Rhodomyrtone Accumulates in Bacterial Cell Wall and Cell Membrane and Inhibits the Synthesis of Multiple Cellular Macromolecules in Epidemic Methicillin-Resistant Staphylococcus aureus
Abstract
1. Introduction
2. Results and Discussion
2.1. Antibacterial Effects of Rhodomyrtone and Other Antimicrobial Agents
2.2. Localisation of Rhodomyrtone in Pathogenic Bacteria
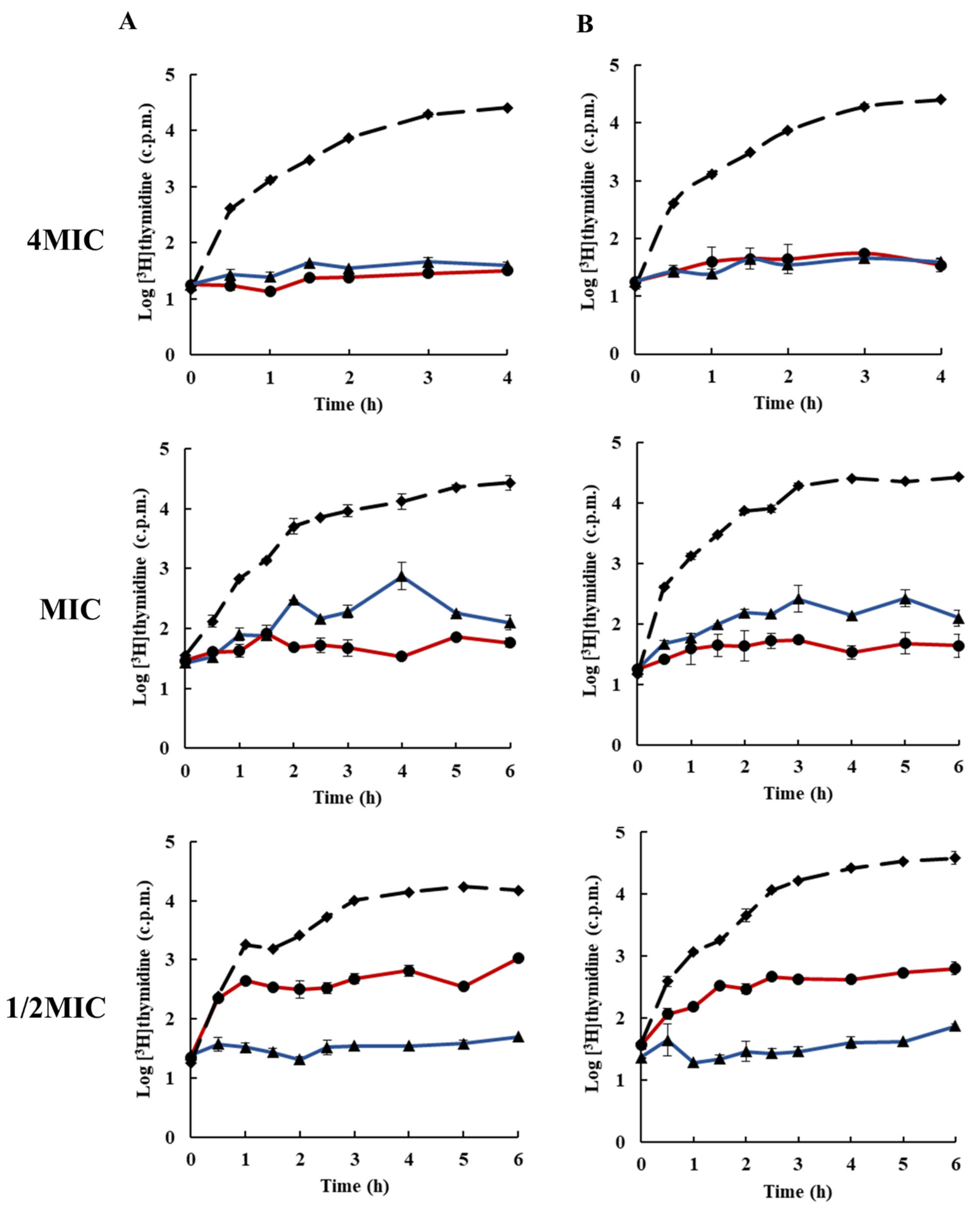
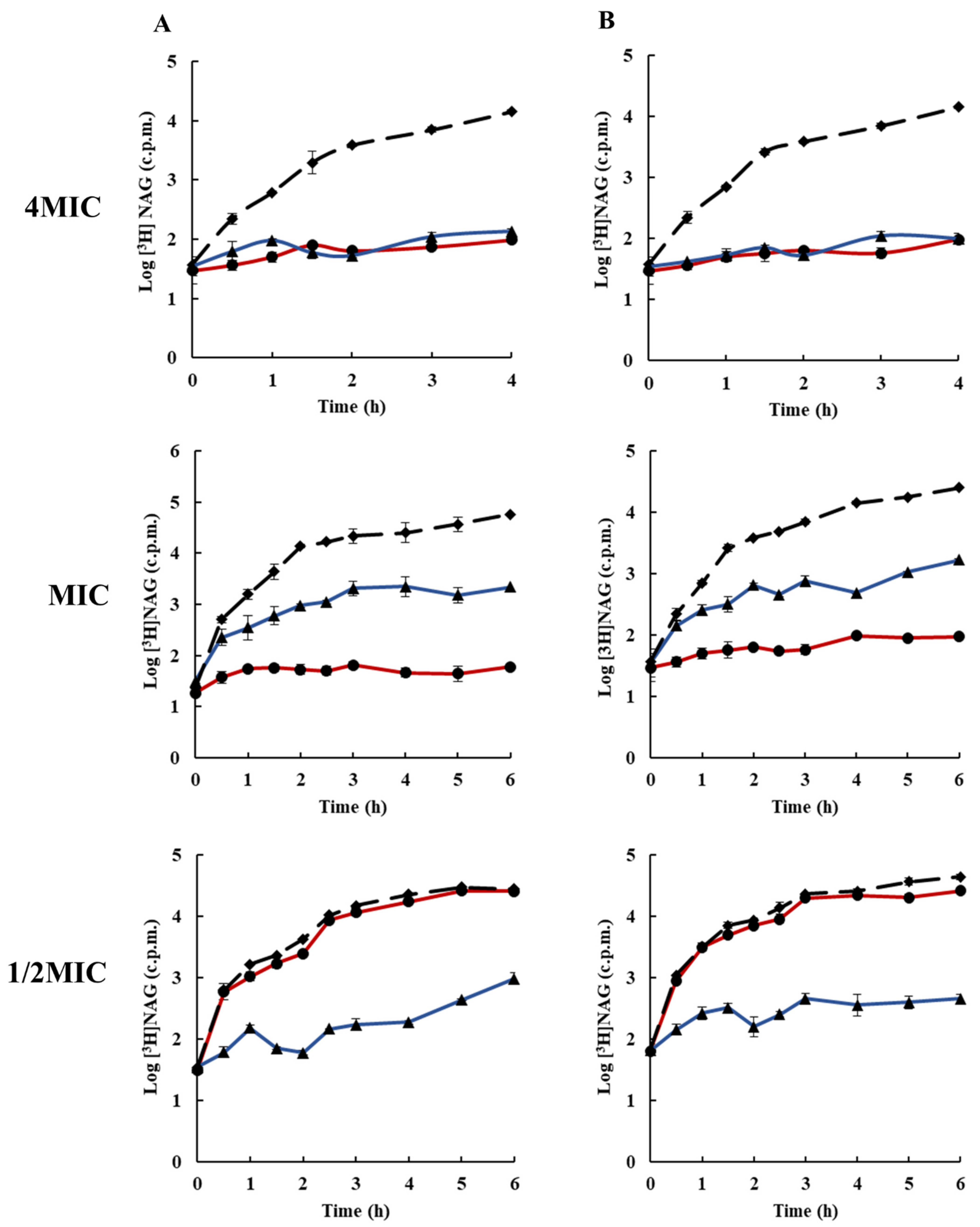
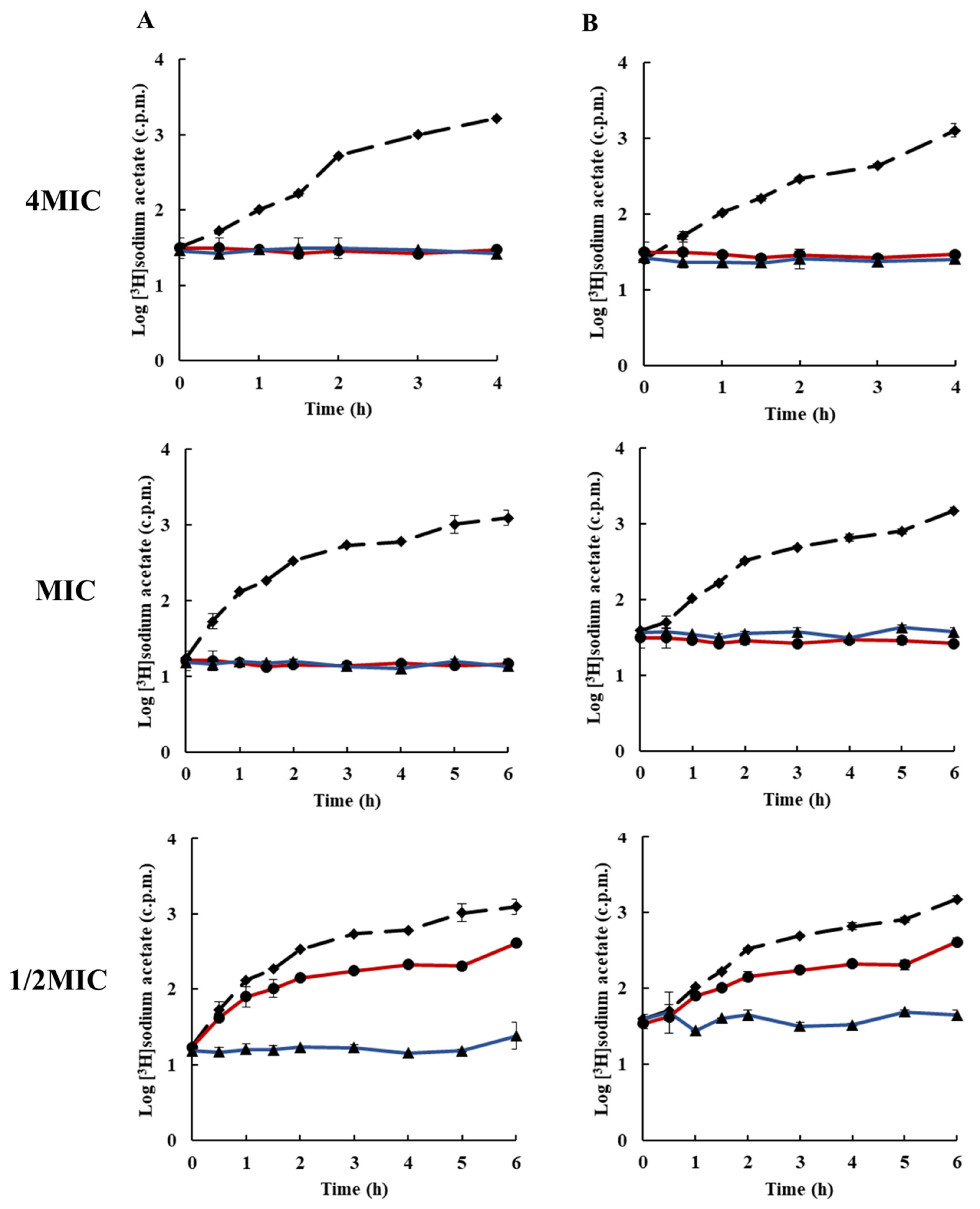
2.3. Effects of Rhodomyrtone on Macromolecular Biosynthesis
2.4. Effect of Rhodomyrtone on the Extracellular Lipase Activity in Staphylococcus aureus
3. Materials and Methods
3.1. Antibiotics and Chemicals
3.2. Bacterial Strains and Growth Conditions
3.3. Antibacterial Assay
3.4. Primary Screening for Rhodomyrtone Target in Staphylococcus aureus
3.5. Inhibition of Macromolecular Biosynthesis
3.6. Lipase Activity Assay
3.7. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Haysom, L.; Cross, M.; Anastasas, R.; Moore, E.; Hampton, S. Prevalence and risk factors for methicillin-resistant Staphylococcus aureus (MRSA) infections in custodial populations: A systematic review. J. Correct. Health Care 2018, 24, 197–213. [Google Scholar] [CrossRef] [PubMed]
- Purrello, S.; Garau, J.; Giamarellos, E.; Mazzei, T.; Pea, F.; Soriano, A.; Stefani, S. Methicillin-resistant Staphylococcus aureus infections: A review of the currently available treatment options. J. Glob. Antimicrob. Resist. 2016, 7, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Magrini, N.; Kahlmeter, G.; Singh, N. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organ. 2017, 27, 318–327. [Google Scholar]
- Holmes, N.E.; Tong, S.Y.; Davis, J.S.; Van Hal, S.J. Treatment of methicillin-resistant Staphylococcus aureus: Vancomycin and beyond. In Seminars in Respiratory and Critical Care Medicine; Thieme Medical Publishers: New York, NY, USA, 2015; pp. 17–30. [Google Scholar]
- Choo, E.J.; Chambers, H.F. Treatment of methicillin-resistant Staphylococcus aureus bacteremia. Infect. Chemother. 2016, 48, 267. [Google Scholar] [CrossRef] [PubMed]
- Wilsey, H.A.; Burgess, D.R.; Burgess, D.S. Focusing the lens on the CAMERA concepts: Early combination β-lactam and vancomycin therapy in methicillin-resistant Staphylococcus aureus bacteremia. Antimicrob. Agents Chemother. 2020, 64. [Google Scholar] [CrossRef]
- Tong, S.Y.; Lye, D.C.; Yahav, D.; Sud, A.; Robinson, J.O.; Nelson, J.; Archuleta, S.; Roberts, M.A.; Cass, A.; Paterson, D.L. Effect of vancomycin or daptomycin with vs without an antistaphylococcal β-lactam on mortality, bacteremia, relapse, or treatment failure in patients with MRSA bacteremia: A randomized clinical trial. JAMA 2020, 323, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Ontong, J.C.; Ozioma, N.F.; Voravuthikunchai, S.P.; Chusri, S. Synergistic antibacterial effects of colistin in combination with aminoglycoside, carbapenems, cephalosporins, fluoroquinolones, tetracyclines, fosfomycin, and piperacillin on multidrug resistant Klebsiella pneumoniae isolates. PLoS ONE 2021, 16, e0244673. [Google Scholar] [CrossRef] [PubMed]
- Nwabor, O.F.; Terbtothakun, P.; Voravuthikunchai, S.P.; Chusri, S. Evaluation of the synergistic antibacterial effects of fosfomycin in combination with selected antibiotics against carbapenem–resistant Acinetobacter baumannii. Pharmaceuticals 2021, 14, 185. [Google Scholar] [CrossRef]
- Umaarasu, T.; Padmavathy, K.; Thirunavukkarasu, D.; Rajesh, S.; Govindaraj, J.; Shanmugam, G. Evaluation of the antimicrobial activity and phytochemical investigation of the leaf extracts of Ricinus communis Linn. against drug-resistant bacterial pathogens. Drug Invent. Today 2019, 11, 1299–1303. [Google Scholar]
- Ali, M.; Yahaya, A.; Zage, A.; Yusuf, Z. In-vitro antibacterial activity and phytochemical screening of Psidium guajava on some enteric bacterial isolates of public health importance. J. Adv. Med Pharm. Sci. 2017. [Google Scholar] [CrossRef]
- Akanmu, A.O.; Bulama, Y.A.; Balogun, S.T.; Musa, S. Antibacterial activities of aqueous and methanol leaf extracts of Solanum incanum linn.(Solanaceae) against multi-drug resistant bacterial isolates. Afr. J. Microbiol. Res. 2019, 13, 70–76. [Google Scholar]
- Ramachandran, G.; Rajivgandhi, G.; Maruthupandy, M.; Manoharan, N. Extraction and partial purification of secondary metabolites from endophytic actinomycetes of marine green algae Caulerpa racemosa against multi drug resistant uropathogens. Biocatal. Agric. Biotechnol. 2019, 17, 750–757. [Google Scholar] [CrossRef]
- Rajivgandhi, G.; Ramachandran, G.; Maruthupandy, M.; Saravanakumar, S.; Manoharan, N.; Viji, R. Antibacterial effect of endophytic actinomycetes from marine algae against multi drug resistant gram negative bacteria. Exam Mar. Biol. Oceanogr. 2018, 1, 1–8. [Google Scholar]
- Srisuwan, S.; Voravuthikunchai, S.P. Rhodomyrtus tomentosa leaf extract inhibits methicillin-resistant staphylococcus aureus adhesion, invasion, and intracellular survival in human HaCaT keratinocytes. Microb. Drug Resist. 2017, 23, 1002–1012. [Google Scholar] [CrossRef] [PubMed]
- Saising, J.; Hiranrat, A.; Mahabusarakam, W.; Ongsakul, M.; Voravuthikunchai, S.P. Rhodomyrtone from Rhodomyrtus tomentosa (Aiton) Hassk. as a natural antibiotic for staphylococcal cutaneous infections. J. Health Sci. 2008, 54, 589–595. [Google Scholar] [CrossRef]
- Bach, Q.N.; Hongthong, S.; Quach, L.T.; Pham, L.V.; Pham, T.V.; Kuhakarn, C.; Reutrakul, V.; Nguyen, P.T. Antimicrobial activity of rhodomyrtone isolated from Rhodomyrtus tomentosa (Aiton) Hassk. Nat. Prod. Res. 2018, 34, 2518–2523. [Google Scholar] [CrossRef] [PubMed]
- Limsuwan, S.; Hesseling-Meinders, A.; Voravuthikunchai, S.P.; Van Dijl, J.M.; Kayser, O. Potential antibiotic and anti-infective effects of rhodomyrtone from Rhodomyrtus tomentosa (Aiton) Hassk. on Streptococcus pyogenes as revealed by proteomics. Phytomedicine 2011, 18, 934–940. [Google Scholar] [CrossRef]
- Srisuwan, S.; Mackin, K.E.; Hocking, D.; Lyras, D.; Bennett-Wood, V.; Voravuthikunchai, S.P.; Robins-Browne, R.M. Antibacterial activity of rhodomyrtone on Clostridium difficile vegetative cells and spores in vitro. Int. J. Antimicrob. Agents 2018, 52, 724–729. [Google Scholar] [CrossRef]
- Sianglum, W.; Srimanote, P.; Wonglumsom, W.; Kittiniyom, K.; Voravuthikunchai, S.P. Proteome analyses of cellular proteins in methicillin-resistant Staphylococcus aureus treated with rhodomyrtone, a novel antibiotic candidate. PLoS ONE 2011, 6, e16628. [Google Scholar] [CrossRef]
- Saeloh, D.; Tipmanee, V.; Jim, K.K.; Dekker, M.P.; Bitter, W.; Voravuthikunchai, S.P.; Wenzel, M.; Hamoen, L.W. The novel antibiotic rhodomyrtone traps membrane proteins in vesicles with increased fluidity. PLoS Pathog. 2018, 14, e1006876. [Google Scholar] [CrossRef]
- Sianglum, W.; Srimanote, P.; Taylor, P.W.; Rosado, H.; Voravuthikunchai, S.P. Transcriptome analysis of responses to rhodomyrtone in methicillin-resistant Staphylococcus Aureus. PLoS ONE 2012, 7, e45744. [Google Scholar] [CrossRef]
- Leejae, S.; Taylor, P.W.; Voravuthikunchai, S.P. Antibacterial mechanisms of rhodomyrtone against important hospital-acquired antibiotic-resistant pathogenic bacteria. J. Med. Microbiol. 2013, 62, 78–85. [Google Scholar] [CrossRef]
- Saising, J.; Ongsakul, M.; Voravuthikunchai, S.P. Rhodomyrtus tomentosa (Aiton) Hassk. ethanol extract and rhodomyrtone: A potential strategy for the treatment of biofilm-forming staphylococci. J. Med Microbiol. 2011, 60, 1793–1800. [Google Scholar] [CrossRef]
- Saising, J.; Voravuthikunchai, S.P. Anti Propionibacterium acnes activity of rhodomyrtone, an effective compound from Rhodomyrtus tomentosa (Aiton) Hassk. leaves. Anaerobe 2012, 18, 400–404. [Google Scholar] [CrossRef]
- Mitsuwan, W.; Wintachai, P.; Voravuthikunchai, S.P. Rhodomyrtus tomentosa leaf extract and rhodomyrtone combat Streptococcus pneumoniae biofilm and inhibit invasiveness to human lung epithelial and enhance pneumococcal phagocytosis by macrophage. Curr. Microbiol. 2020, 77, 3546–3554. [Google Scholar] [CrossRef]
- Liu, H.-X.; Tan, H.-B.; Qiu, S.-X. Antimicrobial acylphloroglucinols from the leaves of Rhodomyrtus tomentosa. J. Asian Nat. Prod. Res. 2016, 18, 535–541. [Google Scholar] [CrossRef]
- Traithan, A.; Tongtawe, P.; Thanongsaksrikul, J.; Voravuthikunchai, S.; Srimanote, P. Antibacterial mechanism of rhodomyrtone involves the disruption of nucleoid segregation checkpoint in Streptococcus suis. AMB Express 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Saising, J.; Nguyen, M.-T.; Härtner, T.; Ebner, P.; Bhuyan, A.A.M.; Berscheid, A.; Muehlenkamp, M.; Schäkermann, S.; Kumari, N.; Maier, M.E. Rhodomyrtone (Rom) is a membrane-active compound. Biochim. Et Biophys. Acta (Bba)-Biomembr. 2018, 1860, 1114–1124. [Google Scholar] [CrossRef]
- Odedina, G.F.; Vongkamjan, K.; Voravuthikunchai, S.P. Potential bio-control agent from Rhodomyrtus tomentosa against Listeria monocytogenes. Nutrients 2015, 7, 7451–7468. [Google Scholar] [CrossRef]
- Saising, J.; Götz, F.; Dube, L.; Ziebandt, A.K.; Voravuthikunchai, S.P. Inhibition of staphylococcal biofilm-related gene transcription by rhodomyrtone, a new antibacterial agent. Ann. Microbiol. 2015, 65, 659–665. [Google Scholar] [CrossRef]
- Sianglum, W.; Saeloh, D.; Tongtawe, P.; Wootipoom, N.; Indrawattana, N.; Voravuthikunchai, S.P. Early effects of rhodomyrtone on membrane integrity in methicillin-resistant Staphylococcus aureus. Microb. Drug Resist. 2018, 24, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Mitsuwan, W.; Jiménez-Munguía, I.; Visutthi, M.; Sianglum, W.; Jover, A.; Barcenilla, F.; García, M.; Pujol, M.; Gasch, O.; Domínguez, M.A.; et al. Rhodomyrtone decreases Staphylococcus aureus SigB activity during exponentially growing phase and inhibits haemolytic activity within membrane vesicles. Microb. Pathog. 2019, 128, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Mitsuwan, W.; Olaya-Abril, A.; Calderón-Santiago, M.; Jiménez-Munguía, I.; González-Reyes, J.A.; Priego-Capote, F.; Voravuthikunchai, S.P.; Rodríguez-Ortega, M.J. Integrated proteomic and metabolomic analysis reveals that rhodomyrtone reduces the capsule in Streptococcus pneumoniae. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wunnoo, S.; Saising, J.; Voravuthikunchai, S.P. Rhodomyrtone inhibits lipase production, biofilm formation, and disorganizes established biofilm in Propionibacterium acnes. Anaerobe 2017, 43, 61–68. [Google Scholar] [CrossRef]
- Hiranrat, A.; Mahabusarakam, W. New acylphloroglucinols from the leaves of Rhodomyrtus tomentosa. Tetrahedron 2008, 64, 11193–11197. [Google Scholar] [CrossRef]
- Limsuwan, S.; Trip, E.N.; Kouwen, T.R.; Piersma, S.; Hiranrat, A.; Mahabusarakam, W.; Voravuthikunchai, S.P.; van Dijl, J.M.; Kayser, O. Rhodomyrtone: A new candidate as natural antibacterial drug from Rhodomyrtus tomentosa. Phytomedicine 2009, 16, 645–651. [Google Scholar] [CrossRef]
- Mohamed, G.A.; Ibrahim, S.R. Eucalyptone G, a new phloroglucinol derivative and other constituents from Eucalyptus globulus Labill. Arkivoc 2007, 15, 281–291. [Google Scholar] [CrossRef]
- Salni, D.; Sargent, M.V.; Skelton, B.W.; Soediro, I.; Sutisna, M.; White, A.H.; Yulinah, E. Rhodomyrtone, an antibotic from Rhodomyrtus tomentosa. Aust. J. Chem. 2002, 55, 229–232. [Google Scholar] [CrossRef]
- CLSI. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; CLSI: Wayne, PA, USA, 2017. [Google Scholar]
- Hobbs, J.K.; Miller, K.; O’neill, A.J.; Chopra, I. Consequences of daptomycin-mediated membrane damage in Staphylococcus aureus. J. Antimicrob. Chemother. 2008, 62, 1003–1008. [Google Scholar] [CrossRef]
- Nostro, A.; Bisignano, G.; Cannatelli, M.A.; Crisafi, G.; Germano, M.P.; Alonzo, V. Effects of Helichrysum italicum extract on growth and enzymatic activity of Staphylococcus aureus. Int. J. Antimicrob. Agents 2001, 17, 517–520. [Google Scholar] [CrossRef]
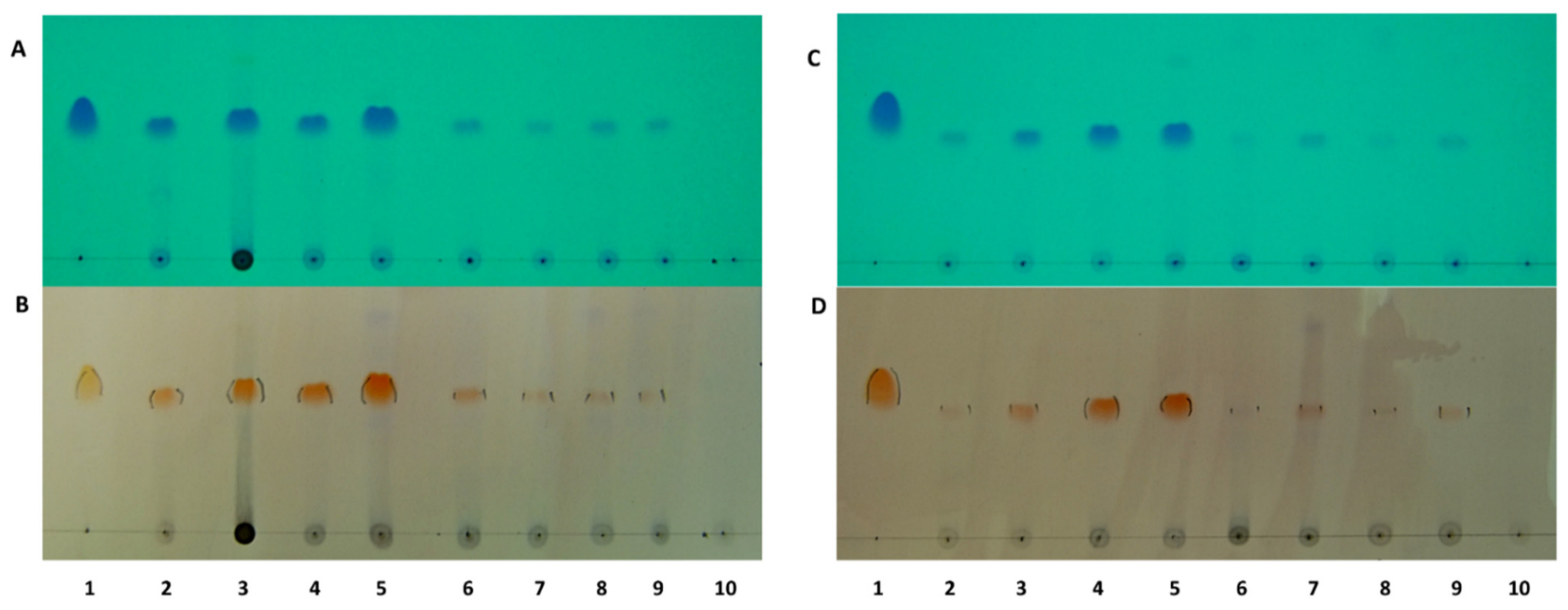


| Antimicrobial Agent | MIC/MBC (µg/mL) | |
|---|---|---|
| S. aureus ATCC 29213 | EMRSA-16 | |
| Rhodomyrtone | 0.5/1 | 0.5/0.5 |
| Chloramphenicol | 16/32 | 8/128 |
| ELB21 | 0.0625/0.125 | 0.0625/0.125 |
| Platensimycin | 1/>8 | 1/>8 |
| Rifampicin | 0.015/0.06 | 0.0625/0.125 |
| Vancomycin | 1/1 | 1/1 |
| Time (h) | Zone Diameter (mm) | |||
|---|---|---|---|---|
| Untreated | 0.5× MIC | 1× MIC | 4× MIC | |
| 0 | 9.0 | 8.5 | 7.0 | 7.7 |
| 1 | 17.0 | 16.7 | 17.3 | 19.0 |
| 2 | 18.6 | 16.3 | 15.5 | 16.7 |
| 3 | 16.7 | 15.7 | 15.7 | 16.0 |
| 4 | 17.0 | 14.7 | 16.7 | 17.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nwabor, O.F.; Leejae, S.; Voravuthikunchai, S.P. Rhodomyrtone Accumulates in Bacterial Cell Wall and Cell Membrane and Inhibits the Synthesis of Multiple Cellular Macromolecules in Epidemic Methicillin-Resistant Staphylococcus aureus. Antibiotics 2021, 10, 543. https://doi.org/10.3390/antibiotics10050543
Nwabor OF, Leejae S, Voravuthikunchai SP. Rhodomyrtone Accumulates in Bacterial Cell Wall and Cell Membrane and Inhibits the Synthesis of Multiple Cellular Macromolecules in Epidemic Methicillin-Resistant Staphylococcus aureus. Antibiotics. 2021; 10(5):543. https://doi.org/10.3390/antibiotics10050543
Chicago/Turabian StyleNwabor, Ozioma F., Sukanlaya Leejae, and Supayang P. Voravuthikunchai. 2021. "Rhodomyrtone Accumulates in Bacterial Cell Wall and Cell Membrane and Inhibits the Synthesis of Multiple Cellular Macromolecules in Epidemic Methicillin-Resistant Staphylococcus aureus" Antibiotics 10, no. 5: 543. https://doi.org/10.3390/antibiotics10050543
APA StyleNwabor, O. F., Leejae, S., & Voravuthikunchai, S. P. (2021). Rhodomyrtone Accumulates in Bacterial Cell Wall and Cell Membrane and Inhibits the Synthesis of Multiple Cellular Macromolecules in Epidemic Methicillin-Resistant Staphylococcus aureus. Antibiotics, 10(5), 543. https://doi.org/10.3390/antibiotics10050543






