Abstract
Relatively little is known about how the corneal epithelium responds to vision-threatening bacteria from the Enterobacterales order. This study investigates the impact of Serratia marcescens on corneal epithelial cell host responses. We also investigate the role of a bacterial transcription factor EepR, which is a positive regulator of S. marcescens secretion of cytotoxic proteases and a hemolytic surfactant. We treated transcriptomic and metabolomic analysis of human corneal limbal epithelial cells with wild-type bacterial secretomes. Our results show increased expression of proinflammatory and lipid signaling molecules, while this is greatly altered in eepR mutant-treated corneal cells. Together, these data support the model that the S. marcescens transcription factor EepR is a key regulator of host-pathogen interactions, and is necessary to induce proinflammatory chemokines, cytokines, and lipids.
1. Introduction
The cornea, the transparent, anterior layer of the eye, is essential for vision and protected by numerous host immune factors, the tear film [1,2], and the corneal epithelium [3,4]. When the epithelium is damaged or compromised, it permits entry of microbes into the stroma where they can multiply and cause damage to the ocular tissues; the progression of infection is rapid, sometimes leading to corneal perforation from bacterial proteases and from the ensuing inflammatory response [5,6,7,8,9].
Serratia marcescens is a gram negative pathogen from the order Enterobacterales frequently isolated from contact lenses, and associated with ocular infections [10,11,12]. Bacteria are linked with chronic infections, non-healing wounds, and are thought to prevent wound closure; however, the impact of bacteria on corneal infection and wound healing is poorly understood [13,14,15]. Our previous study identified S. marcescens LPS as being sufficient to inhibit corneal epithelial wound closure and further identified transposon insertions in genes that rendered the bacterium unable to inhibit corneal cell migration, but the role of these genes in ocular surface host-pathogen interactions was not characterized [16]. One mutation mapped to the eepR-eepS locus, that codes for a hybrid two-component transcription factor system involved in virulence factor secretion, cytotoxicity to mammalian cells, and proliferation in a rabbit keratitis model [17,18,19].
Previous studies have evaluated the impact of bacteria on the global transcriptomic response of corneal cells, but this has only been done with Pseudomonas aeruginosa and Staphylococcus aureus [20,21,22,23]. In this study, the role of the EepR transcriptional regulator in the corneal epithelial cell transcriptional and small molecule response to S. marcescens was evaluated. We report that in contrast to other pathogens, mutation of one bacterial transcription factor in S. marcescens had a broad impact on epithelial cell responses, including reduced expression of inflammatory markers and lipid metabolism genes.
2. Results
2.1. HCLE Cells Exposed to eepR Mutant S. marcescens Secretomes Have an Attenuated Inflammatory Response Compared with Wild-type Treated HCLE Cells
To increase our understanding of the corneal response to an order of bacteria not previously tested, a global transcriptional analysis of the HCLE cells was performed. Here we used a wild-type (WT), low cytotoxicity [24] isolate of S. marcescens (PIC3611), and an isogenic strain with a deletion in the eepR gene that was previously described [19] to further investigate EepR’s role in how bacteria influence corneal biology. In this study, bacterial secretomes were used to stimulate corneal cells because we have previously shown wild-type secretomes to strongly influence the behavior of a human corneal epithelial cell line and because secretomes are less toxic to corneal cells [16,25,26]. Confluent monolayers of the human corneal limbal epithelial (HCLE) cell line were first exposed to S. marcescens WT secretome for 0, 1, 2, 3, 4, and 5 h to determine the time frame for maximal stimulation by assessing levels of the cytokine TNFα. The 5 h exposure time point was chosen based upon our preliminary findings (data not shown) and from a previous ELISA-based study of human corneal epithelial cell inflammatory response to S. marcescens [27].
Next, we compared the transcriptomes of mock-treated (LB medium in equal volume as secretomes) corneal cells with those exposed to normalized secretomes from WT or eepR cells. Lower case eepR refers to the mutant strain. As noted in Materials and Methods, 21,932 microarray panels (unique target sequences) yielded reliable data; valid changes between WT secretome-treated and mock-treated cells occurred in only 2510 panels (11.4%), and of those, only 915 (4.2%) were modulated by 2-fold or more (examples in Table 1 and Table 2). In contrast, valid changes between eepR secretome-treated and mock-treated cells occurred in only 798 panels (3.6%), and of those, only 241 (1.1%) were modulated by 2-fold or more (examples in Table 3 and Table 4). Over half of the eepR secretome-modulated panels (138, 57%) were present in the WT-treatment group also (see nine genes in common between Table 1 and Table 3, eight genes in common between Table 2 and Table 4), and the direction of modulation was concordant between treatments for all these panels except SPRY2, which was increased by WT treatment and decreased by eepR. Visual inspection showed that within this group of 138 genes, whatever the direction of change caused by eepR (increase or decrease), its magnitude was always less than that caused by WT. However, some genes outside this group showed greater modulation by eepR than by WT. Accordingly, the scaled eepR response (eepR − control)/|(WT − control)| was also calculated (Table 5 and Table 6).

Table 1.
Twenty-five genes with the greatest expression increase in cells treated with WT vs. mock secretomes.

Table 2.
Twenty-five genes with the greatest expression decrease in cells treated with WT vs. mock secretomes.

Table 3.
Twenty-five genes with the greatest expression increase in cells treated with eepR vs. mock secretomes.

Table 4.
Twenty-five genes with the greatest expression decrease in cells treated with eepR vs. mock secretomes.

Table 5.
Twenty-five genes with the highest scaled eepR values (i.e., relatively little effect of WT, relatively large increase by eepR).

Table 6.
Twenty-five genes with the lowest scaled eepR values (i.e., relatively little effect of WT, relatively large decrease by eepR).
The 915 panels modulated by WT were submitted to Ingenuity Pathway Analysis software (Qiagen, Germantown, MD, USA), yielding 24 significantly enriched (p < 0.05) canonical pathways which had adequate z-scores (|z| > 2; see Table 7). At least nine of these pathways address direct or indirect immune functions. When submitted for analysis separately, the 798 eepR modulated panels only yielded three significantly enriched pathways, two of which were also WT-modulated (see Table 7). The third pathway (GNRH Signaling) was not significantly enriched by WT treatment. In S. marcescens WT secretome-treated HCLEs versus mock-treated cells, the twenty-five most upregulated genes (9.1- to 56.6-fold increase) included genes involved in inflammatory signaling pathways (Table 1). Genes with the greatest decrease (4.9- to 50-fold decrease) in WT secretome-treated HCLEs were those involved in nucleosome assembly, phospholipid metabolic processes, and transcription (Table 2). Moreover, HCLEs-treated with eepR secretome showed decreased upregulation of genes for proinflammatory factors; however, genes involved in cell to cell adhesion, leukocyte chemotaxis, transport, and signaling were upregulated (Table 3). Genes with the greatest decrease in eepR versus mock-treated secretomes were those involved in nucleic acid binding, transport, and transcription (Table 4).

Table 7.
Significantly (p < 0.05) enriched canonical pathways which respond to WT S. marcescens stimulus.
Interestingly, when we examined our results in the context of scaled eepR (eepR—control/WT—control), there were also several genes where the expression difference was greater than 10-fold in eepR-treated cells in comparison to WT. In particular, there were differences in genes involved in intracellular protein transport, protein binding, transcription, nucleic acid binding, and translation (Table 5). Genes with the lowest expression in the scaled eepR response were involved in protein complex assembly, protein binding, signal transduction, actin filament polymerization, inactivation of MAPK activity, and negative regulation of transcription (Table 6).
From our microarray results, we chose genes to validate by qRT-PCR that are known mediators of response to infection and corneal wound healing, involved in cellular signaling, motility, actin binding, and cellular division/membrane organization, and had at least a 2-fold difference when comparing WT to eepR-treated HCLEs [17,20,21,27]. Overall, our qRT-PCR results validated changes observed with the microarray, including that the eepR-treated HCLEs in most cases had a lower fold change in proinflammatory gene expression (Figure 1, Table 1). We note that, when assayed by qRT-PCR, nine out of the twelve genes show a greater response to WT treatment than they do by microarray analysis, consistent with the greater sensitivity and wider dynamic range of qRT-PCR.
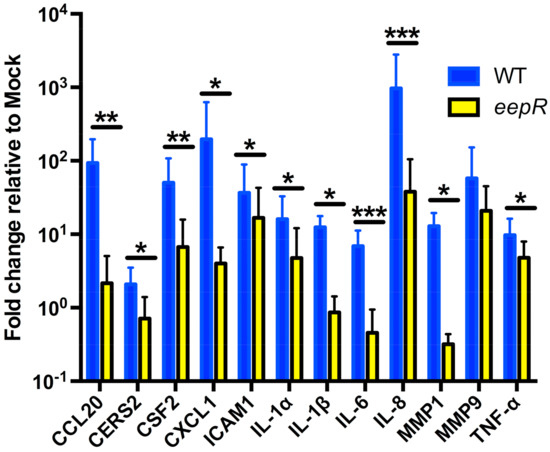
Figure 1.
qRT-PCR of pathway markers confirmed microarray analysis. Graph represents the fold change in gene expression relative to mock (LB) treatment. HCLE cells were exposed to LB, WT, and eepR transcriptomes of 5 h. Gene expression was normalized to GAPDH expression. Means (n = 4–8, n = 3 for IL-1α) and SD are shown. ∆∆CT values were compared by ANOVA with Bonferroni’s post-test, one asterisk (*) indicate p < 0.05, two indicate (**) p < 0.01, and three (***) indicate p < 0.001.
2.2. Bacterial Secretomes Influence Corneal Epithelial CellLipid Metabolism
In addition to evidence of EepR playing a role in producing inflammatory markers, microarray analysis revealed alterations in pathways associated with lipid metabolism and signaling. These pathways include ceramide biosynthesis, ceramide signaling, and Sphingosine-1-phosphate receptor signaling with avalid increases in CERS2 (1.5-fold), S1PR3 (2.3-fold), SPHK1 (1.8-fold), and SPTLC2 (2.5-fold) genes by cells treated with wild-type, but not eepR secretomes. Increased CERS2 expression observed in the microarray was confirmed by qRT-PCR (Figure 1).
To further verify the alteration in producing compounds associated with the lipid pathways implicated in the microarray data and to gain insight into the corneal epithelial cell response to enteric bacteria, small molecule metabolomic analysis was performed on HCLE cells exposed to secretomes derived from WT and the eepR mutant. Consistent with the results of the microarray analysis and qRT-PCR, the metabolomic analysis identified changes in markers involved in lipid metabolism (Figure 2, Supplementary Tables S1 and S2). There were significant increases in metabolites for lipid metabolism for S. marcescens WT-treated HCLEs, including sphingosine, phosphoethanolamine (Figure 2), as well as linoleate, eicosapentaenoate, docosapentaenoate, docosahexaenoate, and myristate (Table S1). Together, these data indicate that S. marcescens secreted factors have a major impact on human corneal cells, including increased expression of inflammatory and lipid metabolism pathways, and that S. marcescens requires EepR for these effects.
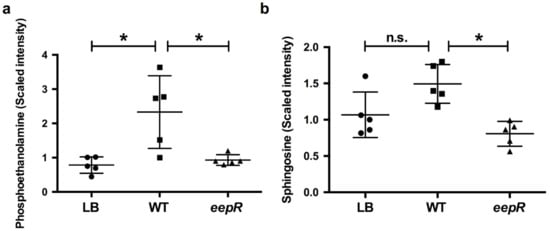
Figure 2.
Metabolomic analysis demonstrates alteration of sphingosine and lipid metabolism in corneal cells challenged by S. marcescens secretomes. HCLE cells were treated with LB, WT, or eepR secretomes for 24 h. Mean and SD (n = 5) of relative amounts of (a) phosphoethanolamine and (b) sphingosine. Circles = LB (mock treatment), squares = WT, and triangles = eepR mutant treated HCLE cells. Asterisks (*) indicate p < 0.05 by one-way ANOVA with Tukey’s post-hoc analysis. n.s., not significant.
3. Discussion
S. marcescens EepR, a master transcriptional regulator of secreted enzymes and secondary metabolites, plays an important role in hemolysis, pigment production, swarming motility, and contributes to bacterial proliferation in the cornea. A previous study demonstrated the importance of the S. marcescens transcription factor EepR in the regulation of protease production, corneal cell-induced cytotoxicity, and its ability to induce the proinflammatory cytokine IL-1β [17]. Because of its involvement in ocular host-pathogen response, we sought to determine differences in gene expression profiles in eepR-treated corneal cells in comparison to WT. Interestingly, genes with the greatest expression in eepR mutant-treated corneal cells compared to WT-treated cells were those involved in intracellular transport, protein binding, cellular component movement, cell adhesion, and membrane-related functions (Table 5), suggesting deletion of EepR promotes cell migration and wound healing. Consistently, eepR-treated cells were found to regulate lipid metabolic process, transcription, and intracellular protein transport (Table 5) and activate the MAPK pathway, which has been demonstrated to promote cell migration [28]. In contrast, WT-treated cells were found to inactivate the MAPK pathway (Table 6), which is consistent with its wound inhibitory phenotype [16].
The effect of bacteria on human corneal epithelial cells is of interest because bacteria cause the majority of corneal ulcers [29]. A limited number of studies have examined the impact of P. aeruginosa and S. aureus on the corneal transcriptomic response [20,21,22,23], but these have not been done with bacteria of the Enterobacterales order. Bacteria, such as Klebsiella, Proteus, and Serratia, cause a significant number of ocular infections [30]. There is a unique immunological response of the cornea, being an immune-privileged site. Chidambaram et al. compared gene expression profiles of corneal tissues from microbial keratitis patients infected with Streptococcus pneumoniae, P. aeruginosa, Fusarium sp., and Aspergillus sp. to normal corneal tissue from cadavers [20]. In agreement with our own data, they found increased expression of the proinflammatory markers MMP9, MMP1, IL-1β, and TNF with the greatest expression observed in MMP9. In addition to the previously mentioned markers, they also found increases in MMP7, MMP10, MMP12, TLR2, and TLR4, all markers known to promote inflammation and immune recognition [20]. Our data also found a 2.2-fold increase in expression of TLR2 in WT versus eepR mutant-treated HCLEs, but no significant changes in TLR4 expression. Microarray gene expression levels for TLR4 were low, but detectable for all conditions in our study. However, expression of TLR4 in corneal epithelial cells has been previously demonstrated to be reduced [31,32], and could explain why our results differed from Chidambaram et al.
The S. marcescens-induced proinflammatory gene response reported here was consistent with a study by Hume et al. [27], who used ELISA to explore the cytokine response of human corneal cells and polymorphonuclear monocytes (PMNs) to clinical isolates of S. marcescens. Though they found strain differences in cytokine response, there was an overall positive trend in activation of TNFα, IL-6, and CXCL8 after 4 h of exposure to bacteria which was similar to our results after 5 h of exposure [27].
The impact of living Pseudomonas aeruginosa upon the transcriptome of murine corneas has been explored by Gao et al. [21]. They reported upregulation of Krt16, MMP10, MMP13, S100A8, Stfna111, and S100A9 genes with an even greater increase in the genes involved in antimicrobial peptide production S100A8 and S100A9, when mice were pretreated with flagellin [21]. Our results were not as striking for S100A8 and S100A9, but did demonstrate a 2-fold increase in WT-treated HCLES in comparison to eepR. Huang et al. used murine corneas infected with P. aeruginosa and demonstrated upregulation of proinflammatory markers GM-CSF, ICAM1, IL1α, IL-1β, IL-6, TNFα, MMP9, MMP10, and MMP13 in accordance with our results [22]. In addition to the previously mentioned genes, we also observed upregulation of proinflammatory markers CCL20, CERS2, CXCL1, CXCL8, and MMP1.
An elegant study by Heimer et al. used a well-defined reference strain of the gram positive bacteria S. aureus to examine corneal epithelial cell responses to bacteria [23]. They evaluated the effect of an isogenic agr sarA double mutant of S. aureus that has similar defects as our eepR mutant in reduced secretion of virulence factors [23]. After treating human corneal cells with S. aureus, highly increased expression of proinflammatory markers CCL20, CSF2, CXCL1, IL-6, CXCL8, and TNFα was observed. These results are in agreement with our own, with the only major notable difference being that the gene most induced by S. marcescens WT bacteria was CXCL8, a neutrophil chemoattractant important for neutrophil migration to the site of infection and clearance of bacteria, whereas S. aureus most induced CCL20 a chemokine with antibacterial properties [33]—the third most highly induced gene in our study. In sharp contrast to their study, while the S. aureus agr sarA double mutant caused relatively little change in host response compared to the WT S. aureus, the S. marcescens eepR mutant was strikingly less able than the WT to induce expression of proinflammatory genes. Another notable difference is that some of the signal transduction factors upregulated by S. aureus were not affected by S. marcescens, notably the plasminogen activator inhibitor SERPINB2 that is involved in macrophage function and cell migration [34], and the glycoprotein STC1 that is involved in angiogenesis and wound healing [35].
Matrix metalloproteinases (MMPs) are enzymes that function in immune responses to infection in addition to numerous other roles. MMPs are involved in recruiting white blood cells, chemokine and cytokine responses, and cell matrix remodeling [36]. In our study, numerous matrix metalloproteases were upregulated >2-fold by S. marcescens, including MMP1, 9, 10, 13, 14, 16, 19, 28, but a similar trend was not described in S. aureus challenged cells [23].The different pathogen associated molecular patterns produced by the bacteria and the challenge with whole S. aureus versus S. marcescens secretomes (which include flagella and LPS) may account for some of the differences observed. Nevertheless, the S. marcescens EepR protein had a much larger role than the S. aureus SarA transcription factor and Agr quorum sensing system in affecting the corneal epithelial cell transcriptional response.
The reason for which eepR mutants confer such a different transcriptional response compared to the WT is not clear at this time. The eepR mutant is defective in the secretion of metalloproteases, such as serralysin and SlpB [17]. Serralysin, also called the 56-kDa protease, was shown in experimental models to have an impact on the immune system, rendering mouse lungs much more susceptible to influenza infection [37]. The protease was shown to increase vascular permeability by activation of the Hageman factor-kallikrein-kinin system [38]. Further studies will evaluate the role of EepR regulated bacterial metalloproteases in corneal wound healing.
Our microarray and qRT-PCR data suggested differences for expression of genes involved in the lipid metabolism pathway for corneal cells exposed to WT, but not eepR mutant secretomes. This data was validated using metabolomics approaches and indicated that the changes in transcription yielded measurable differences in the molecules involved in the altered pathways. Bioactive sphingolipids, such as those with altered expression shown here, like ceramide and sphingosine 1-phosphate, are known signaling molecules that mediate wound healing in many tissues [39], and likely play a different role in corneal responses. These data indicate the importance of a single bacterial transcription factor in dictating the corneal cell response as measured through transcriptomic and metabolomic analysis. The findings and their implications should be discussed in the broadest context possible.
4. Materials and Methods
4.1. Bacterial Growth Conditions and Media
S. marcescens cultures were grown in lysogeny broth (LB) [40] at 30 °C with shaking. Bacteria free secretomes of S. marcescens WT and eepR were prepared by normalizing overnight cultures to OD600 = 2.0 and removing the bacteria by centrifugation at 14,000 rpm for two minutes followed by filtration through a 0.22 μm filter.
4.2. Microarray
HCLE cell line was obtained from Ilene Gipson [41], and were maintained in KSFM media as previously described [16]. Cells were seeded into 12 well plates at a density of 1.5 × 105 cells per well. Secretomes were prepared as described above and added to HCLE cells at the same dosage (500 μL into 1 mL KSFM) and incubated for 5 h at 37 °C + 5% CO2. HCLE cells were washed 3 times with phosphate buffered saline (PBS) and stored in 5 volumes of RNAlater (Sigma-Aldrich, St. Louis, MO, USA) at 4 °C until used. RNA was extracted with a GenElute Mammalian total RNA miniprep kit (Sigma-Aldrich), treated with 1 unit of RQ1 Dnase (Promega, Madison, WI, USA) for 30 min at 37 °C, and quantified by Nanodrop (Thermo Scientific|Thermo Fisher Scientific, Waltham, MA, USA). 500 ng samples of total RNA were processed using an Affymetrix 3′-IVT Express kit (Affymetrix, Santa Clara, CA, USA) and yielded 43.2 ± 14.4 μg of biotinylated cRNA (mean ± SD, n = 5), with one outlier of 7 μg. Twenty μg of biotinylated cRNA was hybridized to Affymetrix U133 Plus 2.0 GeneChips (catalog #900470). The GeneChips were developed and scanned using an Affymetrix GeneChip 3000 Array Scanner.
The resultant DAT files were consolidated to CEL files, which were analyzed with Affymetrix GCOS v1.4 software, using default parameters. Numerical data and the software flags for Presence/Absence and for significant pairwise changes were transferred to Microsoft Excel. Of the 54,675 panels (unique sequence targets) on the microarray, 26,162 showed no detectable expression in any sample and omitted further consideration. Of the remaining 28,513 panels, the 22,553 (79%) which showed consistent detectable expression in the duplicate samples of at least one experimental group were taken for analysis. Of these, 621 panels (2.8%) showed a significant 2-fold difference between duplicates and were rejected as unreliable. For the reliable 21,932 panels, the ratio (mean (WT − treated)/mean (untreated)) was calculated. This ratio represented a valid change if:Both samples in the higher-expressing group reported Present (i.e., detectable target sequence), all four pairwise comparisons between groups showed significant changes using the GCOS software, and the groups did not overlap.
4.3. Quantitative Reverse Transcriptase PCR (qPCR)
RNA was extracted as described above and concentrated using an RNA Clean and Concentration kit (Zymo Research, Irvine, CA, USA). All samples were normalized with nuclease free water to a concentration of 50 ng/µL. 250 µg of RNA was synthesized into cDNA using Superscript III reverse transcriptase (Invitrogen|Thermo Fisher Scientific, Waltham, MA, USA) as previously described [19]. To identify any genomic DNA contamination, non-template controls of each RNA sample were also prepared and verified by reverse transcriptase PCR (RT-PCR) using GAPDH primers [42]. All contaminated samples were discarded. Quantitative reverse transcriptase PCR (qRT-PCR) was performed using Sybr green reagent (Applied Biosystems|Thermo Fisher Scientific, Waltham, MA, USA) using primers for CCL20, CERS2, CSF2, ICAM-1, IL-1α, IL-1β, IL-6, IL-8, MMP1, MMP9, TNFα [42,43,44,45,46,47,48,49,50,51,52]. All gene reactions were normalized to GAPDH [42], and analyzed using the ΔΔCT method. All experiments were performedat least three independent times.
4.4. Metabolomics
One sample containing 100 μL of LB (mock) and five 100 μL samples each of WT and eepR mutant were collected and stored at −80 °C. All samples were collected in two independent harvests on two different days and shipped on dry ice to Metabolon Inc. for small molecule analysis. Samples were prepared using an automated MicroLab STAR® system (The Hamilton Company, Allston, MA, USA) using a proprietary series of organic and aqueous extractions. The prepared extract was then divided into two fractions, one for analysis by liquid chromatography and one for analysis by gas chromatography. Samples were then placed in a TurboVap® (Biotage, Uppsala, Sweden) to remove the organic solvent. Each sample was frozen and dried under vacuum and prepared for liquid chromatography mass spectrometry (LC/MS) or gas chromatography mass spectrometry analysis. Library entries of purified standards or recurrent unknown entities were used to identify compounds. Matches for each sample were verified and corrected as needed.
4.5. Statistical Analysis
Student’s t-test and one-way ANOVA with post hoc statistical tests were performed using GraphPad Prism statistical software version 6.0. For metabolomics analysis, Welch’s t-tests using pairwise comparisons were performed for statistical analysis. Significance for all statistical tests was determined at p < 0.05.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/antibiotics10070770/s1, Table S1: Metabolomics analysis of WT and eepR secretome-treated HCLE, Table S2: Metabolomics data.
Author Contributions
Conceptualization, K.M.B. and R.M.Q.S.; methodology, K.M.B., S.A.K.H. and R.M.Q.S.; software, S.A.K.H.; investigation, K.M.B. and S.A.K.H.; data curation, K.M.B., S.A.K.H. and R.M.Q.S.; writing—original draft preparation, K.M.B.; writing—review and editing, K.M.B., S.A.K.H. and R.M.Q.S.; funding acquisition, R.M.Q.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Research to Prevent Blindness (unrestricted funds), the Eye and Ear Foundation of Pittsburgh, National Institute of Health grants P30EY08098 (to Department of Ophthalmology), F32EY024785 (to K.M.B.), T32EY017271 (to K.M.B.), and R01EY027331 (to R.M.Q.S.).
Data Availability Statement
Microarray data was deposited to NCBI gene expression Omnibus (GEO accession number GSM1832614). Metabolomic data is supplied in Table S2.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fleiszig, S.M.; McNamara, N.A.; Evans, D.J. The tear film and defense against infection. Adv. Exp. Med. Biol. 2002, 506, 523–530. [Google Scholar]
- McDermott, A.M. Antimicrobial compounds in tears. Exp. Eye Res. 2013, 117, 53–61. [Google Scholar] [CrossRef]
- Evans, D.J.; Fleiszig, S.M. Why does the healthy cornea resist Pseudomonas aeruginosa infection? Am. J. Ophthalmol. 2013, 155, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Metruccio, M.M.E.; Tam, C.; Evans, D.J.; Xie, A.L.; Stern, M.E.; Fleiszig, S.M.J. Contributions of MyD88-dependent receptors and CD11c-positive cells to corneal epithelial barrier function against Pseudomonas aeruginosa. Sci. Rep. 2017, 7, 13829. [Google Scholar] [CrossRef] [PubMed]
- Callegan, M.C.; O’Callaghan, R.J.; Hill, J.M. Pharmacokinetic considerations in the treatment of bacterial keratitis. Clin. Pharmacokinet. 1994, 27, 129–149. [Google Scholar] [CrossRef] [PubMed]
- Hazlett, L.D. Role of innate and adaptive immunity in the pathogenesis of keratitis. Ocul. Immunol. Inflamm. 2005, 13, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Pearlman, E.; Sun, Y.; Roy, S.; Karmakar, M.; Hise, A.G.; Szczotka-Flynn, L.; Ghannoum, M.; Chinnery, H.R.; McMenamin, P.G.; Rietsch, A. Host defense at the ocular surface. Int. Rev. Immunol. 2013, 32, 4–18. [Google Scholar] [CrossRef] [PubMed]
- Ruan, X.; Chodosh, J.; Callegan, M.C.; Booth, M.C.; Lee, T.D.; Kumar, P.; Gilmore, M.S.; Pereira, H.A. Corneal expression of the inflammatory mediator CAP37. Investig. Ophthalmol. Vis. Sci. 2002, 43, 1414–1421. [Google Scholar]
- Willcox, M.D. Pseudomonas aeruginosa infection and inflammation during contact lens wear: A review. Optom. Vis. Sci. 2007, 84, 273–278. [Google Scholar] [CrossRef]
- Mah-Sadorra, J.H.; Najjar, D.M.; Rapuano, C.J.; Laibson, P.R.; Cohen, E.J. Serratia corneal ulcers: A retrospective clinical study. Cornea 2005, 24, 793–800. [Google Scholar] [CrossRef]
- Voelz, A.; Muller, A.; Gillen, J.; Le, C.; Dresbach, T.; Engelhart, S.; Exner, M.; Bates, C.J.; Simon, A. Outbreaks of Serratia marcescens in neonatal and pediatric intensive care units: Clinical aspects, risk factors and management. Int. J. Hyg. Environ. Health 2010, 213, 79–87. [Google Scholar] [CrossRef]
- Mahlen, S.D. Serratia infections: From military experiments to current practice. Clin. Microbiol. Rev. 2011, 24, 755–791. [Google Scholar] [CrossRef]
- Rahim, K.; Saleha, S.; Zhu, X.; Huo, L.; Basit, A.; Franco, O.L. Bacterial Contribution in Chronicity of Wounds. Microb. Ecol. 2017, 73, 710–721. [Google Scholar] [CrossRef]
- Scales, B.S.; Huffnagle, G.B. The microbiome in wound repair and tissue fibrosis. J. Pathol. 2013, 229, 323–331. [Google Scholar] [CrossRef]
- Scali, C.; Kunimoto, B. An update on chronic wounds and the role of biofilms. J. Cutan. Med. Surg. 2013, 17, 371–376. [Google Scholar] [CrossRef]
- Brothers, K.M.; Stella, N.A.; Hunt, K.M.; Romanowski, E.G.; Liu, X.; Klarlund, J.K.; Shanks, R.M. Putting on the brakes: Bacterial impediment of wound healing. Sci. Rep. 2015, 5, 14003. [Google Scholar] [CrossRef]
- Brothers, K.M.; Stella, N.A.; Romanowski, E.G.; Kowalski, R.P.; Shanks, R.M. EepR Mediates Secreted-Protein Production, Desiccation Survival, and Proliferation in a Corneal Infection Model. Infect. Immun. 2015, 83, 4373–4382. [Google Scholar] [CrossRef][Green Version]
- Shanks, R.M.Q.; Stella, N.A.; Lahr, R.M.; Aston, M.A.; Brothers, K.M.; Callaghan, J.D.; Sigindere, C.; Liu, X. Suppressor analysis of eepR mutant defects reveals coordinate regulation of secondary metabolites and serralysin biosynthesis by EepR and HexS. Microbiology 2017, 163, 280–288. [Google Scholar] [CrossRef]
- Stella, N.A.; Lahr, R.M.; Brothers, K.M.; Kalivoda, E.J.; Hunt, K.M.; Kwak, D.H.; Liu, X.; Shanks, R.M. Serratia marcescens cyclic AMP-receptor protein controls transcription of EepR, a novel regulator of antimicrobial secondary metabolites. J. Bacteriol. 2015, 197, 2468–2478. [Google Scholar] [CrossRef]
- Chidambaram, J.D.; Kannambath, S.; Srikanthi, P.; Shah, M.; Lalitha, P.; Elakkiya, S.; Bauer, J.; Prajna, N.V.; Holland, M.J.; Burton, M.J. Persistence of Innate Immune Pathways in Late Stage Human Bacterial and Fungal Keratitis: Results from a Comparative Transcriptome Analysis. Front. Cell Infect. Microbiol. 2017, 7, 193. [Google Scholar] [CrossRef]
- Gao, N.; Sang Yoon, G.; Liu, X.; Mi, X.; Chen, W.; Standiford, T.J.; Yu, F.S. Genome-wide transcriptional analysis of differentially expressed genes in flagellin-pretreated mouse corneal epithelial cells in response to Pseudomonas aeruginosa: Involvement of S100A8/A9. Mucosal Immunol. 2013, 6, 993–1005. [Google Scholar] [CrossRef]
- Huang, X.; Hazlett, L.D. Analysis of Pseudomonas aeruginosa corneal infection using an oligonucleotide microarray. Investig. Ophthalmol. Vis. Sci. 2003, 44, 3409–3416. [Google Scholar] [CrossRef][Green Version]
- Heimer, S.R.; Yamada, A.; Russell, H.; Gilmore, M. Response of corneal epithelial cells to Staphylococcus aureus. Virulence 2010, 1, 223–235. [Google Scholar] [CrossRef]
- Shanks, R.M.; Stella, N.A.; Hunt, K.M.; Brothers, K.M.; Zhang, L.; Thibodeau, P.H. Identification of SlpB, a Cytotoxic Protease from Serratia marcescens. Infect. Immun. 2015, 83, 2907–2916. [Google Scholar] [CrossRef] [PubMed]
- Brothers, K.M.; Kowalski, R.P.; Tian, S.; Kinchington, P.R.; Shanks, R.M.Q. Bacteria induce autophagy in a human ocular surface cell line. Exp. Eye Res. 2018, 168, 12–18. [Google Scholar] [CrossRef]
- Brothers, K.M.; Stella, N.A.; Shanks, R.M.Q. Biologically active pigment and ShlA cytolysin of Serratia marcescens induce autophagy in a human ocular surface cell line. BMC Ophthalmol. 2020, 20, 120. [Google Scholar] [CrossRef]
- Hume, E.; Sack, R.; Stapleton, F.; Willcox, M. Induction of cytokines from polymorphonuclear leukocytes and epithelial cells by ocular isolates of Serratia marcescens. Ocul. Immunol. Inflamm. 2004, 12, 287–295. [Google Scholar] [CrossRef]
- Saika, S.; Okada, Y.; Miyamoto, T.; Yamanaka, O.; Ohnishi, Y.; Ooshima, A.; Liu, C.Y.; Weng, D.; Kao, W.W. Role of p38 MAP kinase in regulation of cell migration and proliferation in healing corneal epithelium. Investig. Ophthalmol. Vis. Sci. 2004, 45, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Al-Mujaini, A.; Al-Kharusi, N.; Thakral, A.; Wali, U.K. Bacterial keratitis: Perspective on epidemiology, clinico-pathogenesis, diagnosis and treatment. Sultan Qaboos Univ. Med. J. 2009, 9, 184–195. [Google Scholar]
- Teweldemedhin, M.; Gebreyesus, H.; Atsbaha, A.H.; Asgedom, S.W.; Saravanan, M. Bacterial profile of ocular infections: A systematic review. BMC Ophthalmol. 2017, 17, 212. [Google Scholar] [CrossRef]
- Zhang, J.; Kumar, A.; Wheater, M.; Yu, F.S. Lack of MD-2 expression in human corneal epithelial cells is an underlying mechanism of lipopolysaccharide (LPS) unresponsiveness. Immunol. Cell Biol. 2009, 87, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Ueta, M.; Nochi, T.; Jang, M.H.; Park, E.J.; Igarashi, O.; Hino, A.; Kawasaki, S.; Shikina, T.; Hiroi, T.; Kinoshita, S.; et al. Intracellularly expressed TLR2s and TLR4s contribution to an immunosilent environment at the ocular mucosal epithelium. J. Immunol. 2004, 173, 3337–3347. [Google Scholar] [CrossRef] [PubMed]
- Schutyser, E.; Struyf, S.; Van Damme, J. The CC chemokine CCL20 and its receptor CCR6. Cytokine Growth Factor Rev. 2003, 14, 409–426. [Google Scholar] [CrossRef]
- Shea-Donohue, T.; Zhao, A.; Antalis, T.M. SerpinB2 mediated regulation of macrophage function during enteric infection. Gut Microbes 2014, 5, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Li, Y.; Wang, J.; Li, Y.; Li, Y.; Li, G. Stanniocalcin1 (STC1) Inhibits Cell Proliferation and Invasion of Cervical Cancer Cells. PLoS ONE 2013, 8, e53989. [Google Scholar] [CrossRef]
- Elkington, P.T.; O’Kane, C.M.; Friedland, J.S. The paradox of matrix metalloproteinases in infectious disease. Clin. Exp. Immunol. 2005, 142, 12–20. [Google Scholar] [CrossRef]
- Akaike, T.; Molla, A.; Ando, M.; Araki, S.; Maeda, H. Molecular mechanism of complex infection by bacteria and virus analyzed by a model using serratial protease and influenza virus in mice. J. Virol. 1989, 63, 2252–2259. [Google Scholar] [CrossRef]
- Kamata, R.; Yamamoto, T.; Matsumoto, K.; Maeda, H. A serratial protease causes vascular permeability reaction by activation of the Hageman factor-dependent pathway in guinea pigs. Infect. Immun. 1985, 48, 747–753. [Google Scholar] [CrossRef]
- Shea, B.S.; Tager, A.M. Sphingolipid regulation of tissue fibrosis. Open Rheumatol. J. 2012, 6, 123–129. [Google Scholar] [CrossRef]
- Bertani, G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [CrossRef]
- Gipson, I.K.; Spurr-Michaud, S.; Argueso, P.; Tisdale, A.; Ng, T.F.; Russo, C.L. Mucin gene expression in immortalized human corneal-limbal and conjunctival epithelial cell lines. Investig. Ophthalmol. Vis. Sci. 2003, 44, 2496–2506. [Google Scholar] [CrossRef]
- Dos Santos, A.; Balayan, A.; Funderburgh, M.L.; Ngo, J.; Funderburgh, J.L.; Deng, S.X. Differentiation Capacity of Human Mesenchymal Stem Cells into Keratocyte Lineage. Investig. Ophthalmol. Vis. Sci. 2019, 60, 3013–3023. [Google Scholar] [CrossRef]
- Nozato, K.; Fujita, J.; Kawaguchi, M.; Ohara, G.; Morishima, Y.; Ishii, Y.; Huang, S.K.; Kokubu, F.; Satoh, H.; Hizawa, N. IL-17F Induces CCL20 in Bronchial Epithelial Cells. J. Allergy 2011, 2011, 587204. [Google Scholar] [CrossRef]
- Erez-Roman, R.; Pienik, R.; Futerman, A.H. Increased ceramide synthase 2 and 6 mRNA levels in breast cancer tissues and correlation with sphingosine kinase expression. Biochem. Biophys. Res. Commun. 2010, 391, 219–223. [Google Scholar] [CrossRef]
- Sun, Y.; Guo, Q.M.; Liu, D.L.; Zhang, M.Z.; Shu, R. In vivo expression of Toll-like receptor 2, Toll-like receptor 4, CSF2 and LY64 in Chinese chronic periodontitis patients. Oral Dis. 2010, 16, 343–350. [Google Scholar] [CrossRef]
- Schultz, K.R.; Klarnet, J.P.; Gieni, R.S.; HayGlass, K.T.; Greenberg, P.D. The role of B cells for in vivo T cell responses to a Friend virus-induced leukemia. Science 1990, 249, 921–923. [Google Scholar] [CrossRef]
- Glushakova, O.; Kosugi, T.; Roncal, C.; Mu, W.; Heinig, M.; Cirillo, P.; Sanchez-Lozada, L.G.; Johnson, R.J.; Nakagawa, T. Fructose induces the inflammatory molecule ICAM-1 in endothelial cells. J. Am. Soc. Nephrol. 2008, 19, 1712–1720. [Google Scholar] [CrossRef]
- Dabkeviciene, D.; Sasnauskiene, A.; Leman, E.; Kvietkauskaite, R.; Daugelaviciene, N.; Stankevicius, V.; Jurgelevicius, V.; Juodka, B.; Kirveliene, V. mTHPC-mediated photodynamic treatment up-regulates the cytokines VEGF and IL-1alpha. Photochem. Photobiol. 2012, 88, 432–439. [Google Scholar] [CrossRef]
- Kulik, T.J. Inhaled nitric oxide in the management of congenital heart disease. Curr. Opin. Cardiol. 1996, 11, 75–80. [Google Scholar] [CrossRef]
- Morris, M.C.; Gilliam, E.A.; Button, J.; Li, L. Dynamic modulation of innate immune response by varying dosages of lipopolysaccharide (LPS) in human monocytic cells. J. Biol. Chem. 2014, 289, 21584–21590. [Google Scholar] [CrossRef]
- Huntington, J.T.; Shields, J.M.; Der, C.J.; Wyatt, C.A.; Benbow, U.; Slingluff, C.L., Jr.; Brinckerhoff, C.E. Overexpression of collagenase 1 (MMP-1) is mediated by the ERK pathway in invasive melanoma cells: Role of BRAF mutation and fibroblast growth factor signaling. J. Biol. Chem. 2004, 279, 33168–33176. [Google Scholar] [CrossRef] [PubMed]
- Safranek, J.; Pesta, M.; Holubec, L.; Kulda, V.; Dreslerova, J.; Vrzalova, J.; Topolcan, O.; Pesek, M.; Finek, J.; Treska, V. Expression of MMP-7, MMP-9, TIMP-1 and TIMP-2 mRNA in lung tissue of patients with non-small cell lung cancer (NSCLC) and benign pulmonary disease. Anticancer Res. 2009, 29, 2513–2517. [Google Scholar] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).