Use of Bacteriophage Amended with CRISPR-Cas Systems to Combat Antimicrobial Resistance in the Bacterial Foodborne Pathogen Listeria monocytogenes
Abstract
1. Introduction
1.1. Bacteriophage as Biocontrol against Listeria monocytogenes and Tool to Mitigate AMR
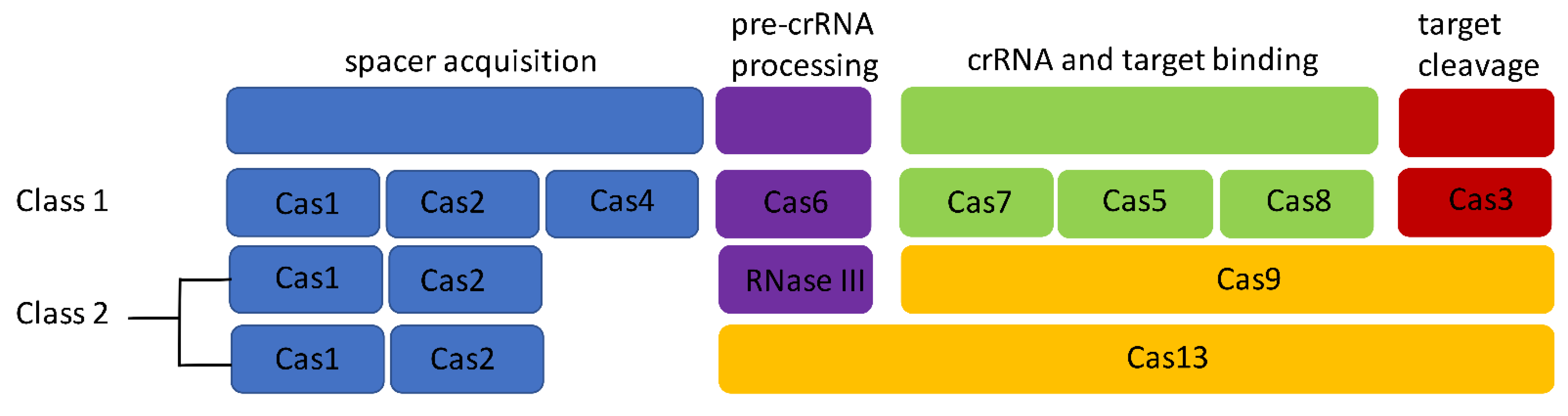
1.2. CRISPR Systems in Listeria
1.3. Phage-Mediated Delivery of Engineered CRISPR-Cas Systems
2. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Gandhi, M.; Chikindas, M.L. Listeria: A foodborne pathogen that knows how to survive. Int. J. Food Microbiol. 2007, 113, 1–15. [Google Scholar] [CrossRef]
- Freitag, N.E.; Port, G.C.; Miner, M.D. Listeria monocytogenes-from saprophyte to intracellular pathogen. Nat. Rev. Microbiol. 2009, 7, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Vivant, A.-L.; Garmyn, D.; Piveteau, P. Listeria monocytogenes, a down-to-earth pathogen. Front. Cell. Infect. Microbiol. 2013, 3, 87. [Google Scholar] [CrossRef] [PubMed]
- Nelson, K.E.; Fouts, D.E.; Mongodin, E.F.; Ravel, J.; DeBoy, R.T.; Kolonay, J.F.; Rasko, D.; Angiuoli, S.V.; Gill, S.R.; Paulsen, I.T.; et al. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 2004, 32, 2386–2395. [Google Scholar] [CrossRef]
- Swaminathan, B.; Gerner-Smidt, P. The epidemiology of human listeriosis. Microbes Infect. 2007, 9, 1236–1243. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States-Major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Schlech, W.F. Epidemiology and Clinical Manifestations of Listeria monocytogenes Infection. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Thønnings, S.; Knudsen, J.D.; Schønheyder, H.C.; Søgaard, M.; Arpi, M.; Gradel, K.O.; Østergaard, C.; Jensen, U.S.; Koch, K.; Smit, J.; et al. Antibiotic treatment and mortality in patients with Listeria monocytogenes meningitis or bacteraemia. Clin. Microbiol. Infect. 2016, 22, 725–730. [Google Scholar] [CrossRef]
- Baquero, F.; Lanza, V.F.; Duval, M.; Coque, T.M. Ecogenetics of antibiotic resistance in Listeria monocytogenes. Mol. Microbiol. 2020, 113, 570–579. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Li, M.; Luque-Sastre, L.; Wang, W.; Hu, Y.; Peng, Z.; Dong, Y.; Gan, X.; Nguyen, S.; Anes, J.; et al. Susceptibility (re)-testing of a large collection of Listeria monocytogenes from foods in China from 2012 to 2015 and WGS characterization of resistant isolates. J. Antimicrob. Chemother. 2019, 74, 1786–1794. [Google Scholar] [CrossRef]
- Troxler, R.; von Graevenitz, A.; Funke, G.; Wiedemann, B.; Stock, I. Natural antibiotic susceptibility of Listeria species: L. grayi, L. innocua, L. ivanovii, L. monocytogenes, L. seeligeri and L. welshimeri strains. Clin. Microbiol. Infect. 2000, 6, 525–535. [Google Scholar] [CrossRef]
- Needle, D.B.; Marr, J.L.; Park, C.J.; Andam, C.P.; Wise, A.G.; Maes, R.K.; Wilkes, R.P.; Anis, E.A.; Sidor, I.F.; Agnew, D.; et al. Concurrent infection of Skunk Adenovirus-1, Listeria monocytogenes, and a regionally specific clade of Canine Distemper Virus in ne Gray Fox (Urocyon cinereoargenteus) and concurrent listeriosis and canine distemper in a second Gray Fox. Pathogens 2020, 9, 591. [Google Scholar] [CrossRef]
- Haubert, L.; Mendonça, M.; Lopes, G.V.; de Itapema Cardoso, M.R.; da Silva, W.P. Listeria monocytogenes isolates from food and food environment harbouring tetM and ermB resistance genes. Lett. Appl. Microbiol. 2016, 62, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Parsons, C.; Chen, Y.; Niedermeyer, J.; Hernandez, K.; Kathariou, S. Draft genome sequence of multidrug-resistant Listeria innocua strain UAM003-1A, isolated from a wild black bear (Ursus americanus). Microbiol. Resour. Announc. 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). Antibiotic/antimicrobial Resistance. 2020. Available online: https://www.cdc.gov/drugresistance/ (accessed on 15 March 2021).
- World Health Organization (WHO). Fact Sheet: Antimicrobial Resistance. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 15 March 2021).
- Kirk, M.D.; Pires, S.M.; Black, R.E.; Caipo, M.; Crump, J.A.; Devleesschauwer, B.; Döpfer, D.; Fazil, A.; Fischer-Walker, C.L.; Hald, T.; et al. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: A data synthesis. PLoS Med. 2015, 12, e1001921. [Google Scholar]
- Bren, L. Bacteria-eating virus approved as food additive. FDA Consum. 2010, 41, 20–22. [Google Scholar]
- Knoll, B.M.; Mylonakis, E. Antibacterial bioagents based on principles of bacteriophage biology: An overview. Clin. Infect. Dis. 2014, 58, 1–7. [Google Scholar] [CrossRef]
- Lang, L.H. FDA approves use of bacteriophages to be added to meat and poultry products. Gastroenterology 2006, 131, 1370. [Google Scholar] [CrossRef] [PubMed]
- Guenther, S.; Huwyler, D.; Richard, S.; Loessner, M.J. Virulent bacteriophage for efficient biocontrol of Listeria monocytogenes in ready-to-eat foods. Appl. Environ. Microbiol. 2009, 75, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Guenther, S.; Loessner, M.J. Bacteriophage biocontrol of Listeria monocytogenes on soft ripened white mold and red-smear cheeses. Bacteriophage 2011, 1, 94–100. [Google Scholar] [CrossRef]
- Iacumin, L.; Manzano, M.; Comi, G. Phage inactivation of Listeria monocytogenes on San Daniele dry-cured ham and elimination of biofilms from equipment and working environments. Microorganisms 2016, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Soni, K.A.; Nannapaneni, R. Removal of Listeria monocytogenes biofilms with bacteriophage P100. J. Food Prot. 2010, 73, 1519–1524. [Google Scholar] [CrossRef] [PubMed]
- Bigot, B.; Lee, W.-J.; McIntyre, L.; Wilson, T.; Hudson, J.A.; Billington, C.; Heinemann, J.A. Control of Listeria monocytogenes growth in a ready-to-eat poultry product using a bacteriophage. Food Microbiol. 2011, 28, 1448–1452. [Google Scholar] [CrossRef] [PubMed]
- Eugster, M.R.; Morax, L.S.; Hüls, V.J.; Huwiler, S.G.; Leclercq, A.; Lecuit, M.; Loessner, M.J. Bacteriophage predation promotes serovar diversification in Listeria monocytogenes. Mol. Microbiol. 2015, 97, 33–46. [Google Scholar] [CrossRef]
- Kim, J.W.; Siletzky, R.M.; Kathariou, S. Host ranges of Listeria-specific bacteriophages from the turkey processing plant environment in the United States. Appl. Environ. Microbiol. 2008, 74, 6623–6630. [Google Scholar] [CrossRef]
- Brown, P.; Chen, Y.; Parsons, C.; Brown, E.; Loessner, M.J.; Shen, Y.; Kathariou, S. Whole genome sequence analysis of phage-resistant Listeria monocytogenes serotype 1/2a strains from turkey processing plants. Pathogens 2021, 10, 199. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1720. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo-Cantabrana, C.; Goh, Y.J.; Barrangou, R. Characterization and repurposing of Type I and Type II CRISPR–Cas systems in bacteria. J. Mol. Biol. 2019, 431, 21–33. [Google Scholar] [CrossRef]
- Roberts, A.; Barrangou, R. Applications of CRISPR-Cas systems in lactic acid bacteria. FEMS Microbiol. Rev. 2020, 44, 523–537. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary classification of CRISPR–Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef]
- Gomaa, A.A.; Klumpe, H.E.; Luo, M.L.; Selle, K.; Barrangou, R.; Beisel, C.L. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. MBio 2014, 5, e00928-13. [Google Scholar] [CrossRef]
- Yosef, I.; Manor, M.; Kiro, R.; Qimron, U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc. Natl. Acad. Sci. USA 2015, 112, 7267–7272. [Google Scholar] [CrossRef]
- Bikard, D.; Euler, C.W.; Jiang, W.; Nussenzweig, P.M.; Goldberg, G.W.; Duportet, X.; Fischetti, V.A.; Marraffini, L.A. Exploiting CRISPR-cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 2014, 32, 1146–1150. [Google Scholar] [CrossRef]
- Citorik, R.J.; Mimee, M.; Lu, T.K. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol. 2014, 32, 1141–1145. [Google Scholar] [CrossRef]
- Kim, J.S.; Cho, D.H.; Park, M.; Chung, W.J.; Shin, D.; Ko, K.S.; Kweon, D.H. Crispr/cas9-mediated re-sensitization of antibiotic-resistant Escherichia coli harboring extended-spectrum β-lactamases. J. Microbiol. Biotechnol. 2015, 26, 394–401. [Google Scholar] [CrossRef]
- Kang, Y.K.; Kwon, K.; Ryu, J.S.; Lee, H.N.; Park, C.; Chung, H.J. Nonviral genome editing based on a polymer-derivatized CRISPR nanocomplex for targeting bacterial pathogens and antibiotic resistance. Bioconjug. Chem. 2017, 28, 957–967. [Google Scholar] [CrossRef]
- Hale, C.R.; Majumdar, S.; Elmore, J.; Pfister, N.; Compton, M.; Olson, S.; Resch, A.M.; Glover, C.V.C.; Graveley, B.R.; Terns, R.M.; et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell 2012, 45, 292–302. [Google Scholar] [CrossRef]
- Crawley, A.B.; Henriksen, E.D.; Stout, E.; Brandt, K.; Barrangou, R. Characterizing the activity of abundant, diverse and active CRISPR-Cas systems in lactobacilli. Sci. Rep. 2018, 8, 11544. [Google Scholar] [CrossRef]
- Cobb Id, L.H.; Park, J.; Swanson, E.A.; Beard, M.C.; Mccabe, E.M.; Rourke, A.S.; Seo, K.S.; Olivier, A.K.; Priddy, L.B. CRISPR-Cas9 modified bacteriophage for treatment of Staphylococcus aureus induced osteomyelitis and soft tissue infection. PLoS ONE 2019, 14, e0220421. [Google Scholar] [CrossRef]
- Li, Y.; Peng, N. Endogenous CRISPR-Cas system-based genome editing and antimicrobials: Review and prospects. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Pyne, M.E.; Bruder, M.R.; Moo-Young, M.; Chung, D.A.; Chou, C.P. Harnessing heterologous and endogenous CRISPR-Cas machineries for efficient markerless genome editing in Clostridium. Sci. Rep. 2016, 6, 25666. [Google Scholar] [CrossRef] [PubMed]
- Selle, K.; Fletcher, J.R.; Tuson, H.; Schmitt, D.S.; McMillan, L.; Vridhambal, G.S.; Rivera, A.J.; Montgomery, S.A.; Fortier, L.C.; Barrangou, R.; et al. In vivo targeting of Clostridioides difficile using phage- delivered CRISPR-Cas3 antimicrobials. MBio 2020, 11. [Google Scholar] [CrossRef]
- Kuenne, C.; Billion, A.; Mraheil, M.A.; Strittmatter, A.; Daniel, R.; Goesmann, A.; Barbuddhe, S.; Hain, T.; Chakraborty, T. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genom. 2013, 14, 47. [Google Scholar] [CrossRef] [PubMed]
- Hain, T.; Ghai, R.; Billion, A.; Kuenne, C.; Steinweg, C.; Izar, B.; Mohamed, W.; Mraheil, M.; Domann, E.; Schaffrath, S.; et al. Comparative genomics and transcriptomics of lineages I, II, and III strains of Listeria monocytogenes. BMC Genom. 2012, 13, 144. [Google Scholar] [CrossRef]
- Den Bakker, H.C.; Cummings, C.a.; Ferreira, V.; Vatta, P.; Orsi, R.H.; Degoricija, L.; Barker, M.; Petrauskene, O.; Furtado, M.R.; Wiedmann, M. Comparative genomics of the bacterial genus Listeria: Genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genom. 2010, 11, 688. [Google Scholar] [CrossRef]
- Di, H.; Ye, L.; Yan, H.; Meng, H.; Yamasak, S.; Shi, L. Comparative analysis of CRISPR loci in different Listeria monocytogenes lineages. Biochem. Biophys. Res. Commun. 2014, 454, 399–403. [Google Scholar] [CrossRef]
- Sesto, N.; Touchon, M.; Andrade, J.M.; Kondo, J.; Rocha, E.P.C.; Arraiano, C.M.; Archambaud, C.; Westhof, É.; Romby, P.; Cossart, P. A PNPase dependent CRISPR system in Listeria. PLoS Genet. 2014, 10, e1004065. [Google Scholar] [CrossRef]
- Meeske, A.J.; Marraffini, L.A. RNA guide complementarity prevents self-targeting in type VI CRISPR systems. Mol. Cell 2018, 71, 791–801.e3. [Google Scholar] [CrossRef]
- Hupfeld, M.; Trasanidou, D.; Ramazzini, L.; Klumpp, J.; Loessner, M.J.; Kilcher, S. A functional type II-A CRISPR–Cas system from Listeria enables efficient genome editing of large non-integrating bacteriophage. Nucleic Acids Res. 2018, 46, 6920–6933. [Google Scholar] [CrossRef]
- Maury, M.M.; Tsai, Y.-H.; Charlier, C.; Touchon, M.; Chenal-Francisque, V.; Leclercq, A.; Criscuolo, A.; Gaultier, C.; Roussel, S.; Brisabois, A.; et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 2016, 48, 308–313. [Google Scholar] [CrossRef]
- Vázquez-Boland, J.A.; Wagner, M.; Scortti, M. Why are some Listeria monocytogenes genotypes more likely to cause invasive (Brain, placental) infection? MBio 2020, 11, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Rauch, B.J.; Silvis, M.R.; Hultquist, J.F.; Waters, C.S.; McGregor, M.J.; Krogan, N.J.; Bondy-Denomy, J. Inhibition of CRISPR-Cas9 with bacteriophage proteins. Cell 2017, 168, 150–158.e10. [Google Scholar] [CrossRef]
- Meeske, A.J.; Jia, N.; Cassel, A.K.; Kozlova, A.; Liao, J.; Wiedmann, M.; Patel, D.J.; Marraffini, L.A. A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity. Science 2020, 369, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo-Cantabrana, C.; Goh, Y.J.; Pan, M.; Sanozky-Dawes, R.; Barrangou, R. Genome editing using the endogenous type I CRISPR-Cas system in Lactobacillus crispatus. Proc. Natl. Acad. Sci. USA 2019, 116, 15774–15783. [Google Scholar] [CrossRef] [PubMed]
- Crawley, A.B.; Henriksen, J.R.; Barrangou, R. CRISPRdisco: An automated pipeline for the discovery and analysis of CRISPR-Cas systems. Cris. J. 2018, 1, 171–181. [Google Scholar] [CrossRef]
- Pawluk, A.; Bondy-Denomy, J.; Cheung, V.H.W.; Maxwell, K.L.; Davidson, A.R. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa. MBio 2014, 5. [Google Scholar] [CrossRef]
- Trasanidou, D.; Gerós, A.S.; Mohanraju, P.; Nieuwenweg, A.C.; Nobrega, F.L.; Staals, R.H.J. Keeping CRISPR in check: Diverse mechanisms of phage-encoded anti-CRISPRs. FEMS Microbiol. Lett. 2019, 366, 98. [Google Scholar] [CrossRef]
- Marino, N.D.; Zhang, J.Y.; Borges, A.L.; Sousa, A.A.; Leon, L.M.; Rauch, B.J.; Walton, R.T.; Berry, J.D.; Joung, J.K.; Kleinstiver, B.P.; et al. Discovery of widespread type I and type V CRISPR-Cas inhibitors. Science 2018, 362, 240–242. [Google Scholar] [CrossRef]
- Park, J.Y.; Moon, B.Y.; Park, J.W.; Thornton, J.A.; Park, Y.H.; Seo, K.S. Genetic engineering of a temperate phage-based delivery system for CRISPR/Cas9 antimicrobials against Staphylococcus aureus. Sci. Rep. 2017, 7, 44929. [Google Scholar] [CrossRef]
- Kilcher, S.; Studer, P.; Muessner, C.; Klumpp, J.; Loessner, M.J. Cross-genus rebooting of custom-made, synthetic bacteriophage genomes in L-form bacteria. Proc. Natl. Acad. Sci. USA 2018, 115, 201714658. [Google Scholar] [CrossRef]
- Meile, S.; Sarbach, A.; Du, J.; Schuppler, M.; Saez, C.; Loessner, M.J.; Kilcher, S. Engineered reporter phages for rapid bioluminescence-based detection and differentiation of viable Listeria cells. Appl. Environ. Microbiol. 2020, 86, e00442-20. [Google Scholar] [CrossRef] [PubMed]
- Dunne, M.; Rupf, B.; Tala, M.; Qabrati, X.; Ernst, P.; Shen, Y.; Sumrall, E.; Heeb, L.; Plückthun, A.; Loessner, M.J.; et al. Reprogramming bacteriophage host range through structure-guided design of chimeric receptor binding proteins. Cell Rep. 2019, 29, 1336–1350.e4. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Muñoz, S.L.; Koskella, B. Bacteria-Phage interactions in natural environments. In Advances in Applied Microbiology; Academic Press Inc.: Cambridge, MA, USA, 2014; Volume 89, pp. 135–183. [Google Scholar]
- Olszak, T.; Latka, A.; Roszniowski, B.; Valvano, M.A.; Drulis-Kawa, Z. Phage Life Cycles Behind Bacterial Biodiversity. Curr. Med. Chem. 2017, 24, 3987–4001. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parsons, C.; Brown, P.; Kathariou, S. Use of Bacteriophage Amended with CRISPR-Cas Systems to Combat Antimicrobial Resistance in the Bacterial Foodborne Pathogen Listeria monocytogenes. Antibiotics 2021, 10, 308. https://doi.org/10.3390/antibiotics10030308
Parsons C, Brown P, Kathariou S. Use of Bacteriophage Amended with CRISPR-Cas Systems to Combat Antimicrobial Resistance in the Bacterial Foodborne Pathogen Listeria monocytogenes. Antibiotics. 2021; 10(3):308. https://doi.org/10.3390/antibiotics10030308
Chicago/Turabian StyleParsons, Cameron, Phillip Brown, and Sophia Kathariou. 2021. "Use of Bacteriophage Amended with CRISPR-Cas Systems to Combat Antimicrobial Resistance in the Bacterial Foodborne Pathogen Listeria monocytogenes" Antibiotics 10, no. 3: 308. https://doi.org/10.3390/antibiotics10030308
APA StyleParsons, C., Brown, P., & Kathariou, S. (2021). Use of Bacteriophage Amended with CRISPR-Cas Systems to Combat Antimicrobial Resistance in the Bacterial Foodborne Pathogen Listeria monocytogenes. Antibiotics, 10(3), 308. https://doi.org/10.3390/antibiotics10030308






