Sulfaguanidine Hybrid with Some New Pyridine-2-One Derivatives: Design, Synthesis, and Antimicrobial Activity against Multidrug-Resistant Bacteria as Dual DNA Gyrase and DHFR Inhibitors
Abstract
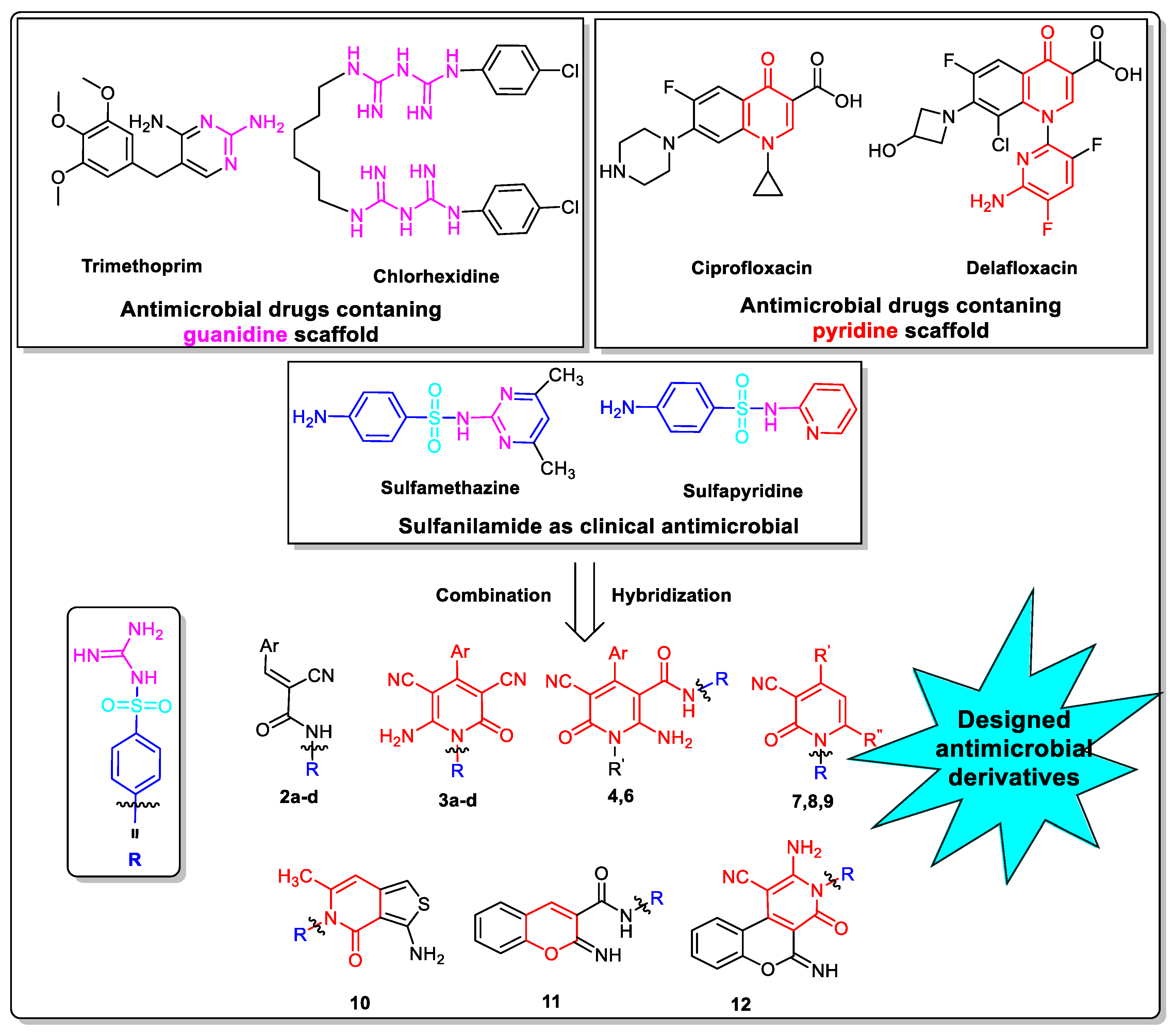
1. Introduction
2. Results and Discussion
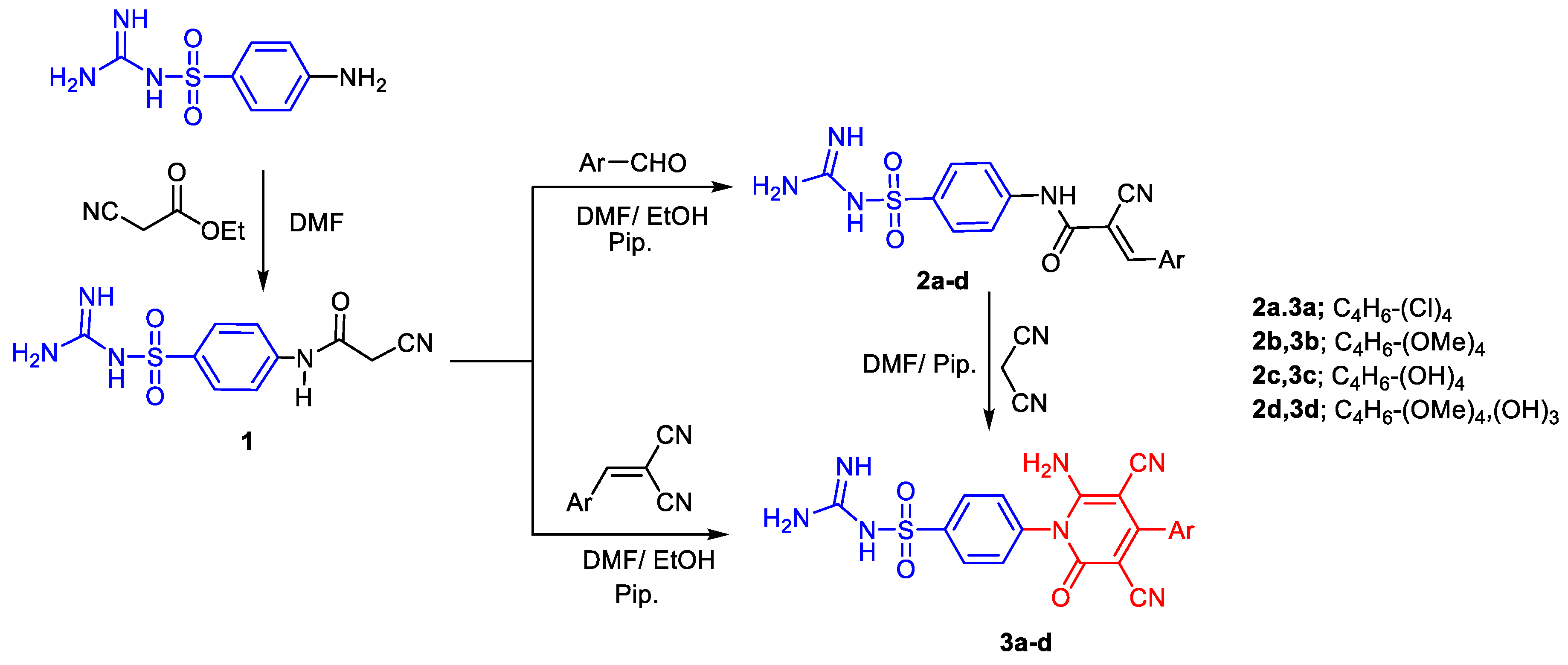
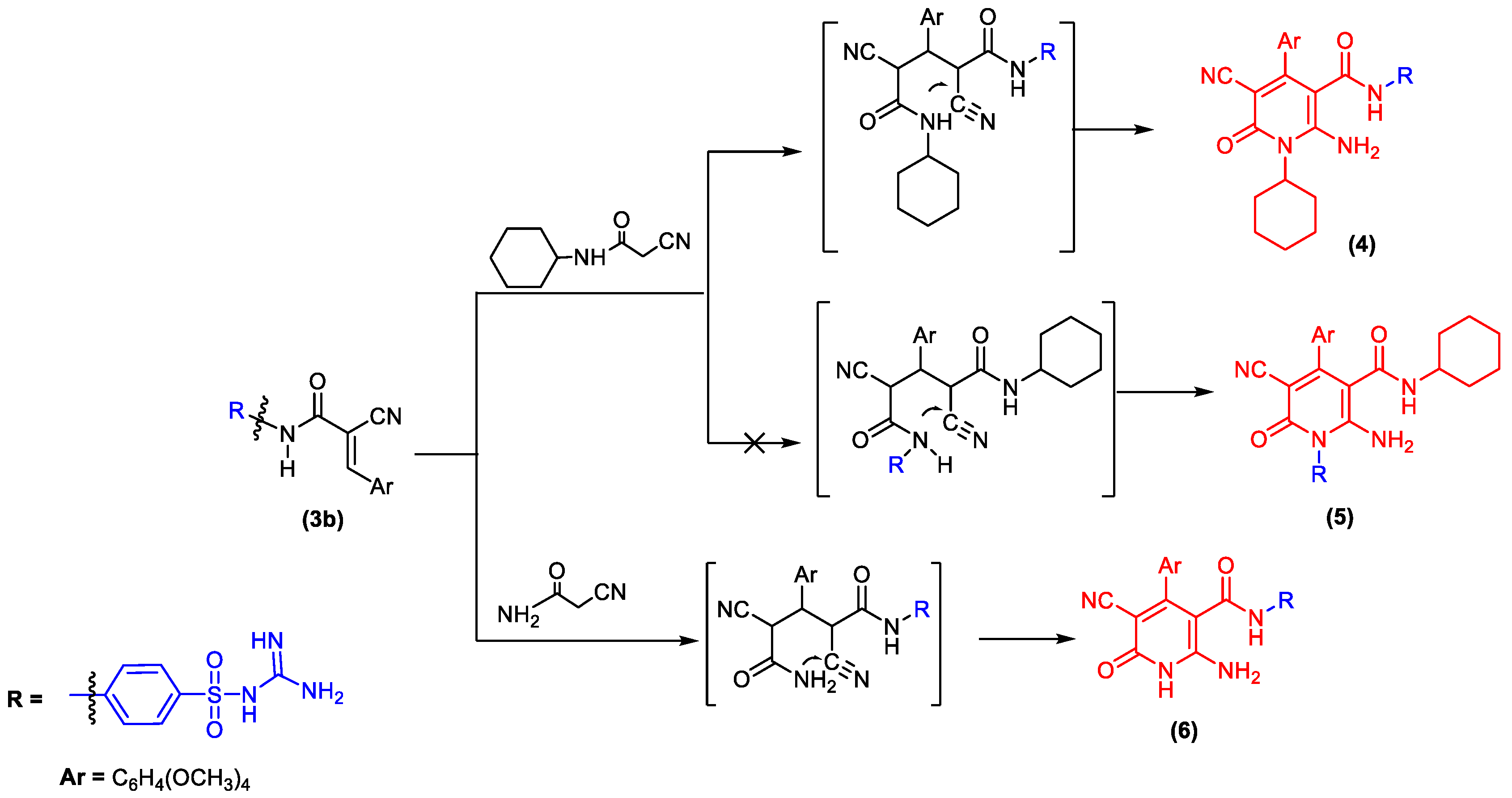
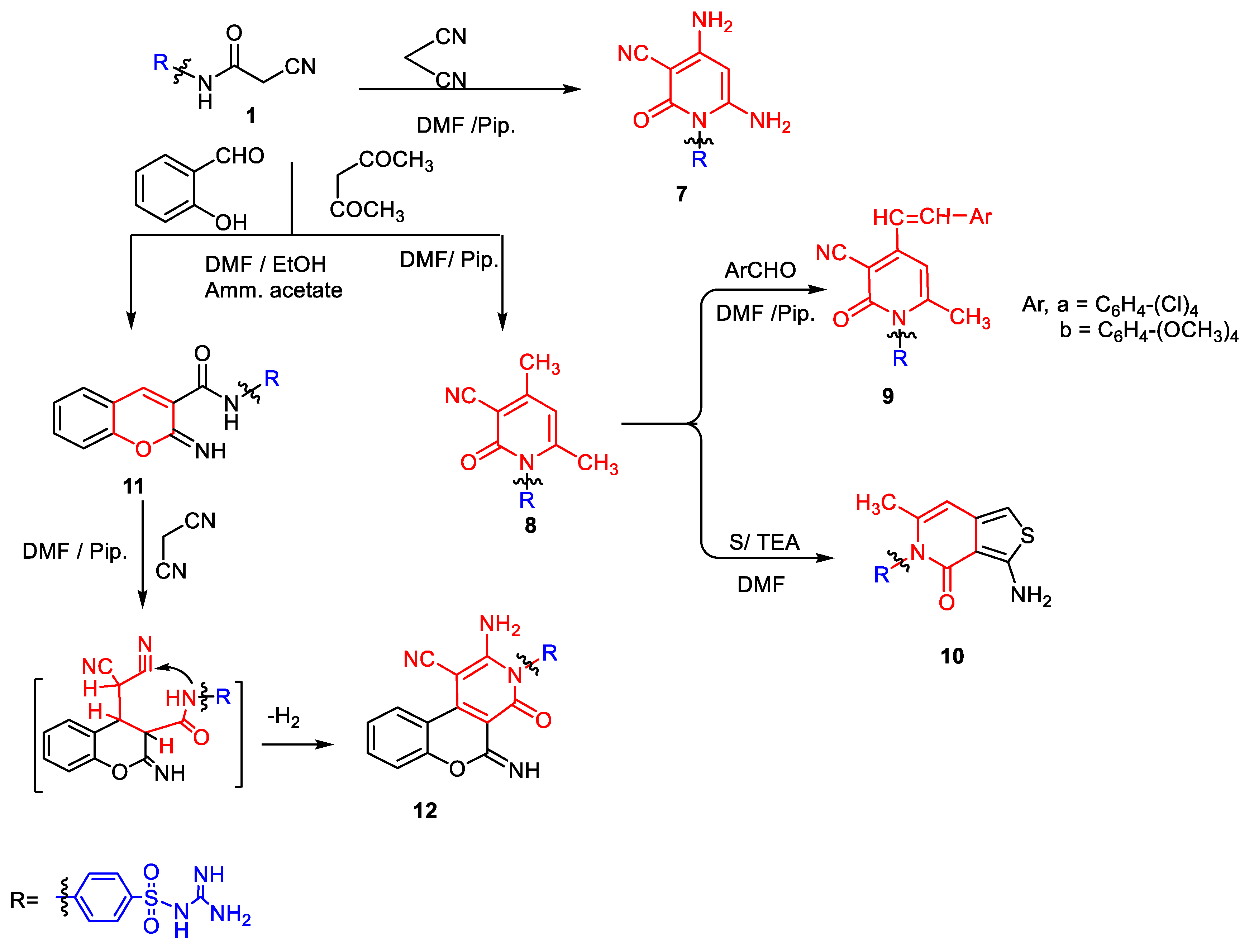
2.1. Chemistry
2.2. Biological Activity
2.2.1. Antimicrobial Activity with Structure Activity Relationship (SAR) Study
2.2.2. Minimal Inhibitory/Bactericidal Concentrations (MIC) and (MBC)
2.2.3. Minimal Inhibitory/Fungicidal Concentrations (MIC) and (MFC)
2.2.4. Multidrug-Resistant Bacteria (MDRB) Study
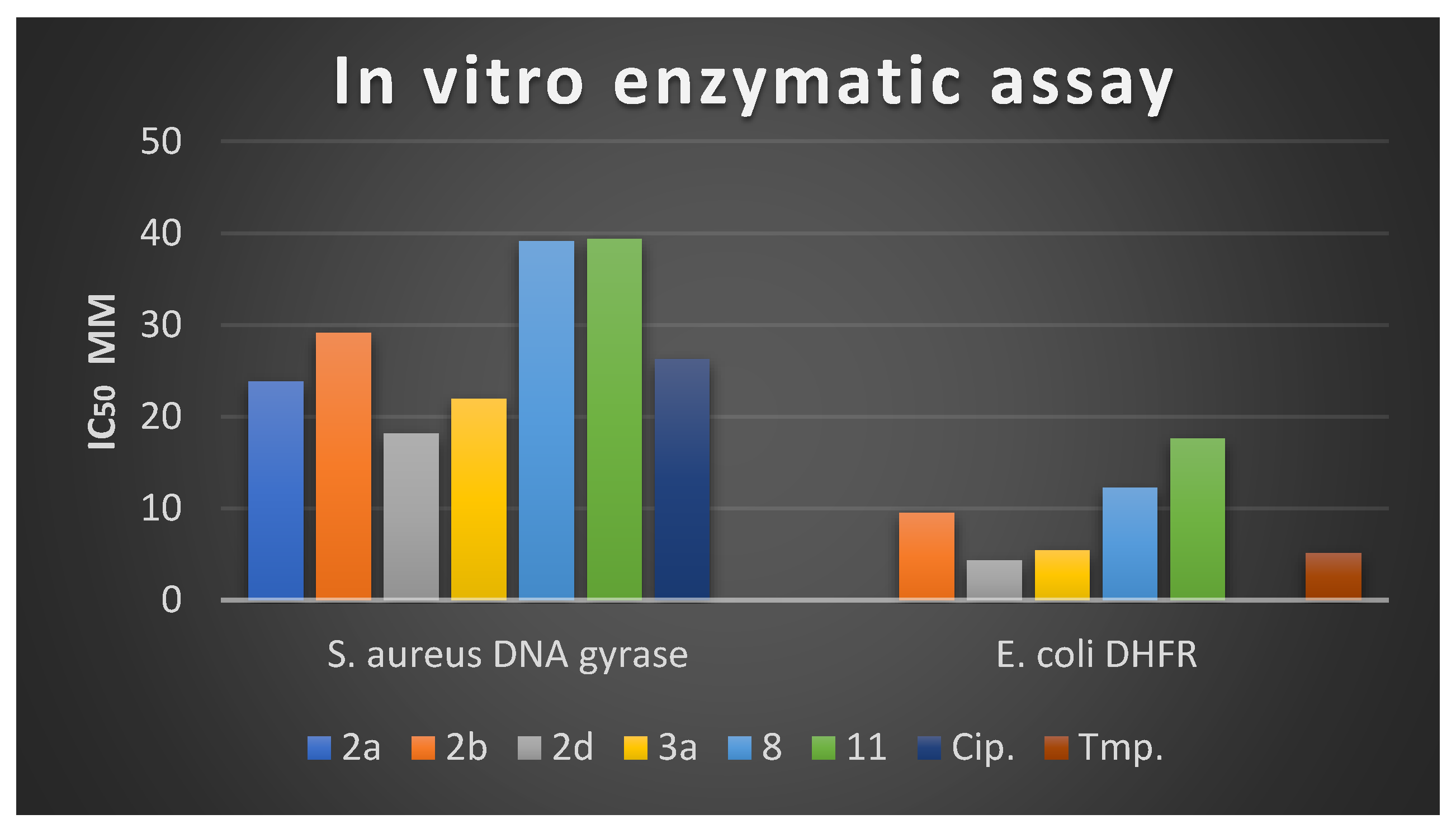
2.2.5. In Vitro S. aureus DNA Gyrase and E. coli DHFR Enzymatic Assay
2.2.6. Immunomodulatory Activity
In Vitro Intracellular Killing Activities
In Vivo Immunomodulatory Investigation
2.3. In Silico Studies
2.3.1. Prediction of Some Physicochemical, Pharmacokinetic, and Toxicity Properties
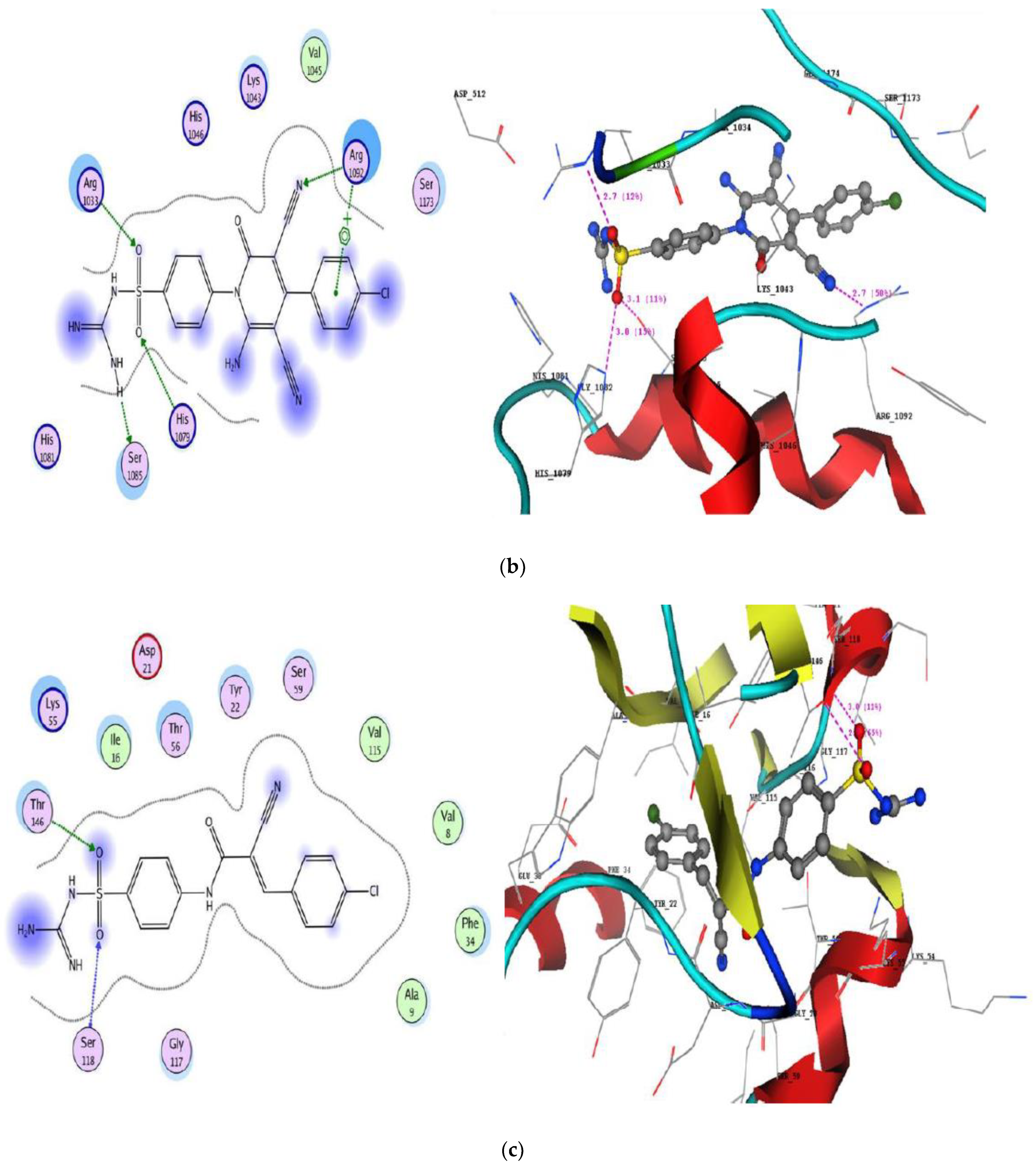
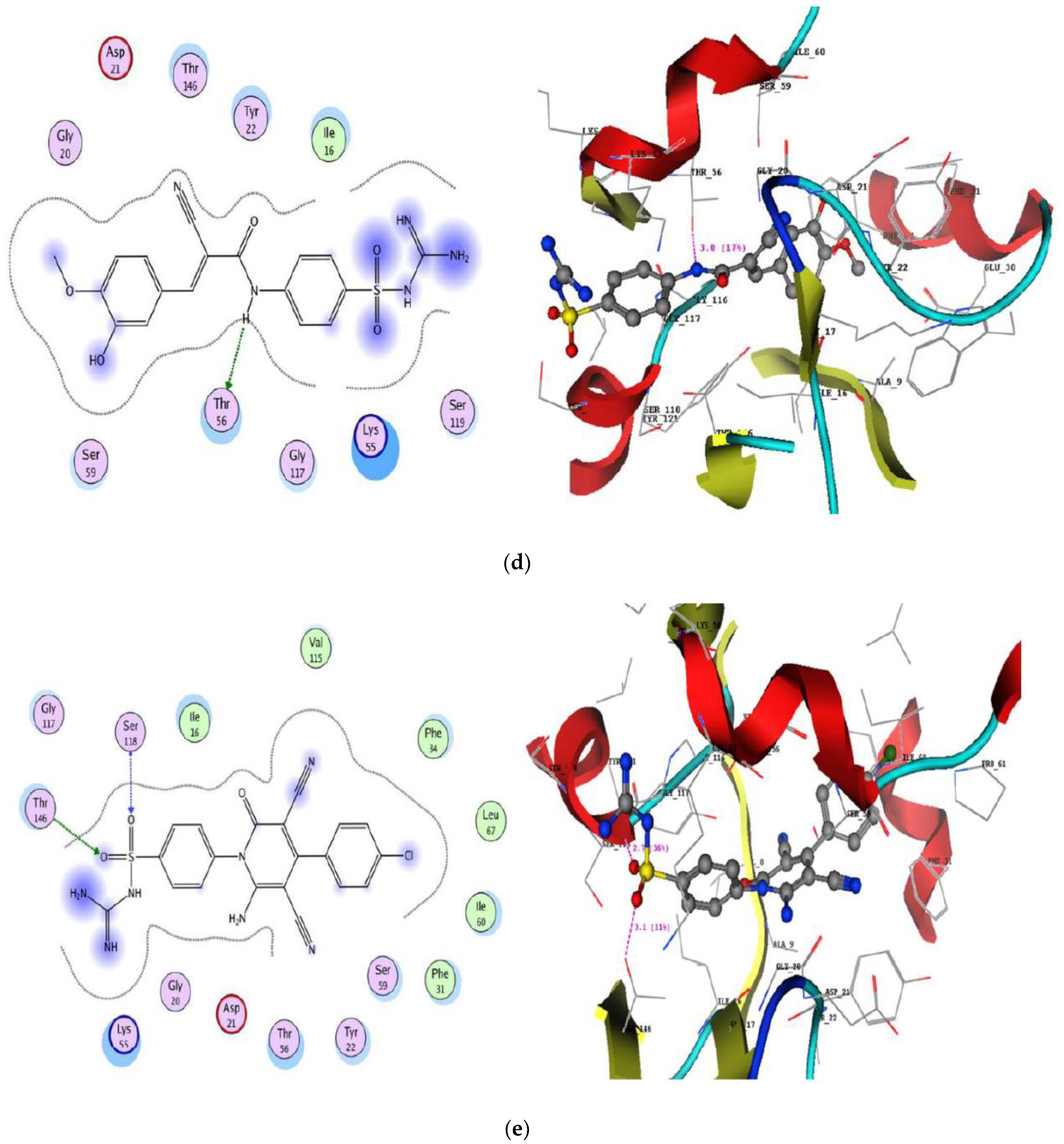
2.3.2. Molecular Docking Studies
3. Materials and Methods
3.1. Chemistry
3.2. In Vitro Antimicrobial Activity (See Supplementary Material)
3.2.1. Bacterial and Fungal Strains Used in the Study
3.2.2. Determination of Inhibition Zones (mm)
3.2.3. Determination the Minimal Inhibition Concentration (MIC) and Minimum Bactericidal Concentration (MBC)
3.2.4. Determination DNA Gyrase and DHFR Inhibitory Assay
3.2.5. Determination Immunomodulatory Activity
3.2.6. Molecular Docking Study
3.2.7. Ethics Statement for Both Animal Models and for Using Volunteer Blood Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Service, R.F. Antibiotics That Resist Resistance. Science 1995, 270, 724–727. [Google Scholar] [CrossRef]
- Fukuda, Y. New approaches to overcoming bacterial resistance. Drugs Future 2009, 34, 127–136. [Google Scholar] [CrossRef]
- O’Neill, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. 2014. Available online: https://www.who.int/antimicrobial-resistance/news/amr-newsletter-no13-july2016.pdf (accessed on 21 October 2020).
- Boucher, H.W.; Ambrose, P.G.; Chambers, H.F.; Ebright, R.H.; Jezek, A.; Murray, B.E.; Newland, J.G.; Ostrowsky, B.; Rex, J.H.; Infectious Diseases Society of America. White Paper: Developing Antimicrobial Drugs for Resistant Pathogens, Narrow-Spectrum Indications, and Unmet Needs. J. Infect. Dis. 2017, 216, 228–236. [Google Scholar] [CrossRef]
- Maxson, T.; Mitchell, D.A. Targeted Treatment for Bacterial Infections: Prospects for Pathogen-Specific Antibiotics Coupled with Rapid Diagnostics. Tetrahedron 2016, 72, 3609–3624. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Jang, J.H.; Kim, S.C.; Cho, J.H. Development of a novel hybrid antimicrobial peptide for targeted killing of Pseudomonas aeruginosa. Eur. J. Med. Chem. 2020, 185, 111814. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, P.; Martens, E. Antibiotics in late clinical development. Biochem. Pharmacol. 2017, 133, 152–163. [Google Scholar] [CrossRef]
- McCarthy, K.A.; Kelly, M.A.; Li, K.; Cambray, S.; Hosseini, A.S.; van Opijnen, T.; Gao, J. Phage Display of Dynamic Covalent Binding Motifs Enables Facile Development of Targeted Antibiotics. J. Am. Chem. Soc. 2018, 140, 6137–6145. [Google Scholar] [CrossRef] [PubMed]
- Ammar, Y.A.; Farag, A.A.; Ali, A.M.; Hessein, S.A.; Askar, A.A.; Fayed, E.A.; Elsisi, D.M.; Ragab, A. Antimicrobial evaluation of thiadiazino and thiazolo quinoxaline hybrids as potential DNA gyrase inhibitors; design, synthesis, characterization and morphological studies. Bioorg. Chem. 2020, 99, 103841. [Google Scholar] [CrossRef]
- Ibrahim, H.S.; Eldehna, W.M.; Abdel-Aziz, H.A.; Elaasser, M.M.; Abdel-Aziz, M.M. Improvement of antibacterial activity of some sulfa drugs through linkage to certain phthalazin-1(2H)-one scaffolds. Eur. J. Med. Chem. 2014, 85, 480–486. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.-Q.; Flavin, M.T.; Flavin, J. Combating multidrug-resistant Gram-negative bacterial infections. Expert Opin. Investig. Drugs 2014, 23, 163–182. [Google Scholar] [CrossRef] [PubMed]
- Zakšauskas, A.; Čapkauskaitė, E.; Jezepčikas, L.; Linkuvienė, V.; Paketurytė, V.; Smirnov, A.; Leitans, J.; Kazaks, A.; Dvinskis, E.; Manakova, E.; et al. Halogenated and di-substituted benzenesulfonamides as selective inhibitors of carbonic anhydrase isoforms. Eur. J. Med. Chem. 2020, 185, 111825. [Google Scholar] [CrossRef] [PubMed]
- Ammar, Y.A.; Sh El-Sharief, A.M.; Belal, A.; Abbas, S.Y.; Mohamed, Y.A.; Mehany, A.B.M.; Ragab, A. Design, synthesis, antiproliferative activity, molecular docking and cell cycle analysis of some novel (morpholinosulfonyl) isatins with potential EGFR inhibitory activity. Eur. J. Med. Chem. 2018, 156, 918–932. [Google Scholar] [CrossRef] [PubMed]
- El-sharief, A.M.S.; Ammar, Y.A.; Belal, A.; El-sharief, M.A.M.S.; Mohamed, Y.A.; Mehany, A.B.M.; Elhag, G.A.M.; Ragab, A. Design, synthesis, molecular docking and biological activity evaluation of some novel indole derivatives as potent anticancer active agents and apoptosis inducers. Bioorg. Chem. 2019, 85, 399–412. [Google Scholar] [CrossRef]
- Isik, S.; Kockar, F.; Aydin, M.; Arslan, O.; Guler, O.O.; Innocenti, A.; Scozzafava, A.; Supuran, C.T. Carbonic anhydrase inhibitors: Inhibition of the β-class enzyme from the yeast Saccharomyces cerevisiae with sulfonamides and sulfamates. Bioorg. Med. Chem. 2009, 17, 1158–1163. [Google Scholar] [CrossRef]
- Bouissane, L.; El Kazzouli, S.; Léonce, S.; Pfeiffer, B.; Rakib, E.M.; Khouili, M.; Guillaumet, G. Synthesis and biological evaluation of N-(7- indazolyl) benzenesulfonamide derivatives as potent cell cycle inhibitors. Bioorg. Med. Chem. 2006, 14, 1078–1088. [Google Scholar] [CrossRef]
- Camoutsis, C.; Geronikaki, A.; Ciric, A.; Soković, M.; Zoumpoulakis, P.; Zervou, M. Sulfonamide-1,2,4-thiadiazole Derivatives as Antifungal and Antibacterial Agents: Synthesis, Biological Evaluation, Lipophilicity, and Conformational Studies. Chem. Pharm. Bull. 2010, 58, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.; Casini, A.; Heine, A.; Kuhn, D.; Supuran, C.T.; Scozzafava, A.; Klebe, G. Unexpected Nanomolar Inhibition of Carbonic Anhydrase by COX-2-Selective Celecoxib: New Pharmacological Opportunities Due to Related Binding Site Recognition. J. Med. Chem. 2004, 47, 550–557. [Google Scholar] [CrossRef] [PubMed]
- Ammar, Y.A.; El-Sharief, A.M.; Mohamed, Y.A.; Mehany, A.B.; Ragab, A. Synthesis, spectral characterization and pharmacological evaluation of novel thiazole-oxoindole hybrid compounds as potential anticancer agents. Al-Azhar Bull. Sci. 2018, 29, 25–37. [Google Scholar]
- Penning, T.D.; Talley, J.J.; Bertenshaw, S.R.; Carter, J.S.; Collins, P.W.; Docter, S.; Graneto, M.J.; Lee, L.F.; Malecha, J.W.; Miyashiro, J.M.; et al. Synthesis and Biological Evaluation of the 1,5-Diarylpyrazole Class of Cyclooxygenase-2 Inhibitors: Identification of 4-[5-(4-Methylphenyl)-3- (trifluoromethyl)-1H-pyrazol-1-yl] benzenesulfonamide (SC-58635, Celecoxib). J. Med. Chem. 1997, 40, 1347–1365. [Google Scholar] [CrossRef]
- Rahimifard, M.; Ziarani, G.M.; Lashkariani, B.M. Application of guanidine and its salts in multicomponent reactions. Turkish J. Chem. 2014, 38, 345–371. [Google Scholar] [CrossRef]
- Khalaf, M.; Zageer, D.; Hussain, Z.; Adil, H.; Mohammed, S.; Yousif, E. Guanidine Group: Definition and Pharmaceutical Applications. Res. J. Pharm. Biol. Chem. Sci. 2016, 7, 1026–1031. [Google Scholar]
- Wang, P.-F.; McLeish, M.J.; Kneen, M.M.; Lee, G.; Kenyon, G.L. An Unusually Low pKa for Cys282 in the Active Site of Human Muscle Creatine Kinase. Biochemistry 2001, 40, 11698–11705. [Google Scholar] [CrossRef]
- Duo, J.; Ma, Y.; Wang, G.; Han, X.; Zhang, C. Metformin Synergistically Enhances Antitumor Activity of Histone Deacetylase Inhibitor Trichostatin a against Osteosarcoma Cell Line. DNA Cell Biol. 2013, 32, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Bahekar, R.H.; Jain, M.R.; Jadav, P.A.; Prajapati, V.M.; Patel, D.N.; Gupta, A.A.; Sharma, A.; Tom, R.; Bandyopadhya, D.; Modi, H.; et al. Synthesis and antidiabetic activity of 2,5-disubstituted-3-imidazol-2-yl-pyrrolo[2,3-b] pyridines and thieno[2,3-b] pyridines. Bioorg. Med. Chem. 2007, 15, 6782–6795. [Google Scholar] [CrossRef]
- Kratzer, C.; Tobudic, S.; Graninger, W.; Buxbaum, A.; Georgopoulos, A. In vitro antimicrobial activity of the novel polymeric guanidine Akacid plus. J. Hosp. Infect. 2006, 63, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Hensler, M.E.; Bernstein, G.; Nizet, V.; Nefzi, A. Pyrrolidine bis-cyclic guanidines with antimicrobial activity against drug-resistant Gram-positive pathogens identified from a mixture-based combinatorial library. Bioorg. Med. Chem. Lett. 2006, 16, 5073–5079. [Google Scholar] [CrossRef] [PubMed]
- Georgopoulos, S.; Mastorakos, D.; Kondi-Pafiti, A.; Katsenis, K.; Arkadopoulos, N.; Kannas, D.; Archontaki, M.; Vestarchis, N.; Kokkalis, G. Hydroxyzine, cimetidine and vitamin C in reducing skin flap necrosis in ischemia-reperfusion injury in rats. A comparative study. J. BU ON. Off. J. Balk. Union Oncol. 2012, 17, 377–382. [Google Scholar]
- Ozeki, T.; Natori, T. The specific inhibition of HepG2 cells proliferation by apoptosis induced by gabexate mesilate. Int. J. Clin. Exp. Pathol. 2010, 3, 710. [Google Scholar] [PubMed]
- Thomas, G.; Gro, R.; Schramm, M. Calcium channel modulation: Ability to inhibit or promote calcium influx resides in the same dihydropyridine molecule. J. Cardiovasc. Pharmacol. 1984, 6, 1170–1176. [Google Scholar] [CrossRef]
- Andreani, A.; Leoni, A.; Locatelli, A.; Morigi, R.; Rambaldi, M.; Pietra, C.; Villetti, G. 4-Aminopyridine derivatives with antiamnesic activity##Presented in part at the Italian–Hungarian–Polish Joint Meeting on Medicinal Chemistry, Giardini Naxos–Taormina, September 28th–October 1st 1999. Eur. J. Med. Chem. 2000, 35, 77–82. [Google Scholar] [PubMed]
- Ammar, Y.A.; Ghorab, M.M.; El-Sharief, A.M.S.; Mohamed, S.I. Naproxen in heterocyclic chemistry: Novel syntheses of triazoles, triazolothiadiazines, triazolothiadiazoles, and triazolothiadiazepine bearing an asymmetric carbon atom and radiostability of the biologically active compounds. Heteroat. Chem. 2002, 13, 199–206. [Google Scholar] [CrossRef]
- Nasr, T.; Bondock, S.; Eid, S. Design, synthesis, antimicrobial evaluation and molecular docking studies of some new thiophene, pyrazole and pyridone derivatives bearing sulfisoxazole moiety. Eur. J. Med. Chem. 2014, 84, 491–504. [Google Scholar] [CrossRef]
- Hagar, M.; Chaieb, K.; Parveen, S.; Ahmed, H.A.; Alnoman, R.B. N-alkyl 2-pyridone versus O-alkyl 2-pyridol: Ultrasonic synthesis, DFT, docking studies and their antimicrobial evaluation. J. Mol. Struct. 2020, 1199, 126926. [Google Scholar] [CrossRef]
- Arnold, M.A.; Gerasyuto, A.I.; Wang, J.; Du, W.; Gorske, Y.J.K.; Arasu, T.; Baird, J.; Almstead, N.G.; Narasimhan, J.; Peddi, S.; et al. 4-Hydroxy-2-pyridones: Discovery and evaluation of a novel class of antibacterial agents targeting DNA synthesis. Bioorg. Med. Chem. Lett. 2017, 27, 5014–5021. [Google Scholar] [CrossRef]
- Li, Q.; Mitscher, L.A.; Shen, L.L. The 2-pyridone antibacterial agents: Bacterial topoisomerase inhibitors. Med. Res. Rev. 2000, 20, 231–293. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Sader, H.S.; Rhomberg, P.R.; Flamm, R.K. Activity of Delafloxacin against Contemporary Bacterial Pathogens from the United States and Europe, 2014. Antimicrob. Agents Chemother. 2017, 61, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Mitscher, L.A. Bacterial topoisomerase inhibitors: Quinolone and pyridone antibacterial agents. Chem. Rev. 2005, 105, 559–592. [Google Scholar] [CrossRef]
- Ammar, Y.A.; El-Gaby, M.S.A.; Salem, M.A. Cyanoacetanilides intermediates in heterocyclic synthesis. Part 6: Preparation of some hitherto unknown 2-oxopyridine, bipyridine, isoquinoline and chromeno[3,4-c] pyridine containing sulfonamide moiety. Arab. J. Chem. 2014, 7, 615–622. [Google Scholar] [CrossRef]
- Fadda, A.A.; Bondock, S.; Rabie, R.; Etman, H.A. Cyanoacetamide derivatives as synthons in heterocyclic synthesis. Turkish J. Chem. 2008, 32, 259–286. [Google Scholar]
- Ammar, Y.A.; Farag, A.A.; Ali, A.M.; Ragab, A.; Askar, A.A.; Elsisi, D.M.; Belal, A. Design, synthesis, antimicrobial activity and molecular docking studies of some novel di-substituted sulfonylquinoxaline derivatives. Bioorg. Chem. 2020, 104, 104164. [Google Scholar] [CrossRef]
- Janakiramudu, D.B.; Subba Rao, D.; Srikanth, C.; Madhusudhana, S.; Sreenivasa Murthy, P.; Nagalakshmidevamma, M.; Chalapathi, P.V.; Naga Raju, C. Sulfonamides and carbamates of 3-fluoro-4-morpholinoaniline (linezolid intermediate): Synthesis, antimicrobial activity and molecular docking study. Res. Chem. Intermed. 2018, 44, 469–489. [Google Scholar] [CrossRef]
- Hussein, E.M.; Al-Rooqi, M.M.; Abd El-Galil, S.M.; Ahmed, S.A. Design, synthesis, and biological evaluation of novel N4-substituted sulfonamides: Acetamides derivatives as dihydrofolate reductase (DHFR) inhibitors. BMC Chem. 2019, 13, 91. [Google Scholar] [CrossRef]
- He, J.; Qiao, W.; An, Q.; Yang, T.; Luo, Y. Dihydrofolate reductase inhibitors for use as antimicrobial agents. Eur. J. Med. Chem. 2020, 195, 112268. [Google Scholar] [CrossRef]
- Parham, P. The immune system. Yale J. Biol. Med. 2015, 88, 99. [Google Scholar]
- Jang, M.; Lim, T.-G.; Ahn, S.; Hong, H.-D.; Rhee, Y.K.; Kim, K.-T.; Lee, E.; Lee, J.H.; Lee, Y.J.; Jung, C.S. Immune-enhancing effects of a high molecular weight fraction of Cynanchum wilfordii Hemsley in macrophages and immunosuppressed mice. Nutrients 2016, 8, 600. [Google Scholar] [CrossRef]
- Zhang, W.-N.; Gong, L.-L.; Liu, Y.; Zhou, Z.-B.; Wan, C.-X.; Xu, J.-J.; Wu, Q.-X.; Chen, L.; Lu, Y.-M.; Chen, Y. Immunoenhancement effect of crude polysaccharides of Helvella leucopus on cyclophosphamide-induced immunosuppressive mice. J. Funct. Foods 2020, 69, 103942. [Google Scholar] [CrossRef]
- Gao, S.; Hong, H.; Zhang, C.; Wang, K.; Zhang, B.; Han, Q.; Liu, H.; Luo, Y. Immunomodulatory effects of collagen hydrolysates from yak (Bos grunniens) bone on cyclophosphamide-induced immunosuppression in BALB/c mice. J. Funct. Foods 2019, 60, 103420. [Google Scholar] [CrossRef]
- Mishra, K.P.; Padwad, Y.S.; Jain, M.; Karan, D.; Ganju, L.; Sawhney, R.C. Aqueous Extract of Rhodiola imbricata Rhizome Stimulates Proinflammatory Mediators via Phosphorylated IκB and Transcription Factor Nuclear Factor-κB. Immunopharmacol. Immunotoxicol. 2006, 28, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jiao, F.; Qiu, Y.; Li, W.; Qu, Y.; Tian, C.; Li, Y.; Bai, R.; Lao, F.; Zhao, Y. Immunostimulatory properties and enhanced TNF-α mediated cellular immunity for tumor therapy by C60 (OH) 20 nanoparticles. Nanotechnology 2009, 20, 415102. [Google Scholar] [CrossRef] [PubMed]
- Wassel, M.M.S.; Ragab, A.; Elhag Ali, G.A.M.; Mehany, A.B.M.; Ammar, Y.A. Novel adamantane-pyrazole and hydrazone hybridized: Design, synthesis, cytotoxic evaluation, SAR study and molecular docking simulation as carbonic anhydrase inhibitors. J. Mol. Struct. 2021, 1223, 128966. [Google Scholar] [CrossRef]
- M. S. Wassel, M.; M. Gamal Eldin, W.; Ragab, A.; A. M. Elhag Ali, G.; A. Ammar, Y. Antiviral Activity Of Adamantane-Pyrazole Derivatives Against Foot And Mouth Disease Virus Infection In Vivo And In Vitro With Molecular Docking Study. J. Appl. Vet. Sci. 2020, 5, 37–46. [Google Scholar] [CrossRef]
- Ammar, Y.A.; Abbas, S.Y.; Ghorab, M.M.; Al Said, M.S. Transmonocyanoacetylation of phenylenediamines: A simple and efficient synthesis of novel N-(aminophenyl)-2-cyanoacetamides and their derivatives. Tetrahedron Lett. 2016, 57, 275–277. [Google Scholar] [CrossRef]
- Ammar, Y.A.; Ali, M.M.; Mohamed, Y.A.; Thabet, H.K.; El-Gaby, M.S.A. Cyanoacetanilide intermediates in heterocyclic synthesis. Part 7: Preparation of some spiro [indoline-3, 4′-pyridine] and chromeno [3, 4-c] pyridine derivatives. Heterocycl. Commun. 2013, 19, 195–200. [Google Scholar] [CrossRef]
- Ammar, Y.A.; Ismail, M.M.F.; El-Sehrawi, H.M.; Noaman, E.; Bayomi, A.H.; Shawer, T.Z. Novel Pirfenidone Analogues: Synthesis of Pyridin-2-ones for the Treatment of Pulmonary Fibrosis. Arch. Pharm. (Weinheim). 2006, 339, 429–436. [Google Scholar] [CrossRef]
- AMMAR, Y.A.; Bondock, S.; EL-SEHMI, A.G.; EL_GABY, M.S.A.; Fouda, A.M.; Thabet, H.K. Facile and convenient synthesis of pyrimidine, 4H-3, 1-benzoxazin-4-one, pyrazolo [5, 1-b] quinazoline, pyrido [1, 2-a] quinazoline, and chromeno [3’, 4’: 4, 5] pyrido [1, 2-a] quinazoline derivatives. Turkish J. Chem. 2011, 35, 893–903. [Google Scholar]
- Rizk, H.F.; El-Borai, M.A.; Ragab, A.; Ibrahim, S.A. Design, synthesis, biological evaluation and molecular docking study based on novel fused pyrazolothiazole scaffold. J. Iran. Chem. Soc. 2020, 17, 2493–2505. [Google Scholar] [CrossRef]
- Ammar, Y.A.; Abbas, S.Y.; El-Sharief, M.A.M.S.; Salem, M.A.E.-R.; Mohamed, A.R. Synthesis and characterization of new imidazolidineiminothione and bis-imidazolidineiminothione derivatives as potential antimicrobial agents. Eur. J. Chem. 2017, 8, 76–81. [Google Scholar] [CrossRef]
- Fayed, E.A.; Ammar, Y.A.; Ragab, A.; Gohar, N.A.; Mehany, A.B.M.; Farrag, A.M. In vitro cytotoxic activity of thiazole-indenoquinoxaline hybrids as apoptotic agents, design, synthesis, physicochemical and pharmacokinetic studies. Bioorg. Chem. 2020, 100, 103951. [Google Scholar] [CrossRef]
- El-Sharief, M.A.M.S.; Abbas, S.Y.; Zahran, M.A.; Mohamed, Y.A.; Ragab, A.; Ammar, Y.A. New 1,3-diaryl-5-thioxo-imidazolidin-2,4-dione derivatives: Synthesis, reactions and evaluation of antibacterial and antifungal activities. Zeitschrift fur Naturforsch. Sect. B J. Chem. Sci. 2016, 71, 875–881. [Google Scholar] [CrossRef]
- Salem, M.A.; Ragab, A.; El-Khalafawy, A.; Makhlouf, A.H.; Askar, A.A.; Ammar, Y.A. Design, synthesis, in vitro antimicrobial evaluation and molecular docking studies of indol-2-one tagged with morpholinosulfonyl moiety as DNA gyrase inhibitors. Bioorg. Chem. 2020, 96, 103619. [Google Scholar] [CrossRef]
- Hassan, A.S.; Askar, A.A.; Naglah, A.M.; Almehizia, A.A.; Ragab, A. Discovery of New Schiff Bases Tethered Pyrazole Moiety: Design, Synthesis, Biological Evaluation, and Molecular Docking Study as Dual Targeting DHFR/DNA Gyrase Inhibitors with Immunomodulatory Activity. Molecules 2020, 25, 2593. [Google Scholar] [CrossRef] [PubMed]
- Ammar, Y.A.; AM, S.; El-Sharief, M.; Ghorab, M.M.; Mohamed, Y.A.; Ragab, A.; Abbas, S.Y. New imidazolidineiminothione, imidazolidin-2-one and imidazoquinoxaline derivatives: Synthesis and evaluation of antibacterial and antifungal activities. Curr. Org. Synth. 2016, 13, 466–475. [Google Scholar] [CrossRef]
- Dias, F.R.F.; Novais, J.S.; Devillart, T.A.; da Silva, W.A.; Ferreira, M.O.; Loureiro, R.D.; Campos, V.R.; Ferreira, V.F.; de Souza, M.C.; Castro, H.C.; et al. Synthesis and antimicrobial evaluation of amino sugar-based naphthoquinones and isoquinoline-5,8-diones and their halogenated compounds. Eur. J. Med. Chem. 2018, 156, 1–12. [Google Scholar] [CrossRef]
- Salem, M.A.; Ragab, A.; Askar, A.A.; El-khalafawy, A.; Makhlouf, A.H. European Journal of Medicinal Chemistry One-pot synthesis and molecular docking of some new spiropyranindol-2-one derivatives as immunomodulatory agents and in vitro antimicrobial potential with DNA gyrase inhibitor. Eur. J. Med. Chem. 2020, 188, 111977. [Google Scholar] [CrossRef]
- Cai, S.; Yuan, W.; Li, Y.; Huang, X.; Guo, Q.; Tang, Z.; Fang, Z.; Lin, H.; Wong, W.; Wong, K.; et al. Antibacterial activity of indolyl-quinolinium derivatives and study their mode of action. Bioorg. Med. Chem. 2019, 27, 1274–1282. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). M07-A10 Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard—Tenth Edition. 2015. Available online: https://clsi.org/media/1632/m07a10_sample.pdf (accessed on 21 October 2020).
- Akbay, P.; Calis, I.; Ündeger, Ü.; Basaran, N.; Basaran, A.A. In vitro Immunomodulatory activity of verbascoside from Nepeta ucrainica L. Phyther. Res. 2002, 16, 593–595. [Google Scholar] [CrossRef]
- Ayeka, P.A.; Bian, Y.; Githaiga, P.M.; Zhao, Y. The immunomodulatory activities of licorice polysaccharides (Glycyrrhiza uralensis Fisch.) in CT 26 tumor-bearing mice. BMC Complement. Altern. Med. 2017, 17, 536. [Google Scholar] [CrossRef]
- Lee, K.Y.; Jeon, Y.J. Macrophage activation by polysaccharide isolated from Astragalus membranaceus. Int. Immunopharmacol. 2005, 5, 1225–1233. [Google Scholar] [CrossRef]
- Cheng, A.; Wan, F.; Wang, J.; Jin, Z.; Xu, X. Macrophage immunomodulatory activity of polysaccharides isolated from Glycyrrhiza uralensis fish. Int. Immunopharmacol. 2008, 8, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Nie, S.; Huang, D.; Li, W.; Xie, M. Immunomodulatory effect of Ganoderma atrum polysaccharide on CT26 tumor-bearing mice. Food Chem. 2013, 136, 1213–1219. [Google Scholar] [CrossRef]
- Brush, J.; Mendenhall, E.; Guggenheim, A.; Chan, T.; Connelly, E.; Soumyanath, A.; Buresh, R.; Barrett, R.; Zwickey, H. The effect of Echinacea purpurea, Astragalus membranaceus and Glycyrrhiza glabra on CD69 expression and immune cell activation in humans. Phyther. Res. 2006, 20, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Asiri, A.M.; Basisi, H.M.; Asad, M.; Zayed, M.E.M.; Sharma, K.; Wani, M.Y. Synthesis and evaluation of Quinoline-3-carbonitrile derivatives as potential antibacterial agents. Bioorg. Chem. 2019, 88, 102968. [Google Scholar] [CrossRef]
- Kartsev, V.; Shikhaliev, K.S.; Geronikaki, A.; Medvedeva, S.M.; Ledenyova, I.V.; Krysin, M.Y.; Petrou, A.; Ciric, A.; Glamoclija, J.; Sokovic, M. dithioloquinolinethiones as new potential multitargeted antibacterial and antifungal agents: Synthesis, biological evaluation and molecular docking studies. Eur. J. Med. Chem. 2019, 175, 201–214. [Google Scholar] [CrossRef]
- Hassan, A.S.; Masoud, D.M.; Sroor, F.M.; Askar, A.A. Synthesis and biological evaluation of pyrazolo[1,5-a] pyrimidine-3-carboxamide as antimicrobial agents. Med. Chem. Res. 2017, 26, 2909–2919. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, H.; Luo, X.; Wang, Y.; Liu, Y.; Jin, H.; Liu, Z.; Yang, W.; Yu, P.; Zhang, L.; et al. Design, synthesis and biological activities of 2,3-dihydroquinazolin-4(1H)-one derivatives as TRPM2 inhibitors. Eur. J. Med. Chem. 2018, 152, 235–252. [Google Scholar] [CrossRef]
- Sun, J.; Zhao, R.; Zeng, J.; Li, G.; Li, X. Characterization of Destrins with Different Dextrose Equivalents. Molecules 2010, 15, 5162–5173. [Google Scholar] [CrossRef]
- Alt, S.; Mitchenall, L.A.; Maxwell, A.; Heide, L. Inhibition of DNA gyrase and DNA topoisomerase IV of Staphylococcus aureus and Escherichia coli by aminocoumarin antibiotics. J. Antimicrob. Chemother. 2011, 66, 2061–2069. [Google Scholar] [CrossRef] [PubMed]
- Akbay, P.; Basaran, A.A.; Undeger, U.; Basaran, N. In vitro immunomodulatory activity of flavonoid glycosides from Urtica dioica L. Phyther. Res. 2003, 17, 34–37. [Google Scholar] [CrossRef] [PubMed]
- The Twinned 3.35A Structure of S. Aureus Gyrase Complex with Ciprofloxacin and DNA. Available online: https://www.rcsb.org/structure/2XCT (accessed on 21 October 2020).
- Methotrexate-Resistant Variants of Human Dihydrofolate Reductase with Substitution of Leucine 22: Kinetics, Crystallography and Potential as Selectable Markers. Available online: https://www.rcsb.org/structure/1DLS (accessed on 21 October 2020).

| Cpd. No. | Mean Diameter of the Inhibition Zone (mm) * | |||||||
|---|---|---|---|---|---|---|---|---|
| Gram-Positive Bacteria | Gram-Negative Bacteria | Fungal Strains | ||||||
| B. subtilis | S. aureus | E. faecalis | E. coli | P. aeruginosa | S. typhi | C. albicans | F. oxysporum | |
| 1 | 14 ± 0.70 | 17 ± 0.50 | 19 ± 0.70 | 16 ± 0.35 | 20±0.65 | 17 ± 0.54 | 15 ± 0.63 | 13 ± 0.54 |
| 2a | 27 ± 0. 50 | 25 ± 0.77 | 23 ± 0.65 | 26 ± 0.11 | 21 ± 0. 20 | 23 ± 0.65 | 25 ± 0.33 | 21 ± 0.16 |
| 2b | 25 ± 0.87 | 21 ± 0.3 | 24 ± 0.35 | 23 ± 0.2 | 21 ± 0.55 | 23 ± 0. 40 | 19 ± 0. 25 | 17 ± 0.50 |
| 2c | 20 ± 0.38 | 21 ± 0.85 | 17 ± 0.37 | 19 ± 0.75 | 20 ± 0.42 | 17 ± 0.24 | 16 ± 0.52 | 14 ± 0.5 |
| 2d | 32 ± 0. 22 | 33 ± 0.53 | 29 ± 0.17 | 30 ± 0.29 | 27 ± 0.73 | 29 ± 0.20 | 27 ± 0.50 | 22 ± 0.11 |
| 3a | 31 ± 0.40 | 25 ± 0.31 | 21 ± 0.11 | 24 ± 0.20 | 20 ± 0.15 | 18 ± 0.16 | 22 ± 0.35 | 21 ± 0.30 |
| 3b | 22 ± 0. 12 | Na | 19 ± 0.65 | 21 ± 0.11 | Na | 14 ± 0.16 | 12 ± 0.36 | Na |
| 3c | 25 ± 0. 21 | Na | 19 ± 0.33 | 22 ± 0. 18 | 17 ± 0.20 | 20 ± 0.33 | 22 ± 0.19 | 19 ± 0.55 |
| 3d | 20 ± 0.11 | 23 ± 0.29 | 21 ± 0.54 | 22 ± 0.43 | Na | 15 ± 0.36 | 17 ± 0.21 | Na |
| 4 | 23 ± 0.22 | 24 ± 0.33 | 25 ± 0.35 | 27 ± 0. 3 | 23 ± 0.74 | Na | 20 ± 0.50 | Na |
| 6 | 18 ± 0.12 | 16 ± 0.54 | Na | 15 ± 0. 96 | Na | 12 ± 0.61 | 18 ± 0.20 | 14 ± 0.38 |
| 7 | 13 ± 0.41 | Na | 14 ± 0.47 | 19 ± 0. 33 | 12 ± 0.63 | Na | 13.0 ± 0.20 | Na |
| 8 | 27 ± 0.50 | 26 ± 0.14 | 25 ± 0.33 | 23 ± 0. 14 | 25 ± 0.85 | 23 ± 0.11 | 24 ± 0.30 | 20 ± 0.82 |
| 9a | 22 ± 0.41 | 17 ± 0.78 | 25 ± 0.11 | 18 ± 0.30 | Na | 14 ± 0.52 | 19 ± 0.65 | Na |
| 9b | 23 ± 0.50 | 23 ± 0.12 | 21 ± 0.55 | 22 ± 0.81 | 16 ± 0.20 | 20 ± 0.16 | 17 ± 0.56 | 15 ± 0.15 |
| 10 | 14 ± 0.56 | 17 ± 0.48 | 19 ± 0.63 | 20 ± 0.43 | 21 ± 0.54 | 12 ± 0.35 | 14 ± 0.45 | 16 ± 0.86 |
| 11 | 28 ± 0.11 | 24 ± 0.29 | 29± 0.54 | 25 ± 0.43 | 24 ± 0.13 | 18 ± 0.36 | 22 ± 0.21 | 18 ± 0.45 |
| 12 | 15 ± 0.45 | Na | 12 ± 0.74 | 14 ± 0.21 | Na | 15 ± 0. 20 | 12 ± 0.50 | Na |
| S1 | 25 ± 0.22 | 25 ± 0.11 | 22 ± 0.25 | 23 ± 0.20 | 20 ± 0.5 | 21 ± 0.55 | Na | Na |
| S2 | Na | Na | Na | Na | Na | Na | 22 ± 0.20 | 18 ± 0.32 |
| Cpd. No. | Gram-Positive | Gram-Negative | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B. subtilis | S. aureus | E. faecalis | E. coli | P. aeruginosa | Salmonella typhi | |||||||
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | |
| 2a | 22.90 ± 1.02 | 45.81 ± 1.65 | 77.38 ± 2.65 | 131.5 ± 1.35 | 19.33 ± 0.65 | 38.67 ± 0.65 | 45.83 ± 0.98 | 90.38 ± 0.98 | 137.43 ± 2.12 | 219.88 ± 1.54 | 77.38 ± 1.06 | 139.28 ± 1.63 |
| 2b | 19.55 ± 0.39 | 35.17 ± 1.25 | 39.10 ± 1.67 | 78.23 ± 0.87 | 23.15 ± 0.61 | 46.31 ± 0.74 | 156.47 ± 2.65 | 219.06 ± 2.14 | 78.23 ± 1.24 | 132.98 ± 1.02 | 39.10 ± 0.36 | 70.37 ± 0.85 |
| 2d | 4.69 ± 0.98 | 9.38 ± 0.96 | 13.40 ± 0.98 | 13.40 ± 0.54 | 9.38 ± 0.24 | 15.95 ± 0.58 | 2.33 ± 0.32 | 4.66 ± 0.21 | 13.40 ± 0.25 | 25.46 ± 0.65 | 18.80 ± 0.85 | 30.06 ± 0.75 |
| 3a | 9.61 ± 1.65 | 19.6 ± 0.65 | 16.69 ± 0.87 | 33.38 ± 0.65 | 8.33 ± 0.36 | 15.83 ± 0.47 | 16.69 ± 0.84 | 30.02 ± 0.52 | 33.38 ± 0.35 | 66.78 ± 0.87 | 11.90 ± 0.62 | 22.61 ± 0.32 |
| 8 | 16.12 ± 0.54 | 30.63 ± 1.78 | 5.64 ± 0.58 | 10.71 ± 0.36 | 11.29 ± 0.52 | 19.19 ± 0.18 | 22.61 ± 0.42 | 36.16 ± 0.42 | 90.48 ± 1.07 | 171.89 ± 1.95 | 11.29 ± 0.75 | 19.19 ± 0.14 |
| 11 | 10.11 ± 0.68 | 17.20 ± 0.54 | 20.26 ± 0.67 | 40.52 ± 1.02 | 40.52 ± 0.74 | 81.08 ± 0.98 | 20.26 ± 0.36 | 40.52 ± 0.75 | 72.05 ± 0.96 | 144.11 ± 1.47 | 48.02 ± 0.85 | 94.70 ± 0.79 |
| Tetr. | 70.31 ± 2.65 | 91.40 ± 2.89 | 140.63 ± 3.21 | 196.8 ± 3.54 | 140.6 ± 1.98 | 210.9 ± 1.78 | 35.14 ± 0.87 | 42.16 ± 0.92 | 140.6 ± 2.85 | 196.89 ± 2.36 | 70.31 ± 1.06 | 98.44 ± 0.87 |
| Cpd. No. | Fungi | |||||
|---|---|---|---|---|---|---|
| C. albicans | F. oxysporum | |||||
| MIC | MFC | MFC/MIC | MIC | MFC | MFC/MIC | |
| 2a | 77.38 ± 0.85 | 103.13 ± 1.02 | <2 | 137.52 ± 1.69 | 216.67 ± 2.98 | <2 |
| 2b | 78.23 ± 0.95 | 132.98 ± 1.65 | <2 | 138.94 ± 1.53 | 222.31 ± 1.65 | <2 |
| 2d | 18.80 ± 0.65 | 37.60 ± 0.38 | <2 | 37.60 ± 0.25 | 66.84 ± 0.85 | <2 |
| 3a | 16.69 ± 0.75 | 26.69 ± 0.54 | <2 | 33.38 ± 0.74 | 56.74 ± 0.98 | <2 |
| 8 | 45.22 ± 0.54 | 81.38 ± 0.84 | <2 | 90.48 ± 0.84 | 135.70 ±1.98 | <2 |
| 11 | 24.00 ± 0.41 | 45.58 ± 0.47 | <2 | 81.08 ± 0.54 | 145.95 ± 1.25 | <2 |
| Amp. B | 16.81 ± 0.29 | 37.26 ± 0.65 | <2 | 33.63 ± 0.98 | 70.62 ± 0.76 | <2 |
| Cpd. No. | Mean Diameter of Inhibition Zone (mm) | |||
|---|---|---|---|---|
| S. aureus ATCC 43300 | S. aureus ATCC 33591 | E. coli ATCC BAA-196 | P. aeruginosa ATCC BAA-2111 | |
| 2a | 23 ± 0.11 | 25 ± 0.33 | 23 ± 0.94 | 24 ± 0.18 |
| 2b | 20 ± 0.34 | 21 ± 0.11 | 22 ± 0.85 | 19 ± 0.16 |
| 2d | 27 ± 0.3 | 24 ± 0.32 | 25 ± 0.86 | 26 ± 0.41 |
| 3a | 25 ± 0.4 | 23 ± 0.47 | 22 ± 0.23 | 24 ± 0.22 |
| 8 | 22 ± 0.5 | 15 ± 0.33 | 20 ± 0.74 | 21 ± 0.63 |
| 11 | 17 ± 0.3 | 22 ± 0.65 | 19 ± 0.88 | 23 ± 0.25 |
| Tetr. | - | - | - | - |
| Nor. | 25 ± 0.5 | 26 ± 0.5 | 27 ± 0.98 | 24 ± 0.47 |
 | ||||||
|---|---|---|---|---|---|---|
| Cpd. No. | Structure | Test | S. aureus ATCC 43300 | S. aureus ATCC 33591 | E. coli ATCC BAA-196 | P. aeruginosa ATCC BAA-2111 |
| 2a | 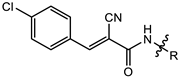 | MIC | 10.99 ± 0.25 | 4.82 ± 0.24 | 15.47 ± 0.26 | 12.87 ± 0.65 |
| MBC | 21.98 ± 0.65 | 8.19 ± 0.26 | 29.39 ± 0.65 | 21.32 ± 0.25 | ||
| 2b |  | MIC | 19.55 ±0.52 | 9.38 ± 0.35 | 4.69 ± 0.45 | 9.38 ± 0.47 |
| MBC | 39.10 ± 0.54 | 18.77 ± 0.21 | 9.38 ± 0.74 | 18.77 ± 0.87 | ||
| 2d | 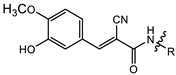 | MIC | 4.69 ± 0.25 | 9.38 ±0.32 | 4.69 ± 0.36 | 9.38 ± 0.49 |
| MBC | 9.38 ± 0.41 | 18.77 ± 0.25 | 9.38 ± 0.54 | 18.77 ± 0.61 | ||
| 3a | 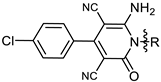 | MIC | 8.33 ± 0.29 | 11.85 ± 0.24 | 16.69 ± 0.41 | 9.48 ± 0.54 |
| MBC | 16.26 ± 0.36 | 23.72 ± 0.28 | 33.38 ± 0.84 | 14.23 ±0.65 | ||
| 8 | 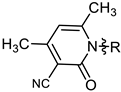 | MIC | 18.09 ± 0.84 | 25.71 ±0.36 | 26.78 ± 0.65 | 22.61 ± 0.98 |
| MBC | 34.36 ± 0.24 | 46.26 ± 0.58 | 53.56 ± 0.41 | 45.22 ± 0.87 | ||
| 11 | 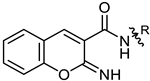 | MIC | 40.52 ± 0.36 | 10.11 ± 0.29 | 24.00 ± 0.49 | 40.52 ± 0.74 |
| MBC | 81.08 ± 0.85 | 19.22 ± 0.65 | 48.00 ±0.81 | 81.08 ± 0.86 | ||
| Nor. | 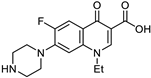 | MIC | 3.91 ± 0.21 | 2.44 ± 0.45 | 4.91 ± 0.69 | 9.80 ± 0.41 |
| MBC | 7.82 ±0.32 | 4.38 ± 0.49 | 8.32 ±0.47 | 16.65 ± 0.19 | ||
| Cpd. No. | IC50 µM | |
|---|---|---|
| S. aureus DNA Gyrase Supercoiling | E. coli DHFR | |
| 2a | 23.87 ± 1.22 | 5.54 ± 0.33 |
| 2b | 29.14 ± 1.93 | 9.51 ± 0. 25 |
| 2d | 18.17 ± 1.18 | 4.33 ± 0.18 |
| 3a | 21.97 ± 1.35 | 5.44 ± 0. 95 |
| 8 | 39.15 ± 1.72 | 12.3 ± 0.73 |
| 11 | 39.41 ± 1.15 | 17.64 ± 0. 54 |
| Cip. | 26.32 ± 1.76 | - |
| Trim. | - | 5.16 ± 0.12 |
| Cpd. No. | Intracellular Killing Activity % |
|---|---|
| 2a | 82.8 ± 0.37 |
| 2b | 98.7 ± 0. 19 |
| 2d | 142.4 ± 0.98 |
| 3a | 113.2 ± 0. 5 |
| 8 | 136.5 ± 0.3 |
| 11 | 116.7 ± 0. 14 |
| Index | Normal | Vitamin C | 2d | 8 | 11 |
|---|---|---|---|---|---|
| Spleen mg | 90 ± 0.02 | 260 ± 0.028 | 250 ± 0.03 | 200 ± 0.025 | 190 ± 0.074 |
| Thymus mg | 19.63 ± 0.35 | 17.2 ± 0.31 | 15.56 ± 0.54 | 13.42 ± 0.46 | 13.67 ± 0.53 |
| Spleen index mg/g | 0.004 ± 0.00 | 0.019 ± 0.00 | 0.018 ± 0.00 | 0.010 ± 0.00 | 0.008 ± 0.00 |
| Thymus index mg/g | 0.29 ± 0.26 | 0.633 ± 0.06 | 0.532 ± 0.06 | 0.537 ± 0.14 | 0.455 ± 0.07 |
| TT Cells, % | Normal | Vitamin C | 2d | 8 | 11 |
|---|---|---|---|---|---|
| CD4+ | 79.82 ± 1.3 | 76.74 ± 0.7 | 79.61 ± 0.3 | 78.70 ± 0.56 | 75.20 ± 0.97 |
| CD8+ | 18.55 ± 1.9 | 19.62 ± 0.21 | 27.05 ± 0.5 | 18.44 ± 0.2 | 24.05 ± 0.32 |
| Cpd. No | MW | MLogP | nHBA | nHBD | nRB | TPSA | Violations from Lipinski’s Rule |
|---|---|---|---|---|---|---|---|
| Rule | <500 g/mol | ≤4.15 | ≤10 | ≤5 | ≤10 | ≤140 Å2 | Yes; 0 or 1 violation |
| 2a | 403.84 | 0.82 | 5 | 4 | 7 | 157.31 | Yes; 0 violation |
| 2b | 399.42 | 0.04 | 6 | 4 | 8 | 166.54 | Yes; 0 violation |
| 2d | 415.42 | −0.46 | 7 | 5 | 8 | 186.77 | Yes; 0 violation |
| 3a | 467.89 | 0.03 | 6 | 4 | 5 | 200.02 | Yes; 0 violation |
| 8 | 345.38 | −0.09 | 5 | 3 | 4 | 150.21 | Yes; 0 violation |
| 11 | 385.40 | 0.39 | 6 | 5 | 6 | 170.51 | Yes; 0 violation |
| Ciprofloxacin | 331.34 | 1.28 | 5 | 2 | 3 | 74.57 | Yes; 0 violation |
| Trimethoprim | 290.32 | 0.41 | 5 | 2 | 5 | 105.51 | Yes; 0 violation |
| Cpd. No. | GI Absorption | BBB Permeant | P-gp Substrate | AMES Toxicity | Carcinogens | Acute Oral Toxicity |
|---|---|---|---|---|---|---|
| 2a | Low | No | No | No | No | III |
| 2b | Low | No | No | No | No | III |
| 2d | Low | No | No | No | No | III |
| 3a | Low | No | No | No | No | III |
| 8 | Low | No | No | No | No | III |
| 11 | Low | No | No | No | No | III |
| Ciprofloxacin | High | No | Yes | Yes | No | III |
| Trimethoprim | High | No | Yes | No | No | IV |
| Cpd. No. | DNA Gyrase (PDB: 2XCT) | DHFR (PDB: IDLS) | ||||
|---|---|---|---|---|---|---|
| S | Interacting Groups | °A | S | Interacting Groups | °A | |
| 2a | −20.27 | Arg1092 with NH (imino) guanidine | 3.2 & 2.8 | −20.26 | Ser118 with oxygen of sulfonyl | 3.0 |
| His1046 with oxygen of sulfonyl | 2.8 | Thr146 with oxygen of sulfonyl | 2.7 | |||
| Arg1033 with cyano group | 3.0 | |||||
| 2b | −17.36 | Ser1085 with two NH of guanidine | 3.0 & 3.2 | −24.44 | Asn64 with oxygen of methoxy | 2.8 |
| Lys1043 with carbonyl of acetamide | 2.8 | Thr146 with oxygen of sulfonyl | 2.9 | |||
| Gln1267 with oxygen of methoxy | 3.0 | Thr56 with NH of guanidine | 3.1 | |||
| His1046 with carbonyl of acetamide | 3.2 | Thr56 with NH (imino) of guanidine | 2.8 | |||
| Arg1092 with cyano group | 3.2 & 2.7 | |||||
| Arg1092 with phenyl of aldehyde | - | |||||
| 2d | −21.67 | His1079 with oxygen of sulfonyl | 2.8 | −24.70 | Thr56 with NH of acetamide derivative | 3.0 |
| Lys1043 with oxygen of acetamide | 2.6 | |||||
| Ser1173 with hydroxyl of aldehyde | 2.5 | |||||
| Gln1267 with oxygen of methoxy | 2.9 | |||||
| Arg1092 with phenyl of aldehyde | - | |||||
| 3a | −20.78 | Arg1033 with oxygen of sulfonyl | 2.7 | −19.54 | Ser118 with oxygen of sulfonyl Thr146 with oxygen of sulfonyl | 2.7 3.1 |
| His1079 with oxygen of sulfonyl | 3.0 | |||||
| Ser1085 with NH2 of guanidine | 3.1 | |||||
| Arg1092 with cyano group | 2.7 | |||||
| Arg1092 with phenyl at position four of pyridine | - | |||||
| 8 | −17.96 | Arg1092 with NH (imino) of guanidine | 2.5 | −23.32 | Glu30 with NH (imino) of guanidine | 2.5 |
| His1046 with oxygen of sulfonyl | 3.0 | Glu30 with NH of guanidine | 2.5 | |||
| Lys1043 with phenyl of sulfonamide | - | |||||
| 11 | −15.98 | Ser1173 with NH of chromene | 2.8 | −18.38 | Ser119 with oxygen of sulfonyl | 2.7 |
| Lys1043 with NH (imino) of guanidine | 2.7 | |||||
| Lys1043 with carbonyl of amide | 2.7 | |||||
| Arg1092 with phenyl of chromene | - | |||||
| STD * | −9.87 | His1081 with oxygen of carboxylate Tyr580 with NH of piperazine | 2.3 2.6 | −24.44 | Arg70 with carbonyl of carboxylate | 3.0 |
| Arg70 with oxygen of carboxylate | 2.3 | |||||
| Asn64 with oxygen of carboxylate | 2.9 | |||||
| Arg28 with carbonyl of carboxylate | 2.9 | |||||
| Asn64 with carbonyl of amide | 2.5 | |||||
| Val115 with NH2 of pyrimidine | 2.8 | |||||
| Lle7 with NH2 of pyrimidine | 3.2 | |||||
| Gln30 with NH2 of pyrimidine | 2.8 & 2.7 | |||||
| Phe34 with pyrazine ring | - - | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ragab, A.; Fouad, S.A.; Ali, O.A.A.; Ahmed, E.M.; Ali, A.M.; Askar, A.A.; Ammar, Y.A. Sulfaguanidine Hybrid with Some New Pyridine-2-One Derivatives: Design, Synthesis, and Antimicrobial Activity against Multidrug-Resistant Bacteria as Dual DNA Gyrase and DHFR Inhibitors. Antibiotics 2021, 10, 162. https://doi.org/10.3390/antibiotics10020162
Ragab A, Fouad SA, Ali OAA, Ahmed EM, Ali AM, Askar AA, Ammar YA. Sulfaguanidine Hybrid with Some New Pyridine-2-One Derivatives: Design, Synthesis, and Antimicrobial Activity against Multidrug-Resistant Bacteria as Dual DNA Gyrase and DHFR Inhibitors. Antibiotics. 2021; 10(2):162. https://doi.org/10.3390/antibiotics10020162
Chicago/Turabian StyleRagab, Ahmed, Sawsan A. Fouad, Ola A. Abu Ali, Entsar M. Ahmed, Abeer M. Ali, Ahmed A. Askar, and Yousry A. Ammar. 2021. "Sulfaguanidine Hybrid with Some New Pyridine-2-One Derivatives: Design, Synthesis, and Antimicrobial Activity against Multidrug-Resistant Bacteria as Dual DNA Gyrase and DHFR Inhibitors" Antibiotics 10, no. 2: 162. https://doi.org/10.3390/antibiotics10020162
APA StyleRagab, A., Fouad, S. A., Ali, O. A. A., Ahmed, E. M., Ali, A. M., Askar, A. A., & Ammar, Y. A. (2021). Sulfaguanidine Hybrid with Some New Pyridine-2-One Derivatives: Design, Synthesis, and Antimicrobial Activity against Multidrug-Resistant Bacteria as Dual DNA Gyrase and DHFR Inhibitors. Antibiotics, 10(2), 162. https://doi.org/10.3390/antibiotics10020162







