Antibiotic and Heavy Metal Susceptibility of Non-Cholera Vibrio Isolated from Marine Sponges and Sea Urchins: Could They Pose a Potential Risk to Public Health?
Abstract
1. Introduction
2. Results
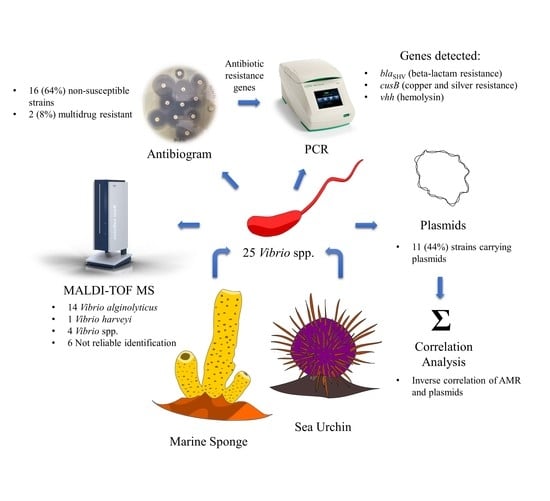
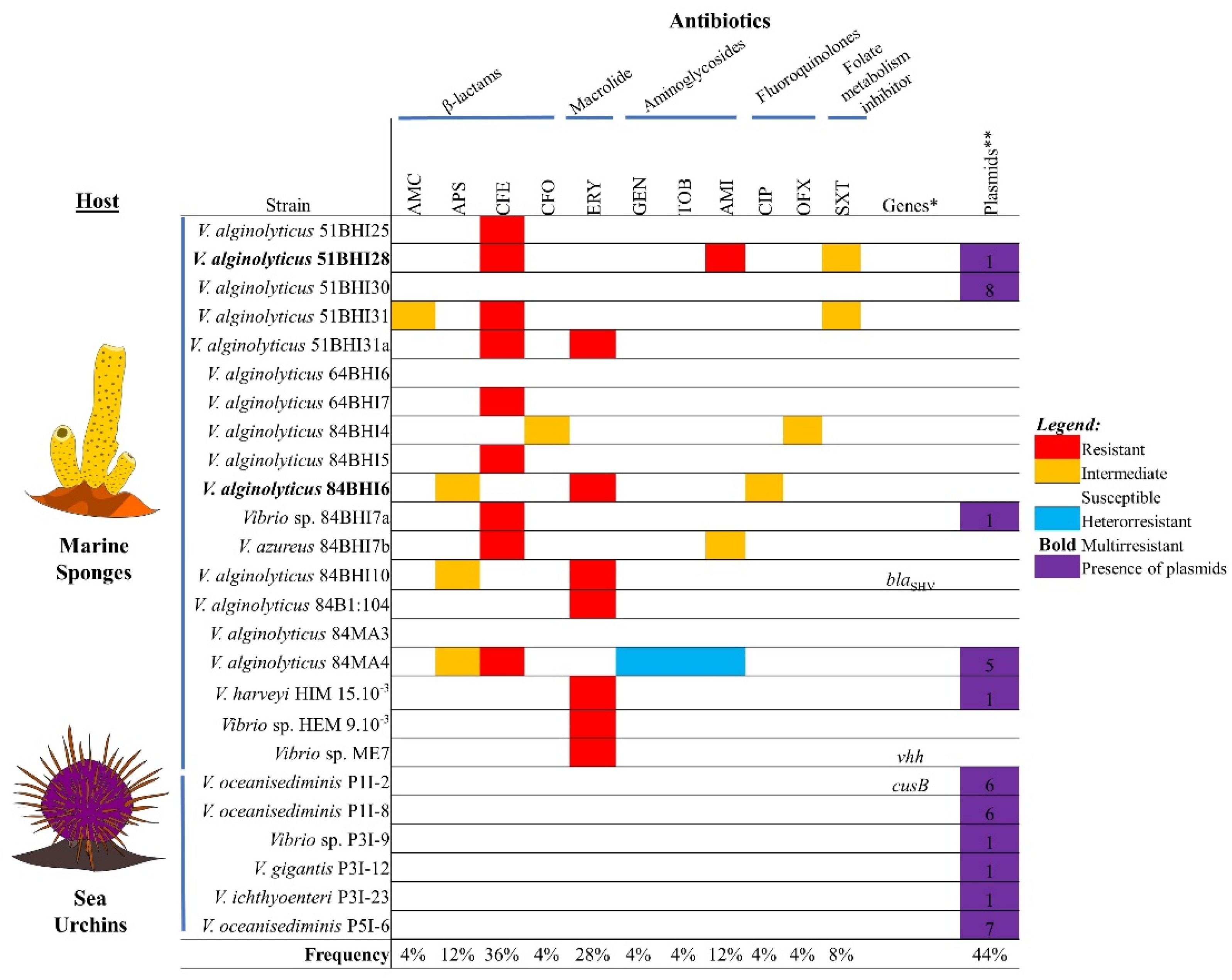
2.1. Identification of Vibrio spp.
2.2. Antibiotic Non-Susceptibility among Vibrio spp. Isolates
2.3. Detection of Genotypes: Antibiotic and Heavy Metal Resistance Genes, Virulence Genes, and Integrons
2.4. Plasmid Profile
2.5. Association between Antibiotic Resistance Phenotypes and Occurrence of Plasmids
3. Discussion
4. Materials and Methods
4.1. Bacterial Isolates
4.2. MALDI-TOF MS Identification
4.3. Antibiotic Susceptibility Profiling
4.4. Genomic and Plasmid DNA Extractions
4.5. PCR Assays
4.6. Verification of Antibiotic Resistance Phenotype and Plasmids Association
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boyd, E.F.; Carpenter, M.R.; Chowdhury, N.; Cohen, A.L.; Haines-Menges, B.L.; Kalburge, S.S.; Kingston, J.J.; Lubin, J.B.; Ongagna-Yhombi, S.Y.; Whitaker, W.B. Post-genomic analysis of members of the family Vibrionaceae. Microbiol. Spectr. 2015, 3, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Oliver, J.D.; Alam, M.; Ali, A.; Waldor, M.K.; Qadri, F.; Martinez-Urtaza, J. Vibrio spp. infections. Nat. Rev. Dis. Primers 2018, 4, 1–19. [Google Scholar] [CrossRef]
- Elmahdi, S.; Da Silva, L.V.; Parveen, S. Antibiotic resistance of Vibrio parahaemolyticus and Vibrio vulnificus in various countries: A review. Food Microbiol. 2016, 57, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Fri, J.; Ndip, R.N.; Njom, H.A.; Clarke, A.M. Antibiotic susceptibility of non-cholera Vibrios isolated from farmed and wild marine fish (Argyrosomus japonicus), implications for public health. Microb. Drug. Resist. 2018, 24, 1296–1304. [Google Scholar] [CrossRef] [PubMed]
- Canellas, A.L.B.; da Costa, W.F.; Paranhos, R.; Laport, M.S. Diving into the unknown: Identification of antimicrobial resistance hotspots in a tropical urban estuary. Lett. Appl. Microbiol. 2021, 73, 270–279. [Google Scholar] [CrossRef]
- Canellas, A.L.B.; Lopes, I.R.; Mello, M.P.; Paranhos, R.; de Oliveira, B.F.R.; Laport, M.S. Vibrio species in an urban tropical estuary: Antimicrobial susceptibility, interaction with environmental parameters, and possible public health outcomes. Microorganisms 2021, 9, 1007. [Google Scholar] [CrossRef]
- Deng, Y.; Xu, H.; Su, Y.; Liu, S.; Xu, L.; Guo, Z.; Wu, J.; Cheng, C.; Feng, J. Horizontal gene transfer contributes to virulence and antibiotic resistance of Vibrio harveyi 345 based on complete genome sequence analysis. BMC Genom. 2019, 20, 761. [Google Scholar] [CrossRef]
- Deng, Y.; Xu, L.; Liu, S.; Wang, Q.; Guo, Z.; Chen, C.; Feng, J. What drives changes in the virulence and antibiotic resistance of Vibrio harveyi in the South China Sea? J. Fish Dis. 2020, 43, 853–862. [Google Scholar] [CrossRef]
- Lloyd, N.A.; Janssen, S.E.; Reinfelder, J.R.; Barkay, T. Co-selection of mercury and multiple antibiotic resistances in bacteria exposed to mercury in the Fundulus heteroclitus gut microbiome. Curr. Microbiol. 2016, 73, 834–842. [Google Scholar] [CrossRef]
- Laport, M.S.; Bauwens, M.; Collard, M.; George, I. Phylogeny and antagonistic activities of culturable bacteria associated with the gut microbiota of the sea urchin (Paracentrotus lividus). Curr. Microbiol. 2018, 75, 359–367. [Google Scholar] [CrossRef]
- Laport, M.S. Isolating bacteria from sponges: Why and how? Curr. Pharm. Biotechnol. 2017, 18, 1224–1236. [Google Scholar] [CrossRef]
- World Health Organization. What Is ‘One Health’? Available online: https://www.who.int/news-room/q-a-detail/one-health (accessed on 27 November 2021).
- Freitas-Silva, J.; Silva-Oliveira, T.; Muricy, G.; Laport, M.S. Bacillus strains associated to Homoscleromorpha sponges are highly active against multidrug resistant bacteria. Curr. Microbiol. 2020, 77, 807–815. [Google Scholar] [CrossRef]
- Eddabra, R.; Prévost, G.; Scheftel, J.-M. Rapid discrimination of environmental Vibrio by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Microbiol. Res. 2012, 167, 226–230. [Google Scholar] [CrossRef]
- Håkonsholm, F.; Lunestad, B.T.; Aguirre Sánchez, J.R.; Martinez-Urtaza, J.; Marathe, N.P.; Svanevik, C.S. Vibrios from the Norwegian marine environment: Characterization of associated antibiotic resistance and virulence genes. Microbiologyopen 2020, 9, 1093. [Google Scholar] [CrossRef] [PubMed]
- Santos, I.C.; Hildenbrand, Z.L.; Schug, K.A. Applications of MALDI-TOF MS in environmental microbiology. Analyst 2016, 141, 2827–2837. [Google Scholar] [CrossRef]
- Hernández-Robles, M.F.; Álvarez-Contreras, A.K.; Juárez-García, P.; Natividad-Bonifacio, I.; Curiel-Quesada, E.; Vázquez-Salinas, C.; Quiñones-Ramírez, E.I. Virulence factors and antimicrobial resistance in environmental strains of Vibrio alginolyticus. Int. Microbiol. 2016, 19, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Li, R.; Ye, L.; Chan, E.W.-C.; Chen, S. Identification and characterization of IncA/C conjugative, blaNDM-1-bearing plasmid in Vibrio alginolyticus of food origin. Antimicrob. Agents Chemother. 2018, 62, e01897-18. [Google Scholar] [CrossRef] [PubMed]
- Cartwright, A.; Arnscheidt, J.; Conwell, M.; Dooley, J.S.G.; McGonigle, C.; Naughton, P.J. Effects of freshwater sponge Ephydatia fluviatilis on conjugative transfer of antimicrobial resistance in Enterococcus faecalis strains in aquatic environments. Lett. Appl. Microbiol. 2020, 71, 39–45. [Google Scholar] [CrossRef]
- Chiou, J.; Li, R.; Chen, S. CARB-17 family of β-lactamases mediates intrinsic resistance to penicillins in Vibrio parahaemolyticus. Antimicrob. Agents Chemother. 2015, 59, 3593–3595. [Google Scholar] [CrossRef] [PubMed]
- Krishnamoorthy, G.; Leus, I.V.; Weeks, J.W.; Wolloscheck, D.; Rybenkov, V.V.; Zgurskaya, H.I. Synergy between active efflux and outer membrane diffusion defines rules of antibiotic permeation into Gram-negative bacteria. MBio 2017, 8, e01172-17. [Google Scholar] [CrossRef]
- Sulca, M.A.; Orozco, R.; Alvarado, D.E. Antimicrobial resistance not related to 1, 2, 3 integrons and Superintegron in Vibrio spp. isolated from seawater sample of Lima (Peru). Mar. Pollut. Bull. 2018, 131, 370–377. [Google Scholar] [CrossRef]
- Prinarakis, E.E.; Miriagou, V.; Tzelepi, E.; Gazouli, M.; Tzouvelekis, L.S. Emergence of an inhibitor-resistant beta-lactamase (SHV-10) derived from an SHV-5 variant. Antimicrob. Agents Chemother. 1997, 41, 838–840. [Google Scholar] [CrossRef] [PubMed]
- da Costa, W.F.; Giambiagi-deMarval, M.; Laport, M.S. Shewanella harboring antimicrobial and copper resistance genes in sea urchins (Paracentrotus lividus) from the Crozon peninsula (Brittany, France). Infect. Genet. Evol. 2020, 85, 104437. [Google Scholar] [CrossRef] [PubMed]
- Conejero, M.J.U.; Hedreyda, C.T. PCR detection of hemolysin (vhh) gene in Vibrio harveyi. J. Gen. Appl. Microbiol. 2004, 50, 137–142. [Google Scholar] [CrossRef][Green Version]
- Ben-Haim, Y.; Zicherman-Keren, M.; Rosenberg, E. Temperature-regulated bleaching and lysis of the coral Pocillopora damicornis by the novel pathogen Vibrio coralliilyticus. Appl. Environ. Microbiol. 2003, 69, 4236–4242. [Google Scholar] [CrossRef]
- Austin, B.; Austin, D.; Sutherland, R.; Thompson, F.; Swings, J. Pathogenicity of vibrios to rainbow trout (Oncorhynchus mykiss, Walbaum) and Artemia nauplii. Environ. Microbiol. 2005, 7, 1488–1495. [Google Scholar] [CrossRef]
- Kim, H.J.; Jun, J.W.; Giri, S.S.; Chi, C.; Yun, S.; Kim, S.G.; Park, S.C. Identification and genome analysis of Vibrio coralliilyticus causing mortality of Pacific oyster (Crassostrea gigas) larvae. Pathogens 2020, 9, 206. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Gil, B.; Soto-Rodríguez, S.; García-Gasca, A.; Roque, A.; Vazquez-Juarez, R.; Thompson, F.L.; Swings, J. Molecular identification of Vibrio harveyi-related isolates associated with diseased aquatic organisms. Microbiology 2004, 150, 1769–1777. [Google Scholar] [CrossRef] [PubMed]
- Bier, N.; Schwartz, K.; Guerra, B.; Strauch, E. Survey on antimicrobial resistance patterns in Vibrio vulnificus and Vibrio cholerae non-O1/non-O139 in Germany reveals carbapenemase-producing Vibrio cholerae in coastal waters. Front. Microbiol. 2015, 6, 1179. [Google Scholar] [CrossRef]
- CLSI. Performance standards for antimicrobial susceptibility testing. In Clinical and Laboratory Standards Institute, 31st ed.; CLSI: Wayne, PA, USA, 2021. [Google Scholar]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, G.C.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 8, 268–281. [Google Scholar] [CrossRef]
- Andersson, D.I.; Nicoloff, H.; Hjort, K. Mechanisms and clinical relevance of bacterial heteroresistance. Nat. Rev. Microbiol. 2019, 17, 479–496. [Google Scholar] [CrossRef]
- Ramos, P.I.P.; Picão, R.C.; Almeida, L.G.P.; Lima, N.C.B.; Girardello, R.; Vivan, A.C.P.; Xavier, D.E.; Barcellos, F.G.; Pelisson, M.; Vespero, E.C.; et al. Comparative analysis of the complete genome of KPC-2-producing Klebsiella pneumoniae Kp13 reveals remarkable genome plasticity and a wide repertoire of virulence and resistance mechanisms. BMC Genom. 2017, 15, 54. [Google Scholar] [CrossRef] [PubMed]
- Santos, L.O.; de Lanna, C.A.; Arcanjo, A.C.C.; Bisch, P.M.; von Krüger, W.M.A. Genotypic diversity and pathogenic potential of clinical and environmental Vibrio parahaemolyticus isolates from Brazil. Front. Microbiol. 2021, 12, 602653. [Google Scholar] [CrossRef]
- Campana, E.H.; Xavier, D.E.; Petrolini, F.V.; Cordeiro-Moura, J.R.; Araujo, M.R.; Gales, A.C. Carbapenem-resistant and cephalosporin-susceptible: A worrisome phenotype among Pseudomonas aeruginosa clinical isolates in Brazil. Braz. J. Infect. Dis. 2017, 21, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Schiwon, K.; Arends, K.; Rogowski, K.M.; Fürch, S.; Prescha, K.; Sakinc, T.; Van Houdt, R.; Werner, G.; Grohmann, K. Comparison of antibiotic resistance, biofilm formation and conjugative transfer of Staphylococcus and Enterococcus isolates from International Space Station and Antarctic Research Station Concordia. Microb. Ecol. 2013, 65, 638–651. [Google Scholar] [CrossRef] [PubMed]
- Park, C.H.; Robicsek, A.; Jacoby, G.A.; Sahm, D.; Hooper, D.C. Prevalence in the United States of aac(6’)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 2006, 50, 3953–3955. [Google Scholar] [CrossRef] [PubMed]
- Heuer, H.; Krögerrecklenfort, E.; Wellington, E.M.H.; Egan, S.; van Elsas, J.D.; van Overbeek, L.; Collard, J.-M.; Guillaume, G.; Karagouni, A.D.; Nikolakopoulou, T.L.; et al. Gentamicin resistance genes in environmental bacteria: Prevalence and transfer. FEMS Microbiol. Ecol. 2002, 42, 289–302. [Google Scholar] [CrossRef]
- Nonaka, L.; Maruyama, F.; Suzuki, S.; Masuda, M. Novel macrolide-resistance genes, mef(C) and mph(G), carried by plasmids from Vibrio and Photobacterium isolated from sediment and seawater of a coastal aquaculture site. Lett. Appl. Microbiol. 2015, 61, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.B.; Frimodt-Møller, N.; Aarestrup, F.M. Presence of erm gene classes in gram-positive bacteria of animal and human origin in Denmark. FEMS Microbiol. Lett. 1999, 170, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Sutcliffe, J.; Grebe, T.; Kamradt, A.T.; Wondrack, L. Detection of erythromycin resistant determinants by PCR. Antimicrob. Agents Chemother. 1996, 40, 2562–2566. [Google Scholar] [CrossRef]
- Luna, V.A.; Cousin, S., Jr.; Whittington, W.L.H.; Roberts, M.C. Identification of the conjugative mef gene in clinical Acinetobacter junii and Neisseria gonorrhoeae isolates. Antimicrob. Agents Chemother. 2000, 44, 2503–2506. [Google Scholar] [CrossRef][Green Version]
- Kraychete, G.B.; Botelho, L.A.B.; Campana, E.H.; Picão, R.C.; Bonelli, R.R. Updated multiplex PCR for detection of all six plasmid-mediated qnr gene families. Antimicrob. Agents Chemother. 2016, 60, 7524–7526. [Google Scholar] [CrossRef] [PubMed]
- De, J.; Ramaiah, N.; Vardanyan, L. Detoxification of toxic heavy metals by marine bacteria highly resistant to mercury. Mar. Biotechnol. 2008, 10, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Deredjian, A.; Colinon, C.; Brothier, E.; Favre-Bonté, S.; Cournoyer, B.; Nazaret, S. Antibiotic and metal resistance among hospital and outdoor strain of Pseudomonas aeruginosa. Res. Microbiol. 2011, 162, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.; Nisar, M.A.; Hussain, S.Z.; Arshad, M.N.; Rehman, A. Cadmium resistance mechanism in Escherichia coli P4 and its potential use to bioremediate environmental cadmium. Appl. Microbiol. Biotechnol. 2015, 99, 10745–10757. [Google Scholar] [CrossRef]
- Oger, C.; Bertheb, T.; Quilleta, L.; Barraya, S.; Chiffoleauc, J.F.; Petit, F. Estimation of the abundance of the cadmium resistance gene cadA in microbial communities in polluted estuary water. Res. Microbiol. 2001, 152, 671–678. [Google Scholar] [CrossRef]
- Borremans, B.; Hobman, J.L.; Provoost, A.; Brown, N.L.; van-Der-Lelie, D. Cloning and functional analysis of the pbr lead resistance determinant of Ralstonia metallidurans CH34. J. Bacteriol. 2001, 183, 5651–5658. [Google Scholar] [CrossRef] [PubMed]
- Guin, S.; Saravanan, M.; Chowdhury, G.; Pazhani, G.P.; Ramamurthy, T.; Das, S.C. Pathogenic Vibrio parahaemolyticus indiarrhoeal patients, fish and aquatic environments and their potential for inter-source transmission. Heliyon 2019, 5, 01743. [Google Scholar] [CrossRef]
- Chao, G.; Jiao, X.; Zhou, X.; Wang, F.; Yang, Z.; Huang, Z.; Pan, Z.; Zhou, L.; Qian, X. Distribution of genes encoding four pathogenicity islands (VPaIs), T6SS, biofilm, and type I pilus in food and clinical strains of Vibrio parahaemolyticus in China. Foodborne Pathog. Dis. 2010, 7, 649–658. [Google Scholar] [CrossRef]
- Jiang, H.; Yu, T.; Yang, Y.; Yu, S.; Wu, J.; Lin, R.; Li, Y.; Fang, J.; Zhu, C. Co-occurrence of antibiotic and heavy metal resistance and sequence type diversity of Vibrio parahaemolyticus isolated from Penaeus vannamei at freshwater farms, seawater farms, and markets in Zhejiang Province, China. Front. Microbiol. 2020, 11, 1294. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa, W.F.; Giambiagi-deMarval, M.; Laport, M.S. Antibiotic and Heavy Metal Susceptibility of Non-Cholera Vibrio Isolated from Marine Sponges and Sea Urchins: Could They Pose a Potential Risk to Public Health? Antibiotics 2021, 10, 1561. https://doi.org/10.3390/antibiotics10121561
Costa WF, Giambiagi-deMarval M, Laport MS. Antibiotic and Heavy Metal Susceptibility of Non-Cholera Vibrio Isolated from Marine Sponges and Sea Urchins: Could They Pose a Potential Risk to Public Health? Antibiotics. 2021; 10(12):1561. https://doi.org/10.3390/antibiotics10121561
Chicago/Turabian StyleCosta, Wellington Felipe, Marcia Giambiagi-deMarval, and Marinella Silva Laport. 2021. "Antibiotic and Heavy Metal Susceptibility of Non-Cholera Vibrio Isolated from Marine Sponges and Sea Urchins: Could They Pose a Potential Risk to Public Health?" Antibiotics 10, no. 12: 1561. https://doi.org/10.3390/antibiotics10121561
APA StyleCosta, W. F., Giambiagi-deMarval, M., & Laport, M. S. (2021). Antibiotic and Heavy Metal Susceptibility of Non-Cholera Vibrio Isolated from Marine Sponges and Sea Urchins: Could They Pose a Potential Risk to Public Health? Antibiotics, 10(12), 1561. https://doi.org/10.3390/antibiotics10121561