? | [134] | |
| Co-infections cause altered inflammatory response | Evasion of the immune system and persistence | [134] | |
| PA dirhamnolipids induce AF ECM production | Inhibits AF growth and facilitates PA binding | [135] | |
| PA phenazines inhibit AF growth by direct contact | Reduced AF abundance in co-infections | [136] | |
| Subinhibitory concentrations of PA phenazines can promote AF growth | Increased AF abundance in co-infections * | [136] | |
| Iron competition | Reduced AF abundance in co-infections | [137,138] | |
| Development of PA SCVs | Reduced AF abundance in co-infections | [139] | |
| Expression of QS molecules | Reduced AF abundance in co-infections | [135,140,141,142] | |
| C. albicans | PA expressed LPS inhibits CA biofilm formation and hyphal development | Reduced CA abundance and virulence in co-infections | [143] |
| PA QS molecule, 3-oxo-C12HSL | Reduced CA abundance in co-infections | [144] | |
| PA 2-heptyl-3-hydroxyl-4-quinolone | Reduced CA abundance in co-infections | [145] | |
| CA secreted Farnesol reduces PA pyocyanin Farnesol inhibits PA haemolysin Farnesol inhibits PA swarming motility | PA less virulent | [146] | |
| PA less virulent | [147] | ||
| PA less virulent | [144] | ||
| CA secreted tyrosol inhibits PA haemolysin and protease production | PA less virulent | [147] |
| Species Interacting | Resistance (Increased/Decreased) | Antibiotic | Mechanism | Ref/s. |
|---|---|---|---|---|
| S. aureus (+PA) | Increased | Aminoglycoside (gentamicin), tetracycline |
| [155] |
| Tetracycline and fluoroquinolone (ciprofloxacin) |
| [27] | ||
| Aminoglycosides |
| [20,160] | ||
| Vancomycin, ampicillin, and ceftriaxone |
| [162] | ||
| Glycopeptide (vancomycin) |
| [47] | ||
| Aminoglycoside (tobramycin) |
| [158] | ||
| MDR tolerance |
| [154] | ||
| Decreased | Aminoglycoside (tobramycin), glycopeptide (vancomycin) |
| [154] | |
| Fluoroquinolones, membrane targeting antimicrobials, antiseptics |
| [163] | ||
| P. aeruginosa (+SA) | Unchanged | Aminoglycoside (gentamicin), tetracycline |
| [155] |
| Increased | Aminoglycoside (Inhaled tobramycin) |
| [28] | |
| β-lactams |
| [112] | ||
| Aminoglycoside (tobramycin) |
| [157] | ||
| S. maltophilia (+PA) | Increased | Aminoglycoside (tobramycin) |
| [18] |
| P. aeruginosa (+SM) | Increased | B-lactams (imipenem) Cephalosporin (ceftazidime) |
| [168] |
| Increased | Polymyxins |
| [111] | |
| Prevotella (+PA) | Increased | β-lactams (ceftazidime) |
| [169] |
| P. aeruginosa (+AF) | Increased | NA |
| [170] |
| Increased | Cephalosporin (Cefepime) |
| [21] | |
| P. aeruginosa (+CA) | Increased | NA |
| [171] |
| Increased | β-lactams (Meropenem) |
| [172] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reece, E.; Bettio, P.H.d.A.; Renwick, J. Polymicrobial Interactions in the Cystic Fibrosis Airway Microbiome Impact the Antimicrobial Susceptibility of Pseudomonas aeruginosa. Antibiotics 2021, 10, 827. https://doi.org/10.3390/antibiotics10070827
Reece E, Bettio PHdA, Renwick J. Polymicrobial Interactions in the Cystic Fibrosis Airway Microbiome Impact the Antimicrobial Susceptibility of Pseudomonas aeruginosa. Antibiotics. 2021; 10(7):827. https://doi.org/10.3390/antibiotics10070827
Chicago/Turabian StyleReece, Emma, Pedro H. de Almeida Bettio, and Julie Renwick. 2021. "Polymicrobial Interactions in the Cystic Fibrosis Airway Microbiome Impact the Antimicrobial Susceptibility of Pseudomonas aeruginosa" Antibiotics 10, no. 7: 827. https://doi.org/10.3390/antibiotics10070827
APA StyleReece, E., Bettio, P. H. d. A., & Renwick, J. (2021). Polymicrobial Interactions in the Cystic Fibrosis Airway Microbiome Impact the Antimicrobial Susceptibility of Pseudomonas aeruginosa. Antibiotics, 10(7), 827. https://doi.org/10.3390/antibiotics10070827






