Strawberry and Ginger Silver Nanoparticles as Potential Inhibitors for SARS-CoV-2 Assisted by In Silico Modeling and Metabolic Profiling
Abstract
1. Introduction
2. Material and Methods
2.1. Plant Material
2.1.1. Strawberry (Fragaria ananassa Duch.)
2.1.2. Ginger (Zingiber officinale)
2.2. Chemicals
2.3. Extraction Procedure
2.4. Synthesis of AgNPs
2.4.1. Synthesis of Strawberry AgNPs Using a Methanolic Extract
2.4.2. Synthesis of Ginger AgNPs Using Methanolic Extract
2.4.3. Metabolic Profiling of Strawberry and Ginger Methanolic Extracts
2.5. Antiviral Activity
2.6. Characterization of the Synthesized AgNPs by TEM
2.7. Zeta Potential Characterization of the Synthesized AgNPs of Strawberry and Ginger Methanolic Extracts
2.8. Molecular Docking
2.9. Molecular Dynamic Simulations
3. Results and Discussion
3.1. TEM Characterization of the Synthesized AgNPs
3.2. UV-Visible Characterization of the Synthesized AgNPs of Strawberry and Ginger Methanolic Extracts
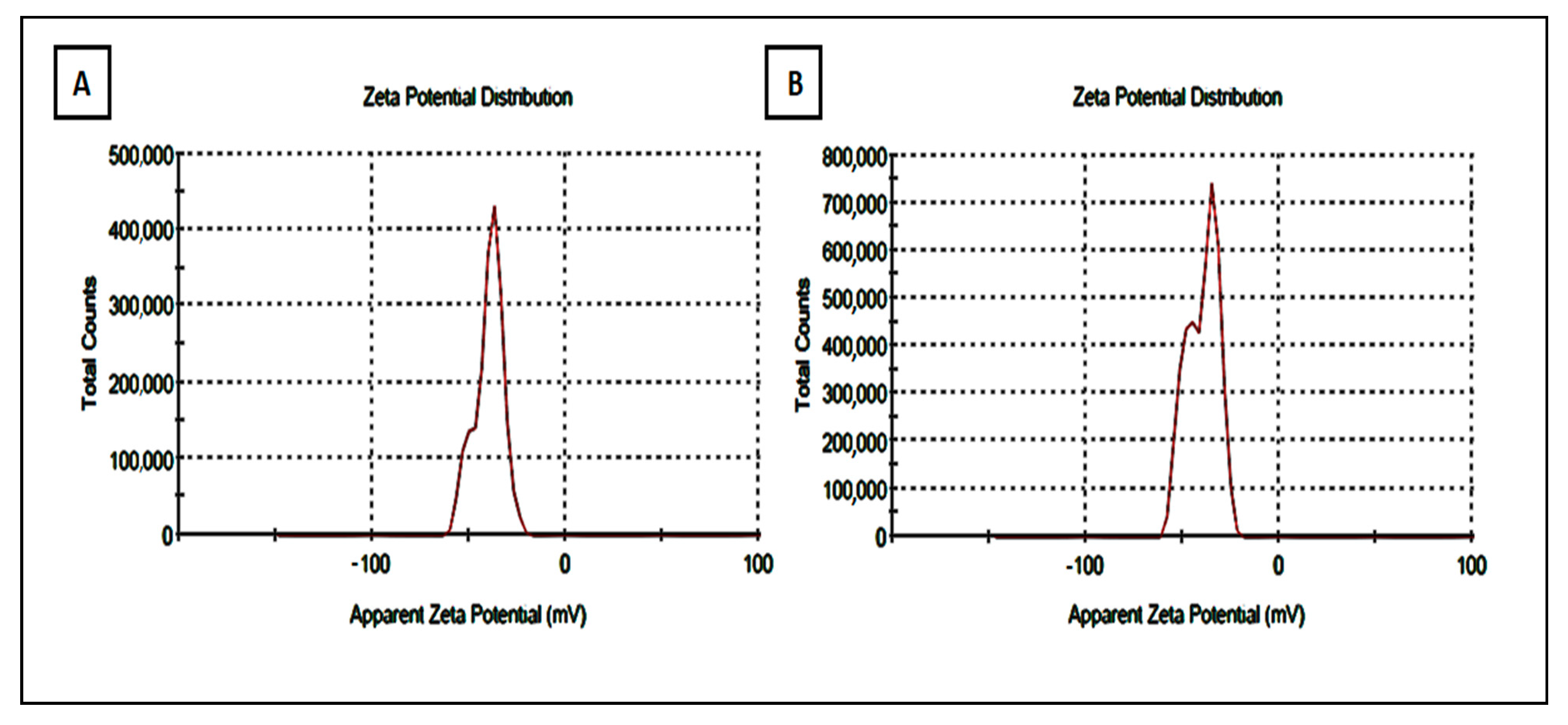
3.3. Determination of Zeta Potential of Strawberry and Ginger Methanolic Extract AgNPs
3.4. Metabolomic Profiling of the Crude Methanolic Extracts of Strawberry and Ginger
3.5. Anti-SARS-CoV-2 Activity
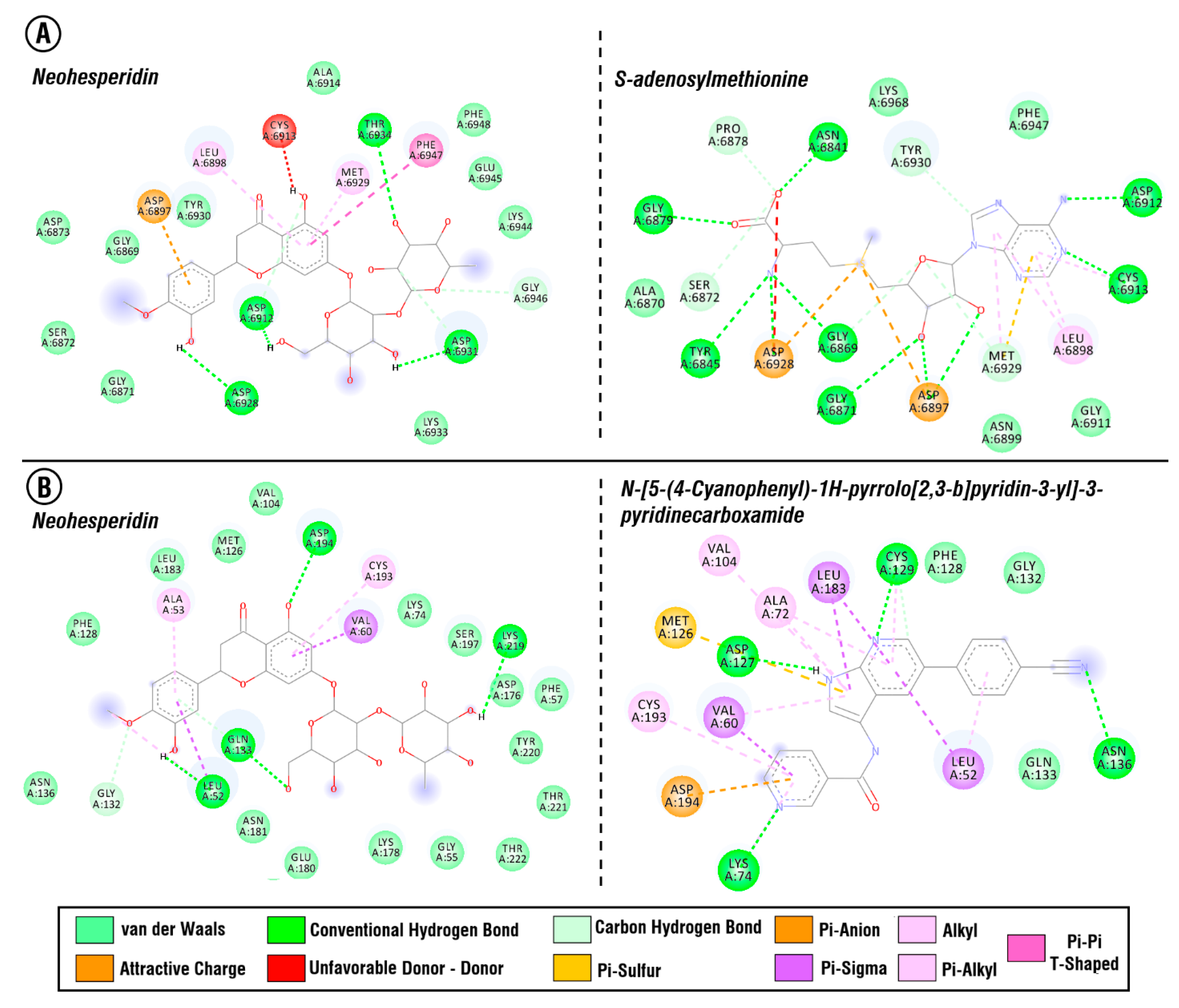
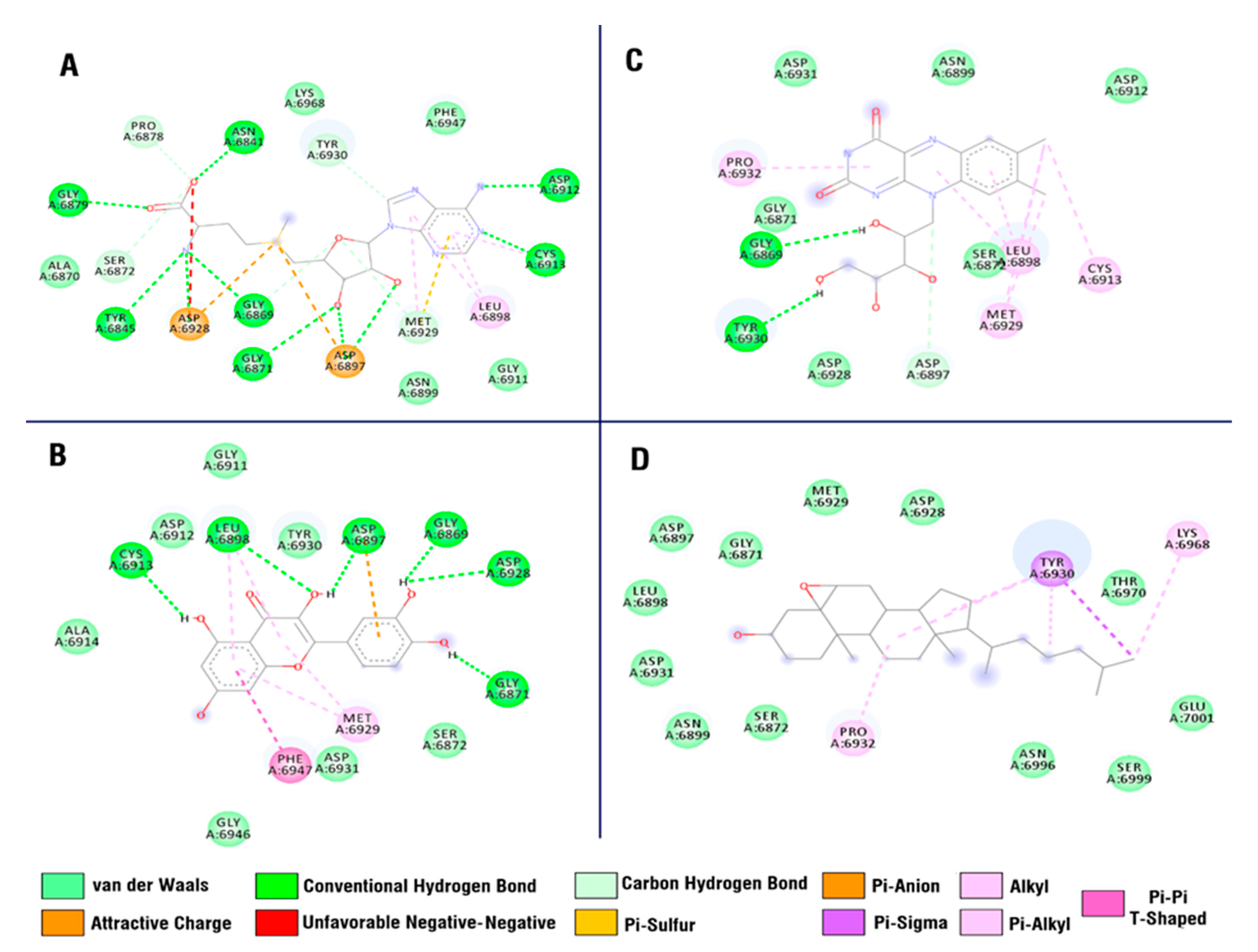
3.6. Molecular Docking Study
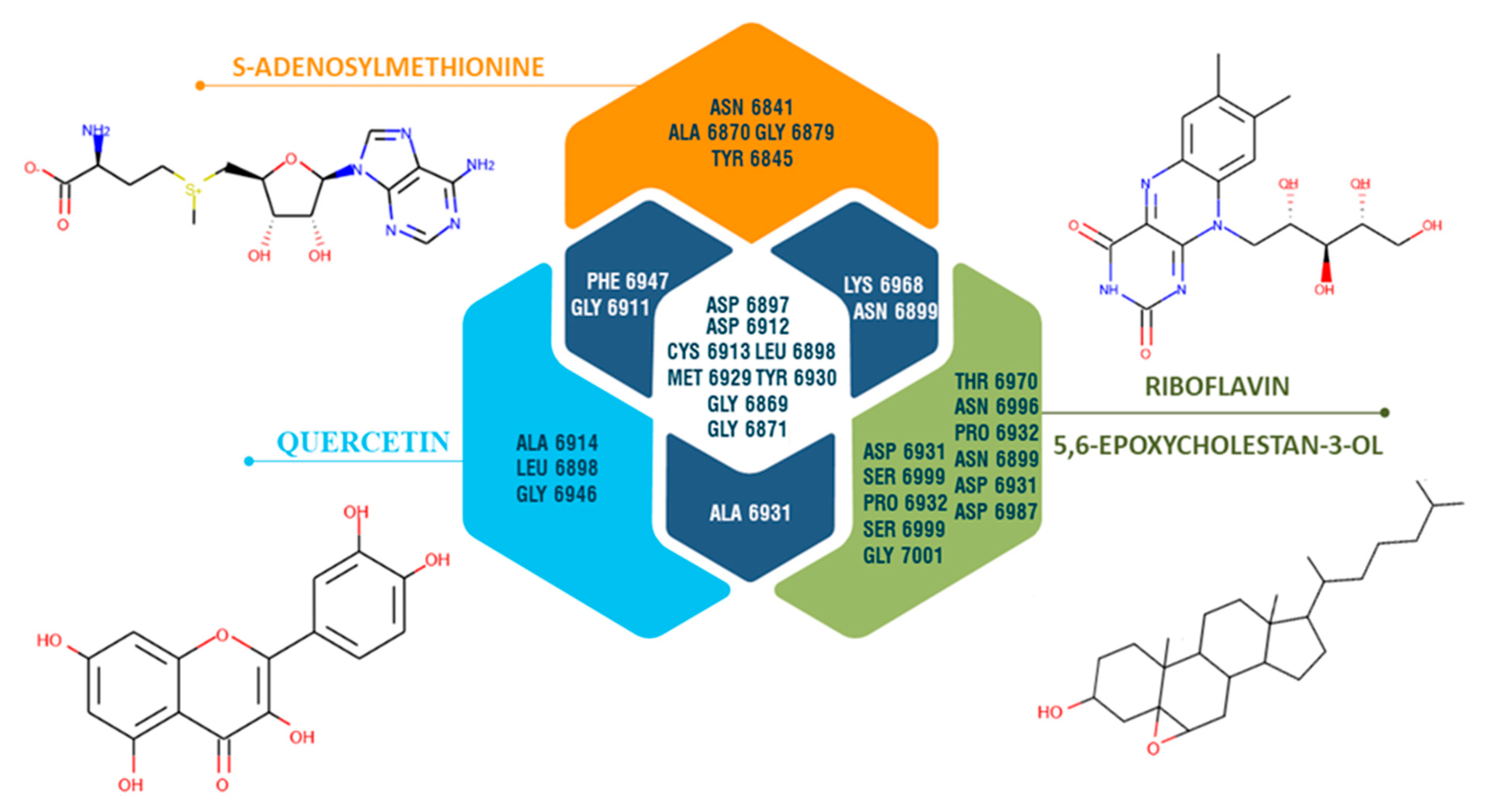
3.7. Molecular Simulation Study
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Wang, D.; Huang, J.; Yeung, A.W.K.; Tzvetkov, N.T.; Horbańczuk, J.O.; Willschke, H.; Gai, Z.; Atanasov, A.G. The significance of natural product derivatives and traditional medicine for COVID-19. Processes 2020, 8, 937. [Google Scholar] [CrossRef]
- Boozari, M.; Hosseinzadeh, H. Natural products for COVID-19 prevention and treatment regarding to previous coronavirus infections and novel studies. Phytother. Res. 2021, 35, 864–876. [Google Scholar] [CrossRef]
- Xian, Y.; Zhang, J.; Bian, Z.; Zhou, H.; Zhang, Z.; Lin, Z.; Xu, H. Bioactive natural compounds against human coronaviruses: A review and perspective. Acta Pharm. Sin. B 2020, 10, 1163–1174. [Google Scholar] [CrossRef]
- Hirano, T.; Murakami, M. COVID-19: A new virus, but a familiar receptor and cytokine release syndrome. Immunity 2020, 52, 731–733. [Google Scholar] [CrossRef]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Wu, Z.; McGoogan, J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in china: Summary of a report of 72 314 cases from the chinese center for disease control and prevention. Jama 2020, 323, 1239–1242. [Google Scholar] [CrossRef] [PubMed]
- Sanders, J.M.; Monogue, M.L.; Jodlowski, T.Z.; Cutrell, J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): A review. Jama 2020, 323, 1824–1836. [Google Scholar] [CrossRef] [PubMed]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-em structure of the 2019-ncov spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016, 24, 490–502. [Google Scholar] [CrossRef]
- Kurokawa, M.; Ochiai, H.; Nagasaka, K.; Neki, M.; Xu, H.; Kadota, S.; Sutardjo, S.; Matsumoto, T.; Namba, T.; Shiraki, K. Antiviral traditional medicines against herpes simplex virus (hsv-1), poliovirus, and measles virus in vitro and their therapeutic efficacies for hsv-1 infection in mice. Antivir. Res. 1993, 22, 175–188. [Google Scholar] [CrossRef]
- Xu, H.-X.; Kadota, S.; Kurokawa, M.; Shiraki, K.; Matsumoto, T.; Namba, T. Isolation and structure of woodorien, a new glucoside having antiviral activity, from woodwardia orientalis. Chem. Pharm. Bull. 1993, 41, 1803–1806. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Calland, N.; Dubuisson, J.; Rouillé, Y.; Séron, K. Hepatitis c virus and natural compounds: A new antiviral approach? Viruses 2012, 4, 2197–2217. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; He, Z.-D.; Jiang, R.-W.; Ye, W.-C.; Xu, H.-X.; But, P.P.-H. Antiviral flavonoids from the root bark of Morus alba L. Phytochemistry 2003, 62, 1235–1238. [Google Scholar] [CrossRef]
- Xu, H.-X.; Lee, S.H.; Lee, S.F.; White, R.L.; Blay, J. Isolation and characterization of an anti-hsv polysaccharide from prunella vulgaris. Antivir. Res. 1999, 44, 43–54. [Google Scholar] [CrossRef]
- XU, H.-X.; Kadota, S.; Wang, H.; Kurokawa, M.; Shiraki, K. A new hydrolyzable tannin from geum japonicum and its antiviral activity. Heterocycles (Sendai) 1994, 38, 167–175. [Google Scholar]
- Kannan, S.; Kolandaivel, P. Antiviral potential of natural compounds against influenza virus hemagglutinin. Comput. Biol. Chem. 2017, 71, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Luganini, A.; Terlizzi, M.E.; Catucci, G.; Gilardi, G.; Maffei, M.E.; Gribaudo, G. The cranberry extract oximacro® exerts in vitro virucidal activity against influenza virus by interfering with hemagglutinin. Front. Microbiol. 2018, 9, 1826. [Google Scholar] [CrossRef]
- Xu, H.-X.; Zeng, F.-Q.; Wan, M.; Sim, K.-Y. Anti-hiv triterpene acids from geum japonicum. J. Nat. Prod. 1996, 59, 643–645. [Google Scholar] [CrossRef]
- Xu, H.-X.; Ming, D.-S.; Dong, H.; BUT, P.P.-H. A new anti-hiv triterpene from geum japonicum. Chem. Pharm. Bull. 2000, 48, 1367–1369. [Google Scholar] [CrossRef]
- Xu, H.X.; Wan, M.; Loh, B.N.; Kon, O.L.; Chow, P.W.; Sim, K.Y. Screening of traditional medicines for their inhibitory activity against hiv-1 protease. Phytother. Res. 1996, 10, 207–210. [Google Scholar] [CrossRef]
- Sahuc, M.-E.; Sahli, R.; Rivière, C.; Pène, V.; Lavie, M.; Vandeputte, A.; Brodin, P.; Rosenberg, A.R.; Dubuisson, J.; Ksouri, R. Dehydrojuncusol, a natural phenanthrene compound extracted from juncus maritimus, is a new inhibitor of hepatitis c virus rna replication. J. Virol. 2019, 93, e02009-18. [Google Scholar] [CrossRef]
- Zhang, Y.-B.; Luo, D.; Yang, L.; Cheng, W.; He, L.-J.; Kuang, G.-K.; Li, M.-M.; Li, Y.-L.; Wang, G.-C. Matrine-type alkaloids from the roots of sophora flavescens and their antiviral activities against the hepatitis b virus. J. Nat. Prod. 2018, 81, 2259–2265. [Google Scholar] [CrossRef]
- Li, B.; Li, L.; Peng, Z.; Liu, D.; Si, L.; Wang, J.; Yuan, B.; Huang, J.; Proksch, P.; Lin, W. Harzianoic acids a and b, new natural scaffolds with inhibitory effects against hepatitis c virus. Bioorgan. Med. Chem. 2019, 27, 560–567. [Google Scholar] [CrossRef]
- Cinatl, J.; Morgenstern, B.; Bauer, G.; Chandra, P.; Rabenau, H.; Doerr, H. Glycyrrhizin, an active component of liquorice roots, and replication of sars-associated coronavirus. Lancet 2003, 361, 2045–2046. [Google Scholar] [CrossRef]
- Lin, S.-C.; Ho, C.-T.; Chuo, W.-H.; Li, S.; Wang, T.T.; Lin, C.-C. Effective inhibition of MERS-COV infection by resveratrol. BMC Infect. Dis. 2017, 17, 144. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A. SARS-CoV-2 cell entry depends on ace2 and tmprss2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Zumla, A.; Chan, J.F.; Azhar, E.I.; Hui, D.S.; Yuen, K.-Y. Coronaviruses—Drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016, 15, 327–347. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Tao, G.; Liu, J.; Cai, J.; Huang, Z.; Chen, J.-x. Current prevention of COVID-19: Natural products and herbal medicine. Front. Pharmacol. 2020, 11, 588508. [Google Scholar] [CrossRef] [PubMed]
- El Hawary, S.S.; Khattab, A.R.; Marzouk, H.S.; El Senousy, A.S.; Alex, M.G.; Aly, O.M.; Teleb, M.; Abdelmohsen, U.R. In silico identification of SARS-CoV-2 spike (S) protein–ace2 complex inhibitors from eight tecoma species and cultivars analyzed by lc-ms. RSC Adv. 2020, 10, 43103–43108. [Google Scholar] [CrossRef]
- Abd El-Mordy, F.M.; El-Hamouly, M.M.; Ibrahim, M.T.; Abd El-Rheem, G.; Aly, O.M.; Abd El-kader, A.M.; Youssif, K.A.; Abdelmohsen, U.R. Inhibition of SARS-CoV-2 main protease by phenolic compounds from Manilkara hexandra (Roxb.) dubard assisted by metabolite profiling and in silico virtual screening. RSC Adv. 2020, 10, 32148–32155. [Google Scholar] [CrossRef]
- Hassan, H.A.; Abdelmohsen, U.R.; Aly, O.M.; Desoukey, S.Y.; Mohamed, K.M.; Kamel, M.S. Potential of Ficus microcarpa metabolites against SARS-CoV-2 main protease supported by docking studies. Nat. Prod. Res. 2020, 1–5. [Google Scholar] [CrossRef]
- Sayed, A.M.; Alhadrami, H.A.; El-Gendy, A.O.; Shamikh, Y.I.; Belbahri, L.; Hassan, H.M.; Abdelmohsen, U.R.; Rateb, M.E. Microbial natural products as potential inhibitors of SARS-CoV-2 main protease (mpro). Microorganisms 2020, 8, 970. [Google Scholar] [CrossRef] [PubMed]
- Albohy, A.; Zahran, E.M.; Abdelmohsen, U.R.; Salem, M.A.; Al-Warhi, T.; Al-Sanea, M.M.; Abelyan, N.; Khalil, H.E.; Desoukey, S.Y.; Fouad, M.A. Multitarget in silico studies of Ocimum menthiifolium, family Lamiaceae against SARS-CoV-2 supported by molecular dynamics simulation. J. Biomol. Struct. Dyn. 2020, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Abdelhafez, O.H.; Othman, E.M.; Fahim, J.R.; Desoukey, S.Y.; Pimentel-Elardo, S.M.; Nodwell, J.R.; Schirmeister, T.; Tawfike, A.; Abdelmohsen, U.R. Metabolomics analysis and biological investigation of three malvaceae plants. Phytochem. Anal. 2020, 31, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Haggag, E.G.; Elshamy, A.M.; Rabeh, M.A.; Gabr, N.M.; Salem, M.; Youssif, K.A.; Samir, A.; Muhsinah, A.B.; Alsayari, A.; Abdelmohsen, U.R. Antiviral potential of green synthesized silver nanoparticles of lampranthus coccineus and malephora lutea. Int. J. Nanomed. 2019, 14, 6217. [Google Scholar] [CrossRef]
- Youssif, K.A.; Elshamy, A.M.; Rabeh, M.A.; Gabr, N.; Afifi, W.M.; Salem, M.A.; Albohy, A.; Abdelmohsen, U.R.; Haggag, E.G. Cytotoxic potential of green synthesized silver nanoparticles of lampranthus coccineus extracts, metabolic profiling and molecular docking study. ChemistrySelect 2020, 5, 12278–12286. [Google Scholar] [CrossRef]
- Youssif, K.A.; Haggag, E.G.; Elshamy, A.M.; Rabeh, M.A.; Gabr, N.M.; Seleem, A.; Salem, M.A.; Hussein, A.S.; Krischke, M.; Mueller, M.J. Anti-alzheimer potential, metabolomic profiling and molecular docking of green synthesized silver nanoparticles of lampranthus coccineus and malephora lutea aqueous extracts. PLoS ONE 2019, 14, e0223781. [Google Scholar] [CrossRef]
- Elsayed, Y.; Refaat, J.; Abdelmohsen, U.R.; Othman, E.M.; Stopper, H.; Fouad, M.A. Metabolomic profiling and biological investigation of the marine sponge-derived bacterium rhodococcus sp. Ua13. Phytochem. Anal. 2018, 29, 543–548. [Google Scholar] [CrossRef]
- Yan, Y.; Pang, Y.; Lyu, Z.; Wang, R.; Wu, X.; You, C.; Zhao, H.; Manickam, S.; Lester, E.; Wu, T. The COVID-19 vaccines: Recent development, challenges and prospects. Vaccines 2021, 9, 349. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. Autodock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Alibakhshi, A.; Ranjbar, M.M.; Javanmard, S.H.; Yarian, F.; Ahangarzadeh, S. Virtual screening for the identification of potential candidate molecules against envelope (e) and membrane (m) proteins of SARS-CoV-2. J. Comput. Biophys. Chem. 2021, 20, 209–224. [Google Scholar] [CrossRef]
- Khorrami, S.; Zarrabi, A.; Khaleghi, M.; Danaei, M.; Mozafari, M. Selective cytotoxicity of green synthesized silver nanoparticles against the mcf-7 tumor cell line and their enhanced antioxidant and antimicrobial properties. Int. J. Nanomed. 2018, 13, 8013. [Google Scholar] [CrossRef] [PubMed]
- Shameli, K.; Bin Ahmad, M.; Jaffar Al-Mulla, E.A.; Ibrahim, N.A.; Shabanzadeh, P.; Rustaiyan, A.; Abdollahi, Y.; Bagheri, S.; Abdolmohammadi, S.; Usman, M.S. Green biosynthesis of silver nanoparticles using callicarpa maingayi stem bark extraction. Molecules 2012, 17, 8506–8517. [Google Scholar] [CrossRef]
- Jacobs, C.; Müller, R.H. Production and characterization of a budesonide nanosuspension for pulmonary administration. Pharm. Res. 2002, 19, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, A.; Rawat, E. Interplays of nanosilver and medicinal domain: Implications and perspectives. Chem. Sci. Rev. Lett. 2016, 5, 53–71. [Google Scholar]
- Yue, P.-F.; Yuan, H.-L.; Yang, M.; You, R.-H.; Cong, L.-B.; Zhu, J.; Wang, Q.; Zhu, W.-F.; Xiao, X.-H. Preparation, characterization, and pharmacokinetic evaluation of puerarin submicron emulsion. PDA J. Pharm. Sci. Technol. 2008, 62, 32–45. [Google Scholar] [PubMed]
- Prinz, S.; Ringl, A.; Huefner, A.; Pemp, E.; Kopp, B. 4′′′-acetylvitexin-2 ″-o-rhamnoside, isoorientin, orientin, and 8-methoxykaempferol-3-o-glucoside as markers for the differentiation of crataegus monogyna and crataegus pentagyna from crataegus laevigata (rosaceae). Chem. Biodivers. 2007, 4, 2920–2931. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, B.; Rehman, M.U.; Amin, I.; Arif, A.; Rasool, S.; Bhat, S.A.; Afzal, I.; Hussain, I.; Bilal, S. A review on pharmacological properties of zingerone (4-(4-hydroxy-3-methoxyphenyl)-2-butanone). Sci. World J. 2015, 2015, 816364. [Google Scholar] [CrossRef] [PubMed]
- Suk, S.; Kwon, G.T.; Lee, E.; Jang, W.J.; Yang, H.; Kim, J.H.; Thimmegowda, N.; Chung, M.Y.; Kwon, J.Y.; Yang, S. Gingerenone a, a polyphenol present in ginger, suppresses obesity and adipose tissue inflammation in high-fat diet-fed mice. Mol. Nutr. Food Res. 2017, 61, 1700139. [Google Scholar] [CrossRef]
- Tang, S.; Song, H.; Liu, E.; Qi, J. Isolation and purification of gingerols from ginger by high-speed counter-current chromatography. Asian J. Chem. 2014, 26, 3331. [Google Scholar] [CrossRef]
- Charles, R.; Garg, S.; Kumar, S. New gingerdione from the rhizomes of zingiber officinale. Fitoterapia 2000, 71, 716–718. [Google Scholar] [CrossRef]
- Olennikov, D.; Kashchenko, N. 1-dehydro-[14]-gingerdione, a new constituent from zingiber officinale. Chem. Nat. Compd. 2015, 51, 877–881. [Google Scholar] [CrossRef]
- Maltese, F.; Erkelens, C.; van der Kooy, F.; Choi, Y.H.; Verpoorte, R. Identification of natural epimeric flavanone glycosides by nmr spectroscopy. Food Chem. 2009, 116, 575–579. [Google Scholar] [CrossRef]
- Li, P.; Su, W.; Xie, C.; Zeng, X.; Peng, W.; Liu, M. Rapid identification and simultaneous quantification of multiple constituents in nao-shuan-tong capsule by ultra-fast liquid chromatography/diode-array detector/quadrupole time-of-flight tandem mass spectrometry. J. Chromatogr. Sci. 2015, 53, 886–897. [Google Scholar] [CrossRef][Green Version]
- Aldaw, N.; Haroun, M.; Nasser, M.; Mousa, Y. Isolation and ultra-purification of oleic acid extracted from olive oil using urea crystallization. Res. J. Pharm. Technol. 2018, 11, 624–627. [Google Scholar] [CrossRef]
- Qin, Q.-x.; Yang, J.; Chen, D.-z.; Yang, B.; Zhang, J. An alternate preparation of 3, 4, 5-trimethoxyphenol. Org. Prep. Proced. Int. 2013, 45, 321–324. [Google Scholar] [CrossRef]
- Singh, A.; Demont, A.; Goretta, L.A.; Lévêques, A.; Holvoet, S.; Nutten, S. Identification of epicatechin as the bioactive constituent in polyphenol enriched extracts that demonstrate a beneficial effect on allergic symptoms. Clin. Transl. Allergy 2013, 3, P7. [Google Scholar] [CrossRef][Green Version]
- Liu, Y.; Peng, T.-S.; Hu, X.-D.; Li, S.-J.; He, Y.-H.; Hu, F.; Jiang, J. Oxypeucedanin hydrate monoacetate isolated from angelica dahurica induces apoptosis in caco-2 colon carcinoma cells through the mediation of pi3k-signalling pathway and inhibition of cancer cell migration. Bangladesh J. Pharmacol. 2016, 11, 402–407. [Google Scholar] [CrossRef][Green Version]
- Park, J.B. Isolation and quantification of major chlorogenic acids in three major instant coffee brands and their potential effects on h 2 o 2-induced mitochondrial membrane depolarization and apoptosis in pc-12 cells. Food Funct. 2013, 4, 1632–1638. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.F.; Guo, H.J.; Huang, Y.; Wu, C.T.; Zhang, X.F. Eriodictyol, a plant flavonoid, attenuates lps-induced acute lung injury through its antioxidative and anti-inflammatory activity. Exp. Ther. Med. 2015, 10, 2259–2266. [Google Scholar] [CrossRef]
- Kumar, K.; Sabu, V.; Sindhu, G.; Rauf, A.; Helen, A. Isolation, identification and characterization of apigenin from justicia gendarussa and its anti-inflammatory activity. Int. Immunopharmacol. 2018, 59, 157–167. [Google Scholar] [CrossRef]
- Lin, L.-C.; Pai, Y.-F.; Tsai, T.-H. Isolation of luteolin and luteolin-7-o-glucoside from dendranthema morifolium ramat tzvel and their pharmacokinetics in rats. J. Agric. Food Chem. 2015, 63, 7700–7706. [Google Scholar] [CrossRef]
- Kokpol, U.; Wannachet-Isara, N.; Tip-pyang, S.; Chavasiri, W.; Veerachato, G.; Simpson, J.; Weavers, R.T. A c-methylflavone from trianthema portulacastrum. Phytochemistry 1997, 44, 719–722. [Google Scholar] [CrossRef]
- King, L.C.; Ball, C.D.; Riegel, B.; Schweitzer, C.E.; Smith, P.G.; Meyer, E.W. The isolation of β-amyrin from the leaves and seeds of alfalfa. J. Am. Chem. Soc. 1943, 65, 1168–1170. [Google Scholar] [CrossRef]
- Noor, N.F.M.; Sirat, H.M. Isolation, characterization and modification of zerumbone from zingiber zerumbet. eProc. Chem. 2016, 1, 7–10. [Google Scholar]
- Shareef, H.K.; Muhammed, H.J.; Hussein, H.M.; Hameed, I.H. Antibacterial effect of ginger (zingiber officinale) roscoe and bioactive chemical analysis using gas chromatography mass spectrum. Orient. J. Chem. 2016, 32, 20–40. [Google Scholar] [CrossRef]
- Li, W.; Wang, S.; Feng, J.; Xiao, Y.; Xue, X.; Zhang, H.; Wang, Y.; Liang, X. Structure elucidation and nmr assignments for curcuminoids from the rhizomes of curcuma longa. Magn. Reson. Chem. 2009, 47, 902–908. [Google Scholar] [CrossRef] [PubMed]
- El Sohaimy, S.; Abdelwahab, A.; Brennan, C.S.; Aboul-Enein, A. Phenolic content, antioxidant and antimicrobial activities of egyptian date palm (Phoenix dactylifera L.) fruits. Aust. J. Basic Appl. Sci. 2015, 9, 141–147. [Google Scholar]
- Gaire, B.P.; Kwon, O.W.; Park, S.H.; Chun, K.-H.; Kim, S.Y.; Shin, D.Y.; Choi, J.W. Neuroprotective effect of 6-paradol in focal cerebral ischemia involves the attenuation of neuroinflammatory responses in activated microglia. PLoS ONE 2015, 10, e0120203. [Google Scholar] [CrossRef]
- Zahoor, M.; Shafiq, S.; Ullah, H.; Sadiq, A.; Ullah, F. Isolation of quercetin and mandelic acid from aesculus indica fruit and their biological activities. BMC Biochem. 2018, 19, 5. [Google Scholar] [CrossRef]
- Bhattarai, K.; Pokharel, B.; Maharjan, S.; Adhikari, S. Chemical constituents and biological activities of ginger rhizomes from three different regions of nepal. J. Nutr. Diet. Probiotics 2018, 1, 180005. [Google Scholar]
- Jayashree, S.; Jayaraman, K.; Kalaichelvan, G. Isolation, screening and characterization of riboflavin producing lactic acid bacteria from katpadi, vellore district. Recent Res. Sci. Technol. 2010, 2, 83–88. [Google Scholar]
- Terenteva, E.; Apyari, V.; Dmitrienko, S.; Zolotov, Y.A. Formation of plasmonic silver nanoparticles by flavonoid reduction: A comparative study and application for determination of these substances. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 151, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Anjum, S.; Abbasi, B.H. Thidiazuron-enhanced biosynthesis and antimicrobial efficacy of silver nanoparticles via improving phytochemical reducing potential in callus culture of Linum usitatissimum L. Int. J. Nanomed. 2016, 11, 715. [Google Scholar]





| M/Z | Retention Time (min.) | M.wt. | Name | Molecular Formula | References |
|---|---|---|---|---|---|
| 557.27576 | 1.7060625 | 556.26849 | Epicatechin 5-O-beta-D-glucopyranoside-3-benzoate | C28H28O12 | [38] |
| 611.26602 | 1.8878542 | 610.25874 | Neohesperidin | C28H34O15 | [54] |
| 579.27182 | 2.1045125 | 578.26455 | Kaempferol-3,7-dirhamnoside (Kaempferitrin) | C27H30O14 | [37] |
| 595.26792 | 2.1045083 | 594.26064 | Quercetin-3-O-neohesperidoside | C27H30O16 | [55] |
| 283.23742 | 2.1895583 | 282.23015 | Oleic acid | C18H34O2 | [56] |
| 185.12046 | 2.2322833 | 184.11318 | 3,4,5-Trimethoxyphenol | C9HO4 | [57] |
| 291.14876 | 2.4932 | 290.14149 | Epicatechin | C15H14O6 | [58] |
| 305.13074 | 2.6144542 | 304.12347 | Oxypeucedanin hydrate | C16H16O6 | [59] |
| 369.10693 | 2.6189167 | 368.09965 | 3-Feruloylquinic acid | C17H20O9 | [60] |
| 289.13343 | 2.8026875 | 288.12616 | Eriodictyol | C15H12O6 | [61] |
| 271.11876 | 2.8048708 | 270.11148 | Apigenin | C15H10O5 | [62] |
| 287.11667 | 3.1027417 | 286.1094 | Luteolin | C15H10O6 | [63] |
| 295.166 | 4.4977708 | 294.15873 | C-Methyl flavone | C19H18O3 | [64] |
| 427.44564 | 6.6987333 | 426.43837 | β- amyrin | C30H50O | [65] |
| 579.2612 | 8.2327222 | 578.25392 | Isovitexin-2″-O-rhamnoside | C27H30O14 | [48] |
| M/Z. | Retention Time (min.) | M.wt. | Name | Molecular Formula | References |
|---|---|---|---|---|---|
| 195.10038 | 3.470625 | 194.0931 | Zingerone | C11H14O3 | [49] |
| 357.13919 | 5.799025 | 356.13191 | Gingerenone-A | C21H24O5 | [50] |
| 267.15755 | 6.4666167 | 266.15027 | 4-Gingerol | C15H22O4 | [51] |
| 293.17373 | 6.8985917 | 292.16646 | 6-Gingerdione | C17H24O4 | [52] |
| 347.23134 | 7.1113167 | 346.22406 | 1-Dehydro-(10) gingerdione | C21H30O4 | [53] |
| 219.13866 | 8.0879333 | 218.13138 | Zerumbone | C15H22O | [66] |
| 291.19394 | 8.3226583 | 290.18666 | 6-Dehydrogingerdione | C17H22O4 | [52] |
| 333.25208 | 8.4842667 | 332.2448 | 10-Shogaol | C21H32O3 | [52] |
| 237.18401 | 8.8010667 | 236.17673 | Spiro [4.5] decan-7-one, 1,8- Dimethyl-8,9-epoxy-4-isopropyl | C15H24O2 | [67] |
| 369.22559 | 9.3268667 | 368.21831 | 1,7-Bis-(4-Hydroxy-3-methoxyphenyl)-hepta-1,6-diene-3,5-dione | C21H20O6 | [68] |
| 219.17552 | 9.5892 | 218.16825 | Nuciferol | C15H22O | [69] |
| 293.2097 | 9.767 | 292.20242 | 7-Paradol | C18H28O3 | [70] |
| 303.14746 | 9.8031667 | 302.14018 | Quercetin | C15H10O7 | [71] |
| 403.34227 | 12.798483 | 402.33499 | Cholestan-3-ol, 5,6-epoxy-, (3.beta.,5.alpha.,6.alpha.) | C27H46O2 | [72] |
| 377.17929 | 16.717167 | 376.17201 | Riboflavin | C17H20N4O6 | [73] |
| Protein | Ligand | VDWAALS (kcal/mol) | EEL (kcal/mol) | EGB (kcal/mol) | ESURF (kcal/mol) | ΔG Gas (kcal/mol) | ΔG Solv (kcal/mol) | ΔTOTAL (kcal/mol) |
|---|---|---|---|---|---|---|---|---|
| AAK1 | LKB1 (reference) | −46.4 | −27.03 | 36.61 | −5.89 | −73.43 | 30.72 | −42.7 |
| Neohesperidin | −52.33 | −31.55 | 49.01 | −6.56 | −83.88 | 42.45 | −41.43 | |
| NSP16 | SAM (reference) | −46.3 | −88.43 | 88.72 | −5.93 | −134.73 | 82.8 | −51.94 |
| Neohesperidin | −33.74 | −48.01 | 55.07 | −4.83 | −81.75 | 50.24 | −31.51 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Sanea, M.M.; Abelyan, N.; Abdelgawad, M.A.; Musa, A.; Ghoneim, M.M.; Al-Warhi, T.; Aljaeed, N.; Alotaibi, O.J.; Alnusaire, T.S.; Abdelwahab, S.F.; et al. Strawberry and Ginger Silver Nanoparticles as Potential Inhibitors for SARS-CoV-2 Assisted by In Silico Modeling and Metabolic Profiling. Antibiotics 2021, 10, 824. https://doi.org/10.3390/antibiotics10070824
Al-Sanea MM, Abelyan N, Abdelgawad MA, Musa A, Ghoneim MM, Al-Warhi T, Aljaeed N, Alotaibi OJ, Alnusaire TS, Abdelwahab SF, et al. Strawberry and Ginger Silver Nanoparticles as Potential Inhibitors for SARS-CoV-2 Assisted by In Silico Modeling and Metabolic Profiling. Antibiotics. 2021; 10(7):824. https://doi.org/10.3390/antibiotics10070824
Chicago/Turabian StyleAl-Sanea, Mohammad M., Narek Abelyan, Mohamed A. Abdelgawad, Arafa Musa, Mohammed M. Ghoneim, Tarfah Al-Warhi, Nada Aljaeed, Ohoud J. Alotaibi, Taghreed S. Alnusaire, Sayed F. Abdelwahab, and et al. 2021. "Strawberry and Ginger Silver Nanoparticles as Potential Inhibitors for SARS-CoV-2 Assisted by In Silico Modeling and Metabolic Profiling" Antibiotics 10, no. 7: 824. https://doi.org/10.3390/antibiotics10070824
APA StyleAl-Sanea, M. M., Abelyan, N., Abdelgawad, M. A., Musa, A., Ghoneim, M. M., Al-Warhi, T., Aljaeed, N., Alotaibi, O. J., Alnusaire, T. S., Abdelwahab, S. F., Helmy, A., Abdelmohsen, U. R., & Youssif, K. A. (2021). Strawberry and Ginger Silver Nanoparticles as Potential Inhibitors for SARS-CoV-2 Assisted by In Silico Modeling and Metabolic Profiling. Antibiotics, 10(7), 824. https://doi.org/10.3390/antibiotics10070824







