Does an Antibiotic Stewardship Applied in a Pig Farm Lead to Low ESBL Prevalence?
Abstract
1. Introduction
2. Material and Methods
2.1. Swabs Recovery
2.2. Selection of Resistant Isolates
2.3. Identification and Phenotype of Resistance of Isolates
2.4. Identification of β-Lactamase Genes
2.5. Genetic Subtyping
2.6. Plasmid Characterization
3. Results
3.1. Prevalence of β-Lactamase-Producing and Colistin-Resistant Enterobacterales
3.2. Antimicrobial Resistance Features of Colistin-Resistant Isolates
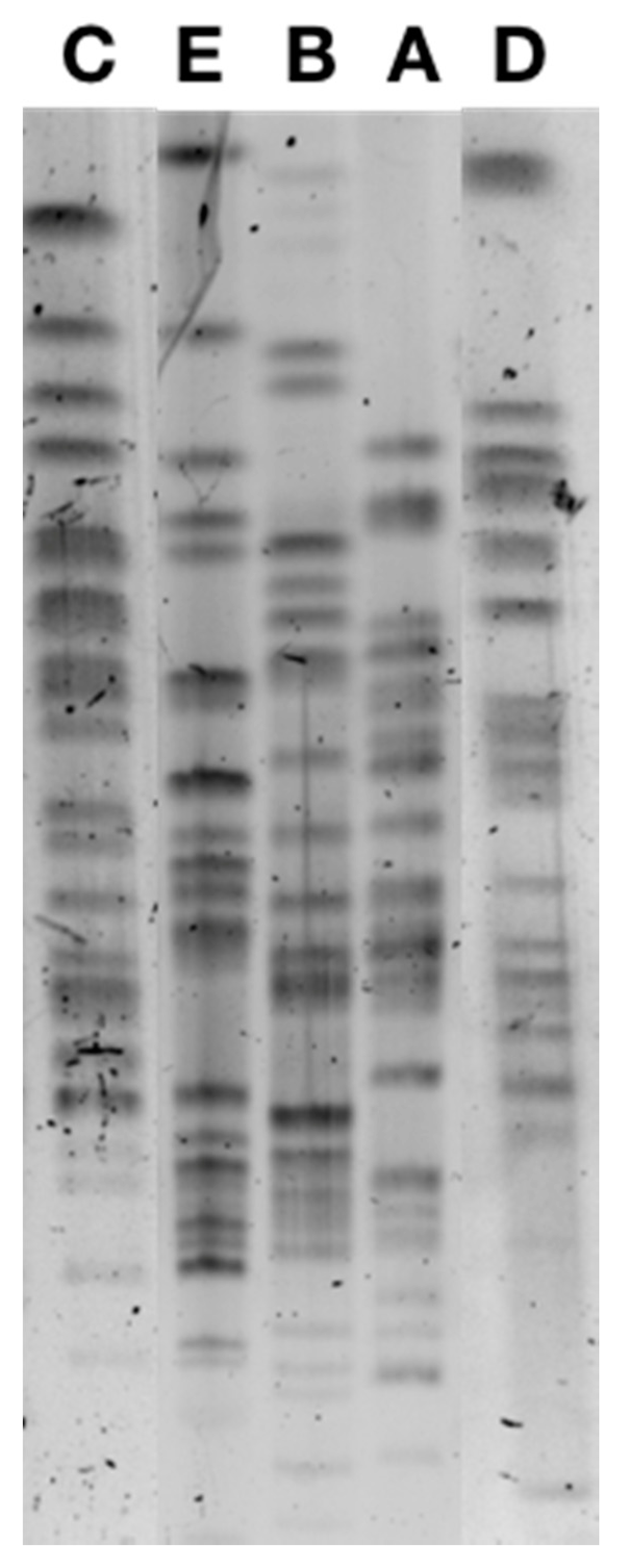
3.3. Clonal Relationship
3.4. Plasmid Analysis
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Amusi, J.; Tamara, L.; Horton, R.; Winkler, A.S. Reconnecting for our future: The Lancet One Health commission. Lancet 2020, 395, 1469–1471. [Google Scholar] [CrossRef]
- Colligon, P.J.; McEwen, S.A. One Health—Its importance in helping to better control antimicrobial resistance. Trop. Med. Infect. Dis. 2019, 4, 1–21. [Google Scholar]
- Aarestrup, F.M.; Wegner, H.C.; Collignon, P. Resistane in bacteria of the food chain: Epidemiology and control strategies. Expert Rev. Anti-Infect. Ther. 2008, 6, 733–750. [Google Scholar] [CrossRef]
- Bergšpica, I.; Kaprou, G.; Alexa, E.A.; Prieto, M.; Alvarez-Ordóñez, A. Extended spectrum β-lactamase (ESBL) Escherichia coli in pigs and pork meat in European Union. Antibiotics 2020, 9, 678. [Google Scholar] [CrossRef] [PubMed]
- Suis, A.G. Available online: https://www.suisag.ch/fr/gesundheit/programme-sante-suissano (accessed on 22 February 2021).
- Nordmann, P.; Jayol, A.; Poirel, L. Rapid detection of polymyxin resistance in Enterobacteriaceae. Emerg. Infect. Dis. 2016, 22, 1038–1043. [Google Scholar] [CrossRef]
- Nordmann, P.; Mazé, A.; Culebras, E.; Dobias, J.; Jayol, A.; Poirel, L. A culture medium screening 16S rRNA methylase-porducing pan-aminoglycoside resistant Gram-negative bacteria. Diag. Microbiol. Infect. Dis. 2018, 91, 118–122. [Google Scholar] [CrossRef]
- Nordmann, P.; Jayol, A.; Poirel, L. An universal culture medium for screening polymyxin-resistant Gram-negative isoaltes. J. Clin. Microbiol. 2016, 54, 1395–1399. [Google Scholar] [CrossRef]
- Dortet, L.; Poirel, L.; Nordmann, P. Rapid detection of ESBL-producing Enterobacteriaceae in blood culture. Emerg. Infect. Dis. 2015, 21, 504–507. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L.; Dortet, L. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 2012, 18, 1503–1507. [Google Scholar] [CrossRef]
- European Comittee on Antimicrobial Susceptiblity Testing. In Breakpoint Tables for Interpretation of MICs and Zone Diameters; Version 10.0; European Comittee on Antimicrobial Susceptiblity Testing: Växjö, Sweden, 2020.
- Fournier, C.; Aires-de-Sousa, M.; Nordmann, P.; Poirel, L. Occurrence of CTX-M-15 and MCR-1-producing Enterobacterales in pigs, Portugal; evidences of direct links with antibiotic selective pressure. Int. J. Antimicrob. Agents 2019, 55, 105802. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cg (accessed on 22 February 2021).
- Centers for Disease Control and Prevention. Available online: https://www.cdc.gov/pulsenet/pdf/ecoli-shigella-salmonella-pfge-protocol-508c.pdf (accessed on 22 February 2021).
- Public Databases for Molecular Typing and Microbial Genome Diversity. Available online: https://pubmlst.org/bigsdb?db=pubmlst_ecoli_achtman_seqdef (accessed on 22 February 2021).
- Kieser, T. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 1984, 12, 19–36. [Google Scholar] [CrossRef]
- Zurfluh, K.; Jakobi, G.; Stephan, R.; Hächler, H.; Nüesch-Inderbinen, M. Replicon typing of plasmids carrying blaCTX-M-1 in Enterobacteriaceae of animal, environmental and human origin. Front. Microb. 2014, 5, 555. [Google Scholar] [CrossRef]
- Carattoli, A.; Bertini, A.; Villa, L.; Falbo, V.; Hopkins, K.L.; Threfall, E.J. Identification of plasmids by PCR-based replicon typing. Microbiol. Methods 2005, 63, 219–228. [Google Scholar] [CrossRef] [PubMed]
- The Comprehensive Antibiotic Resistance Database. Available online: https://card.mcmaster.ca (accessed on 22 February 2021).
- Center for Genomic Epidemiology. Available online: https://cge.cbs.dtu.dk/services/PlasmidFinder/ (accessed on 22 February 2021).
- Kieffer, N.; Aires-de-Sousa, M.; Nordmann, P.; Poirel, L. High rate of MCR-1-producing Escherichia coli and Klebsiella pneumoniae among pigs, Portugal. Emerg. Infect. Dis. 2017, 23, 2023–2029. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Sun, L.; Ding, B.; Yang, Y.; Xu, X.; Liu, W.; Zhu, D.; Yang, F.; Zhang, H.; Hu, F. Outbreak of NDM-1-producing Klebsiella pneumoniae ST76 and ST37 isolates in neonates. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 611–618. [Google Scholar] [CrossRef]
- Su, S.; Zhao, Y.; Yu, L.; Li, C.; Wang, Y.; Wang, Y.; Boa, M.; Fu, Y.; Li, C.; Zang, X. Outbreak of KPC-2 carbapenem-resistant Klebsiella pneumoniae ST76 and carbapenem-resistant K2 hypervirulent Klebsiella pneumoniae ST375 strains in Northeast China: Molecular and virulent characteristics. BMC Infect. Dis. 2020, 20, 472. [Google Scholar] [CrossRef]
- Gong, X.; Zhang, J.; Su, J.; Fu, Y.; Bao, M.; Wang, Y.; Zhang, X. Characterization and epidemiology of carbapenem non-susceptible Enterobacteriacae isolates in the Eastern region of Heilongjiang Province, China. BMC Infect. Dis. 2018, 18, 417. [Google Scholar] [CrossRef]
- Day, M.J.; Hopkins, K.L.; Warenham, D.W.; Toleman, M.A.; Elviss, N.; Randall, L.; Teale, C.; Cleary, P.; Wiuff, C.; Doumith, M.; et al. Extended-spectrum β-lactamase-producing Escherichia coli in human-derived and food chain-derived samples from England, Wales, and Scotland: An epidemiological surveillance and typing study. Lancet Infect. Dis. 2019, 19, 1325–1335. [Google Scholar] [CrossRef]
- Zurfluh, K.; Wang, J.; Klumpp, J.; Nüesch-Inderbinden, M.; Fanning, S.; Stephan, R. Vertical transmission of highly simialr blaCTX-M-1-harboring IncI1 plasmids in Escherichia coli with different MLST types in the poultry production pyramid. Front. Microbiol. 2014, 5, 519. [Google Scholar] [CrossRef]
- Geser, N.; Stephan, R.; Korczak, B.M.; Beutin, L.; Hächler, H. Molecular identification of extended-spectrum-β-lactamase from Enterobacteriaceae isolated from healthy human carriers in Switzerland. Antimicrob. Agents Chemother. 2012, 56, 1609–1612. [Google Scholar] [CrossRef] [PubMed]
- Valcek, A.; Roer, L.; Overballe-Petersen, S.; Hansen, F.; Bortolaia, V.; Leekitcharoenphon, P.; Korsgaard, H.B.; Seyfarth, A.M.; Hendriksen, R.S.; Hasman, H.; et al. IncI1 ST3 and IncI1 ST7 plasmids form CTX-M-1 producing Escherichia coli obtained from patients with bloodstream infections are closely related to plasmids form E. coli of animal origin. J. Antimicrob. Chemother. 2019, 74, 2171–2175. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.A.; Magnuson, R.D. Modular organisatzion of the PhD repressor/antitoxin protein. J. Bacteriol. 2004, 186, 2692–2698. [Google Scholar] [CrossRef]
- Carattoli, A.; Villa, L.; Fortini, D.; García-Fernández, A. Contemporary IncI1 plasmids involved in the transmission and spread of antimicrobial resistance in Enterobacteriaceae. Plasmid 2018. [Google Scholar] [CrossRef] [PubMed]
- Swiss Interior Federal Department Ordinance Regulation the Hygiene in Diary Production (OHyPL). RS 916.351.021.1 (State at the 8 December 2020). Available online: https://www.fedlex.admin.ch/eli/cc/2005/824/fr (accessed on 22 February 2021).
- Darphorn, T.S.; Bel, K.; Koenders-van Sint Anneland, B.B.; Brul, S.; Ter Kuile, B.H. Antibiotic resistance plasmid composition and architecture in Escherichia coli isolates form meat. Sci. Rep. 2021, 11, 2136. [Google Scholar] [CrossRef] [PubMed]
- Rossolini, G.M.; D’Andrea, M.M.; Mugnaioli, C. The spread of CTX-M-type extended-spectrum β-lactamases. Clin. Microbiol. Infect. 2008, 14, 33–41. [Google Scholar] [CrossRef] [PubMed]
| Stable Number | Number of Pigs | Number of Resistant Isolates | Type of Livestock | Weight of Piglets | Clones Present | Rate of Pigs Carrying Resistant Strains | Resistant Determinant | Co-Resistance Phenotype |
|---|---|---|---|---|---|---|---|---|
| 1 | 8 | 7 | Fattening | 40 to 42 kg | A (n = 6) | 87.50% | blaCTX-M-1 | SUL, TET a (n = 1); SUL (n = 4); |
| None (n = 1) | ||||||||
| C (n = 1) | blaCTX-M-1 | NAL, CIP, TET, SUL, CHL, FLO (n = 1) | ||||||
| 2 | 8 | 5 | Fattening | 40 to 42 kg | A (n = 5) | 62.50% | blaCTX-M-1 | SUL (n = 4); NAL, CIP, TET, SUL, CHL, FLO (n = 1) |
| 3 | 6 | 2 | Fattening | 102 kg | A (n = 1) | 33.30% | blaCTX-M-1 | SUL (n = 1) |
| B1 (n = 1) | blaCTX-M-1 | SUL (n = 1) | ||||||
| 4 | 10 | 4 | Weaning | 18 to 20 kg | A (n = 4) | 40% | blaCTX-M-1 | SUL (n = 4) |
| 5 | 10 | 8 | Weaning | 18 to 20 kg | A (n = 3) | 60% | blaCTX-M-1 | SUL (n = 3) |
| B1 (n = 2) | blaCTX-M-1 | SUL, TET (n = 2) | ||||||
| E (n = 1) | blaCTX-M-1 | TET (n = 1) | ||||||
| E. cloacae (n = 1) | blaCTX-M-1 | SUL, TET (n = 1) | ||||||
| K. pneumoniae (n = 1) | mgrB truncation | SUL (n = 1) | ||||||
| 6 | 20 | 4 | Weaning | 9 to 10 kg | A (n = 2) | 20% | blaCTX-M-1 | SUL (n = 1); NAL, CIP, SUL, TET, CHL, FLO (n = 1); |
| E. cloacae (n = 1) | ND d | None (n = 1) | ||||||
| K. pneumoniae (n = 1) | mgrB truncation | None (n = 1) | ||||||
| 7 | 10 | 8 | Reproduction | - | B1 (n = 7) | blaCTX-M-1 | SUL, TET (n = 7) | |
| D (n = 1) | blaCTX-M-1 | GMI, KMN, TMN, SUL, TET (n = 1) | ||||||
| 8 | 3 | 2 | Sow b with 3 piglets | B1 (n = 2) | 66% | blaCTX-M-1 | SUL, TET (n = 2) | |
| 9 | 3 | 2 | Sow c with 3 piglets | B2 (n = 1) | 33% | blaTEM-1 | KMN, SUL, SXT, TET (n = 1) | |
| B1 (n = 1) | 33% | blaCTX-M-1 | SUL, TET (n = 1) | |||||
| 10 | 3 | 1 | Sow d with 3 piglets | A (n = 1) | 33% | blaCTX-M-1 | SUL (n = 1) |
| Number of Isolates | Species | Phylogenic Group | ST | Pulsotype | Resistance Determinants | Incompatibility Group of the Plasmid Carrying blaCTX-M-1 | Coresistance on the Plasmid Carrying blaCTX-M-1 |
|---|---|---|---|---|---|---|---|
| 22 | E. coli | A | ST10 | A | blaCTX-M-1 | Inc I1 | TET, SUL a |
| 12 | E. coli | A | ST10 | B1 b | blaCTX-M-1 | Inc I1 | TET, SUL |
| 1 | E. coli | A | ST10 | B2 b | blaCTX-M-1 | IncI1 | GMI, KMN, TMN, SUL, TET |
| 1 | E. coli | A | ST744 | C | blaCTX-M-1 | Inc I1 | TET, SUL |
| 1 | E. coli | A | ST34 | D | blaCTX-M-1 | Inc I1 | TET, SUL |
| 1 | E. coli | A | ST10 | E | blaCTX-M-1 | Inc I1 | TET, SUL |
| 1 | E. cloacae | ND | blaCTX-M-1 | Inc I1 | TET, SUL |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fournier, C.; Nordmann, P.; Pittet, O.; Poirel, L. Does an Antibiotic Stewardship Applied in a Pig Farm Lead to Low ESBL Prevalence? Antibiotics 2021, 10, 574. https://doi.org/10.3390/antibiotics10050574
Fournier C, Nordmann P, Pittet O, Poirel L. Does an Antibiotic Stewardship Applied in a Pig Farm Lead to Low ESBL Prevalence? Antibiotics. 2021; 10(5):574. https://doi.org/10.3390/antibiotics10050574
Chicago/Turabian StyleFournier, Claudine, Patrice Nordmann, Olivier Pittet, and Laurent Poirel. 2021. "Does an Antibiotic Stewardship Applied in a Pig Farm Lead to Low ESBL Prevalence?" Antibiotics 10, no. 5: 574. https://doi.org/10.3390/antibiotics10050574
APA StyleFournier, C., Nordmann, P., Pittet, O., & Poirel, L. (2021). Does an Antibiotic Stewardship Applied in a Pig Farm Lead to Low ESBL Prevalence? Antibiotics, 10(5), 574. https://doi.org/10.3390/antibiotics10050574






