Evaluation of Antibiotic Resistance of Salmonella Serotypes and Whole-Genome Sequencing of Multiresistant Strains Isolated from Food Products in Russia
Abstract
:1. Introduction
2. Results
2.1. Prevalence of Salmonella spp. in Food Products
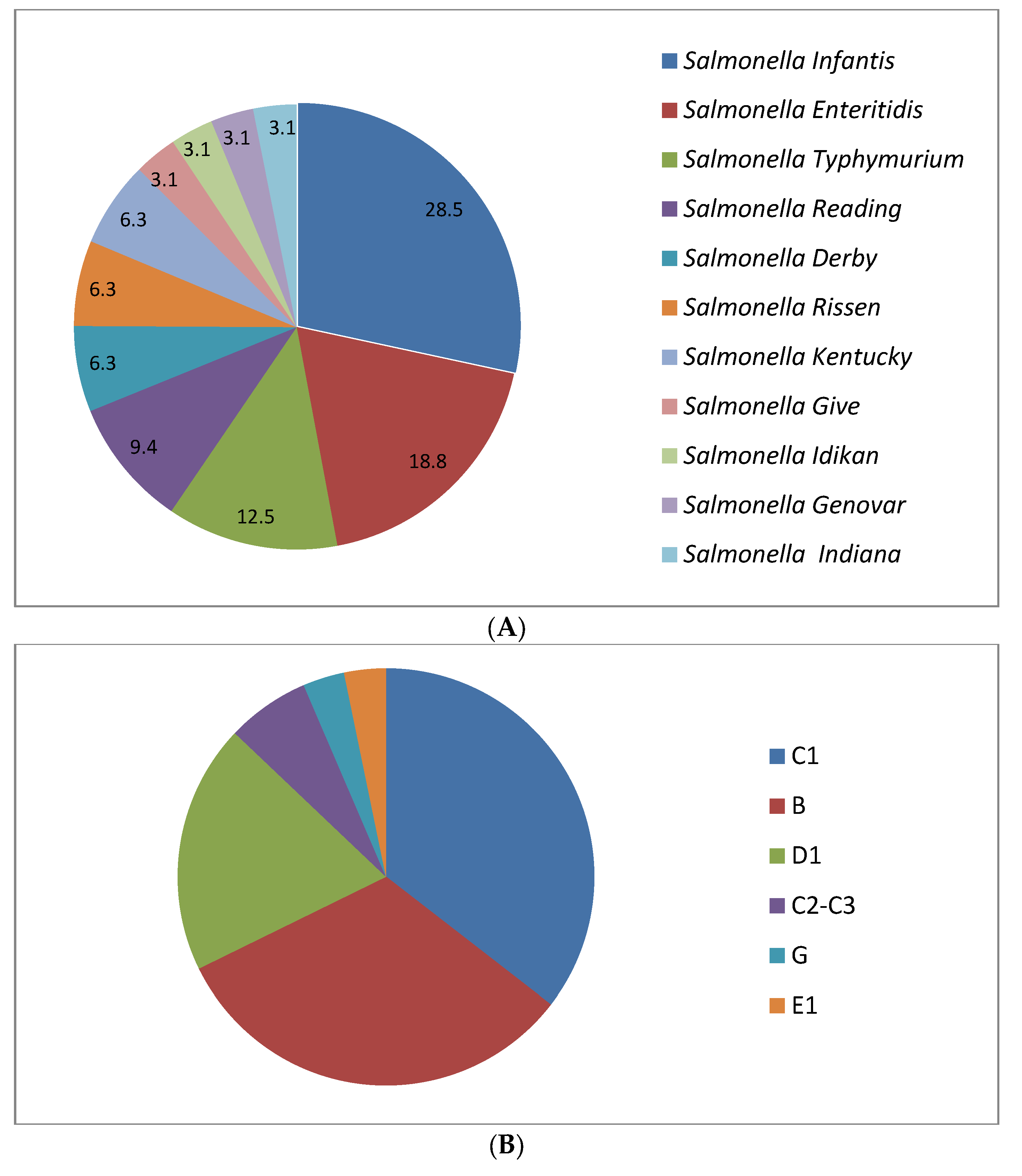
2.2. Serological Identification of Salmonella
2.3. Prevalence of Antimicrobial Resistance
2.4. Sequencing
2.5. General Characterization of the Genomes
2.6. Multilocus Sequence Typing of Strains
2.7. Antibiotic Resistance Genes
2.8. Plasmids
2.9. Pathogenicity Islands
2.10. Prophages
3. Discussion
4. Materials and Methods
4.1. Strains Used in the Work
4.2. Isolation and Confirmation of Salmonella Isolates
4.3. Serotyping of Salmonella Isolates
4.4. Screening of the Isolates for Resistance to Antimicrobial Preparations
4.5. DNA Isolation and Sequencing
4.6. Bioinformatic Techniques
4.7. Nucleotide Sequence Accession Number
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. WHO Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden Epidemiology Reference Group 2007–2015; WHO: Geneva, Switzerland, 2015.
- Lee, H.; Yoon, Y. Etiological Agents Implicated in Foodborne Illness World Wide. Food Sci. Anim. Resour. 2021, 41, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Li, C.; Hsu, C.-H.; Tyson, G.H.; Strain, E.; Tate, H.; Tran, T.-T.; Abbott, J.; McDermott, P.F. Comparative Genomic Analysis of 450 Strains of Salmonella enterica Isolated from Diseased Animals. Genes 2020, 11, 1025. [Google Scholar] [CrossRef]
- State Report “About the State Sanitary and Epidemiological Welfare of the Population in Russian Federation in 2020”. Available online: https://www.rospotrebnadzor.ru/upload/iblock/5fa/gd-seb_02.06-_s-podpisyu_.pdf (accessed on 28 October 2021).
- Nesbitt, A.; Ravel, A.; Murray, R.; McCORMICK, R.; Savelli, C.; Finley, R.; Parmley, J.; Agunos, A.; Majowicz, S.E.; Gilmour, M.; et al. Integrated surveillance and potential sources of Salmonella enteritidis in human cases in Canada from 2003 to 2009. Epidemiol. Infect. 2012, 140, 1757–1772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Layton, A.N.; Galyov, E.E. Salmonella-induced enteritis: Molecular pathogenesis and therapeutic implications. Expert Rev. Mol. Med. 2007, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Angulo, F.J.; Johnson, K.R.; Tauxe, R.V.; Cohen, M.L. Origins and Consequences of Antimicrobial-Resistant Nontyphoidal Salmonella: Implications for the Use of Fluoroquinolones in Food Animals. Microb. Drug Resist. 2000, 1, 77. [Google Scholar] [CrossRef] [PubMed]
- Parry, C.M.; Threlfall, E.J. Antimicrobial resistance in typhoidal and nontyphoidal salmonellae. Curr. Opin. Infect. Dis. 2008, 5, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Patchanee, P.; Tansiricharoenkul, K.; Buawiratlert, T.; Wiratsudakul, A.; Angchokchatchawal, K.; Yamsakul, P.; Yano, T.; Boonkhot, P.; Rojanasatien, S.; Tadee, P. Salmonella in pork retail outlets and dissemination of its pulsotypes through pig production chain in Chiang Mai and surrounding areas, Thailand. Prev. Veter. Med. 2016, 130, 99–105. [Google Scholar] [CrossRef]
- Possebon, F.S.; Casas, R.; Nero, A.; Yamatogi, S.; Araújo, P.; Pinto, P. Prevalence, antibiotic resistance, PFGE and MLST characterization of Salmonella in swine mesenteric lymph nodes. Prev. Veter. Med. 2020, 179, 105024. [Google Scholar] [CrossRef]
- Thai, T.H.; Hirai, T.; Lan, N.T.; Yamaguchi, R. Antibiotic resistance profiles of Salmonella serovars isolated from retail pork and chicken meat in North Vietnam. Int. J. Food Microbiol. 2012, 2, 147–151. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.; Sotomayor, C.; Wang, Q.; Holmes, N.; Furlong, C.; Ward, K.; Howard, P.; Octavia, S.; Lan, R.; Sintchenko, V. Whole genome sequencing of Salmonella typhimurium illuminates distinct outbreaks caused by an endemic multi-locus variable number tandem repeat analysis type in Australia, 2014. BMC Microbiol. 2016, 16, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Cummins, M.L.; Sanderson-Smith, M.; Newton, P.; Carlile, N.; Phalen, D.N.; Maute, K.; Monahan, L.G.; Hoye, B.J.; Djordjevic, S.P. Whole-Genome Sequence Analysis of an Extensively Drug-Resistant Salmonella enterica Serovar Agona Isolate from an Australian Silver Gull (Chroicocephalus novaehollandiae) Reveals the Acquisition of Multidrug Resistance Plasmids. mSphere 2020, 5, 5. [Google Scholar] [CrossRef]
- Cohen, E.; Rahav, G.; Gal-Mor, O. Genome Sequence of an Emerging Salmonella enterica Serovar Infantis and Genomic Comparison with Other S. Infantis Strains. Genome Biol. Evol. 2020, 3, 151–159. [Google Scholar] [CrossRef] [Green Version]
- Mashe, T.; Leekitcharoenphon, P.; Mtapuri-Zinyowera, S.; A Kingsley, R.; Robertson, V.; Tarupiwa, A.; Kock, M.M.; Makombe, E.P.; Chaibva, B.V.; Manangazira, P.; et al. Salmonella enterica serovar Typhi H58 clone has been endemic in Zimbabwe from 2012 to 2019. J. Antimicrob. Chemother. 2021, 5, 1160–1167. [Google Scholar] [CrossRef]
- Wahl, A.; Battesti, A.; Ansaldi, M. Prophages in Salmonella enterica: A driving force in reshaping the genome and physiology of their bacterial host? Mol. Microbiol. 2019, 2, 303–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 2, 1–37. [Google Scholar]
- Wencewicz, T.A. Crossroads of Antibiotic Resistance and Biosynthesis. J. Mol. Biol. 2019, 431, 3370–3399. [Google Scholar] [CrossRef] [PubMed]
- Castro-Vargas, R.E.; Herrera-Sánchez, M.P.; Rodríguez-Hernández, R.; Rondón-Barragán, I.S. Antibiotic resistance in Salmonella spp. isolated from poultry: A global overview. Vet. World 2020, 10, 2070–2084. [Google Scholar] [CrossRef]
- He, D.; Zhu, Y.; Li, R.; Pan, Y.; Liu, J.; Yuan, L.; Hu, G. Emergence of a hybrid plasmid derived from IncN1-F33:A−:B− and mcr-1-bearing plasmids mediated by IS26. J. Antimicrob. Chemother. 2019, 11, 3184–3189. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.-H.; Li, C.; Hoffmann, M.; McDermott, P.; Abbott, J.; Ayers, S.; Tyson, G.H.; Tate, H.; Yao, K.; Allard, M.; et al. Comparative Genomic Analysis of Virulence, Antimicrobial Resistance, and Plasmid Profiles of Salmonella Dublin Isolated from Sick Cattle, Retail Beef, and Humans in the United States. Microb. Drug Resist. 2019, 8, 1238–1249. [Google Scholar] [CrossRef] [PubMed]
- Mansour, M.N.; Yaghi, J.; El Khoury, A.; Felten, A.; Mistou, M.-Y.; Atoui, A.; Radomski, N. Prediction of Salmonella serovars isolated from clinical and food matrices in Lebanon and genomic-based investigation focusing on Enteritidis serovar. Int. J. Food Microbiol. 2020, 333, 108831. [Google Scholar] [CrossRef] [PubMed]
- Hayward, M.R.; Jansen, V.A.; Woodward, M.J. Comparative genomics of Salmonella enterica serovars Derby and Mbandaka, two prevalent serovars associated with different livestock species in the UK. BMC Genom. 2013, 14, 365. [Google Scholar] [CrossRef] [Green Version]
- Antunes, P.; Mourão, J.; Campos, J.; Peixe, L. Salmonellosis: The role of poultry meat. Clin. Microbiol. Infect. 2016, 22, 110–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mechesso, A.F.; Moon, D.C.; Kim, S.-J.; Song, H.-J.; Kang, H.Y.; Na, S.H.; Choi, J.-H.; Kim, H.-Y.; Yoon, S.-S.; Lim, S.-K. Nationwide surveillance on serotype distribution and antimicrobial resistance profiles of non-typhoidal Salmonella serovars isolated from food-producing animals in South Korea. Int. J. Food Microbiol. 2020, 335, 108893. [Google Scholar] [CrossRef] [PubMed]
- Hendriksen, R.S.; Vieira, A.R.; Karlsmose, S.; Lo Fo Wong, D.M.; Jensen, A.B.; Wegener, H.C.; Aarestrup, F.M. Global Monitoring of Salmonella Serovar Distribution from the World Health Organization Global Foodborne Infections Network Country Data Bank: Results of Quality Assured Laboratories from 2001 to 2007. Foodborne Pathog. Dis. 2011, 8, 887–900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Issenhuth-Jeanjean, S.; Roggentin, P.; Mikoleit, M.; Guibourdenche, M.; de Pinna, E.; Nair, S.; Fields, P.I.; Weill, F.-X. Supplement 2008–2010 (no. 48) to the White–Kauffmann–Le Minor scheme. Res. Microbiol. 2014, 165, 526–530. [Google Scholar] [CrossRef] [Green Version]
- Kürekci, C.; Sahin, S.; Iwan, E.; Kwit, R.; Bomba, A.; Wasyl, D. Whole-genome sequence analysis of Salmonella infantis isolated from raw chicken meat samples and insights into pESI-like megaplasmid. Int. J. Food Microbiol. 2021, 337, 108956. [Google Scholar] [CrossRef]
- Yang, X.; Wu, Q.; Zhang, J.; Huang, J.; Chen, L.; Wu, S.; Zeng, H.; Wang, J.; Chen, M.; Wu, H.; et al. Prevalence, Bacterial Load, and Antimicrobial Resistance of Salmonella Serovars Isolated From Retail Meat and Meat Products in China. Front. Microbiol. 2019, 10, 2121. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wu, Q.; Zhang, J.; Huang, J.; Chen, L.; Liu, S.; Yu, S.; Cai, S. Prevalence, enumeration, and characterization of Salmonella isolated from aquatic food products from retail markets in China. Food Control 2015, 57, 308–313. [Google Scholar] [CrossRef]
- Sadkowska-Todys, M.; Czarkowski, M.P. Salmonellosis in Poland in 2012. Przeglad Epidemiol. 2014, 68, 243–248. [Google Scholar]
- Fernandes, S.A.; Tavechio, A.T.; Ghilardi, Â.C.; Dias, Â.M.; De Almeida, I.A.; De Melo, L.C. Salmonella serovars isolated from humans in São Paulo State, Brazil, 1996–2003. Rev. Do Inst. De Med. Trop. De São Paulo 2006, 48, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Puthucheary, S.; Boey, C. Non-typhoid Salmonella gastroenteritis. J. Paediatr. Child Health 1998, 34, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Spiliopoulou, I.; Zografou, S.; Goula, A.; Dimitracopoulos, G.; Christofidou, M. Molecular Epidemiology and Antibiotic Resistance Patterns of Salmonella enterica from Southwestern Greece. Chemotherapy 2007, 53, 392–396. [Google Scholar] [CrossRef]
- Li, Y.; Xie, X.; Xu, X.; Wang, X.; Chang, H.; Wang, C.; Wang, A.; He, Y.; Yu, H.; Wang, X.; et al. Nontyphoidal Salmonella Infection in Children with Acute Gastroenteritis: Prevalence, Serotypes, and Antimicrobial Resistance in Shanghai, China. Foodborne Pathog. Dis. 2014, 11, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Wotzka, S.Y.; Nguyen, B.D.; Hardt, W.-D. Salmonella Typhimurium Diarrhea Reveals Basic Principles of Enteropathogen Infection and Disease-Promoted DNA Exchange. Cell Host Microbe 2017, 21, 443–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Chávez, L.; Cabrera-Diaz, E.; Pérez-Montaño, J.; Garay-Martínez, L.; Varela-Hernández, J.; Castillo, A.; Lucia, L.; Ávila-Novoa, M.; Cardona-Lopez, M.A.; Gutiérrez-González, P.; et al. Quantitative distribution of Salmonella spp. and Escherichia coli on beef carcasses and raw beef at retail establishments. Int. J. Food Microbiol. 2015, 210, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Molina-López, R.A.; Vidal, A.; Obón, E.; Martín, M.; Darwich, L. Multidrug-resistant Salmonella enterica Serovar Typhimurium Monophasic Variant 4,12:i:- Isolated from Asymptomatic Wildlife in a Catalonian Wildlife Rehabilitation Center, Spain. J. Wildl. Dis. 2015, 3, 759–763. [Google Scholar] [CrossRef] [PubMed]
- Arnedo-Pena, A.; Sabater-Vidal, S.; Herrera-León, S.; Bellido-Blasco, J.B.; Silvestre-Silvestre, E.; Meseguer-Ferrer, N.; Yague-Muñoz, A.; Gil-Fortuño, M.; Romeu-García, A.; Moreno-Muñoz, R. An outbreak of monophasic and biphasic Salmonella typhimurium, and Salmonella derby associated with the consumption of dried pork sausage in Castellon (Spain). Enferm. Infecc. Y Microbiol. Clínica 2016, 9, 544–550. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wang, M.; Zhou, C.; Gu, G.; Liang, J.; Hou, X.; Wang, M.; Wei, P. Prevalence and antimicrobial resistance of retail-meat-borne Salmonella in southern China during the years 2009–2016: The diversity of contamination and the resistance evolution of multidrug-resistant isolates. Int. J. Food Microbiol. 2020, 333, 108790. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; M’Ikanatha, N.M.; Nyirabahizi, E.; McDermott, P.F.; Tate, H. Antimicrobial resistance in non-Typhoidal Salmonella from retail poultry meat by antibiotic usage-related production claims–United States, 2008–2017. Int. J. Food Microbiol. 2021, 342, 10904. [Google Scholar] [CrossRef]
- Hu, L.; Cao, G.; Brown, E.W.; Allard, M.W.; Ma, L.M.; Khan, A.A.; Zhang, G. Antimicrobial resistance and related gene analysis of Salmonella from egg and chicken sources by whole-genome sequencing. Poult. Sci. 2020, 99, 7076–7083. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Tao, J.; Jiao, Y.; Fei, X.; Zhou, L.; Wang, Y.; Zheng, H.; Pan, Z.; Jiao, X. Phenotypic characteristics and genotypic correlation between Salmonella isolates from a slaughterhouse and retail markets in Yangzhou, China. Int. J. Food Microbiol. 2016, 222, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhao, S.; White, D.G.; Schroeder, C.M.; Lu, R.; Yang, H.; McDermott, P.F.; Ayers, S.; Meng, J. Characterization of Multiple-Antimicrobial-Resistant Salmonella Serovars Isolated from Retail Meats. Appl. Environ. Microbiol. 2004, 70, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, B.; Qu, D.; Zhang, X.; Shen, J.; Cui, S.; Shi, Y.; Xi, M.; Sheng, M.; Zhi, S.; Meng, J. Prevalence and characterization of Salmonella serovars in retail meats of marketplace in Shaanxi, China. Int. J. Food Microbiol. 2010, 141, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, T.M.; Levy, S.B. The impact of antibiotic use on resistance development and persistence. Drug Resist. Updat. 2000, 3, 303–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, M.; Xie, M.; Qu, Z.; Zhao, S.; Wang, J.; Wang, Y.; He, T.; Wang, H.; Zuo, Z.; Wu, C. Prevalence and antimicrobial resistance of Salmonella isolated from an integrated broiler chicken supply chain in Qingdao, China. Food Control 2016, 62, 270–276. [Google Scholar] [CrossRef]
- Bada-Alambedji, R.; Fofana, A.; Seydi, M.; Akakpo, A.J. Antimicrobial resistance of Salmonella isolated from poultry carcasses in Dakar (Senegal). Braz. J. Microbiol. 2006, 4, 510–515. [Google Scholar] [CrossRef] [Green Version]
- Tibaijuka, B.; Molla, B.; Hildebrandt, G.; Kleer, J.; Salah, W. Antimicrobila resistance to Salmonellae isolated from retail raw chicken meat and giblets in Ethiopia. Bull. Anim. Health Prod. Afr. 2002, 2, 86–95. [Google Scholar] [CrossRef]
- Elkenany, R.; Elsayed, M.M.; Zakaria, A.I.; El-Sayed, S.A.-E.-S.; Rizk, M.A. Antimicrobial resistance profiles and virulence genotyping of Salmonella enterica serovars recovered from broiler chickens and chicken carcasses in Egypt. BMC Veter. Res. 2019, 1, 124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vélez, D.C.; Rodríguez, V.; García, N.V. Phenotypic and Genotypic Antibiotic Resistance of Salmonella from Chicken Carcasses Marketed at Ibague, Colombia. Rev. Bras. De Ciência Avícola 2017, 2, 347–354. [Google Scholar] [CrossRef] [Green Version]
- Shrestha, P.; Cooper, B.S.; Coast, J.; Oppong, R.; Thuy, N.D.T.; Phodha, T.; Celhay, O.; Guerin, P.J.; Wertheim, H.; Lubell, Y. Enumerating the economic cost of antimicrobial resistance per antibiotic consumed to inform the evaluation of interventions affecting their use. Antimicrob. Resist. Infect. Control. 2018, 7, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaw, K.J.; Rather, P.N.; Hare, R.S.; Miller, G.H. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 1993, 1, 98. [Google Scholar] [CrossRef] [PubMed]
- Kor, S.-B.; Choo, Q.-C.; Chew, C.-H. New integron gene arrays from multiresistant clinical isolates of members of the Enterobacteriaceae and Pseudomonas aeruginosa from hospitals in Malaysia. J. Med. Microbiol. 2013, 3, 412–420. [Google Scholar] [CrossRef] [PubMed]
- Kazama, H.; Kizu, K.; Iwasaki, M.; Hamashima, H.; Sasatsu, M.; Arai, T. Isolation and structure of a new integron that includes a streptomycin resistance gene from the R plasmid of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 1995, 134, 137–141. [Google Scholar] [CrossRef]
- Sundin, G.W. Distinct Recent Lineages of the strA-strB Streptomycin-Resistance Genes in Clinical and Environmental Bacteria. Curr. Microbiol. 2002, 1, 63–69. [Google Scholar] [CrossRef]
- Scholz, P.; Haring, V.; Wittmann-Liebold, B.; Ashman, K.; Bagdasarian, M.; Scherzinger, E. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene 1989, 2, 271–288. [Google Scholar] [CrossRef]
- Villa, L.; García-Fernández, A.; Fortini, D.; Carattoli, A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 2010, 65, 2518–2529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alcaine, S.; Warnick, L.D.; Wiedmann, M. Antimicrobial Resistance in Nontyphoidal Salmonella. J. Food Prot. 2007, 70, 3. [Google Scholar] [CrossRef]
- Greenwood, D.; Finch, R.; Davey, P.; Wilcox, M. Antimicrobial Chemotherapy; Oxford University Press: Oxford, UK, 2007. [Google Scholar]
- Stürenburg, E.; Mack, D. Extended-spectrum β-lactamases: Implications for the clinical microbiology laboratory, therapy, and infection control. J. Infect. 2003, 47, 273–295. [Google Scholar] [CrossRef]
- McClelland, M.; Sanderson, K.E.; Spieth, J.; Clifton, S.W.; Latreille, P.; Courtney, L.; Porwollik, S.; Ali, J.; Dante, M.; Du, F.; et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 2001, 413, 852–856. [Google Scholar] [CrossRef] [Green Version]
- Salipante, S.J. Determining the Limits of the Evolutionary Potential of an Antibiotic Resistance Gene. Mol. Biol. Evol. 2003, 4, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Perreten, V.; Boerlin, P. A New Sulfonamide Resistance Gene (sul3) in Escherichia coli Is Widespread in the Pig Population of Switzerland. Antimicrob. Agents Chemother. 2003, 47, 1169–1172. [Google Scholar] [CrossRef] [Green Version]
- Cain, A.; Hall, R.M. Evolution of a multiple antibiotic resistance region in IncHI1 plasmids: Reshaping resistance regions in situ. J. Antimicrob. Chemother. 2012, 12, 2848–2853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ojo, K.K.; Kehrenberg, C.; Schwarz, S.; Odelola, H.A. Identification of a Complete dfrA14 Gene Cassette Integrated at a Secondary Site in a Resistance Plasmid of Uropathogenic Escherichia coli from Nigeria. Antimicrob. Agents Chemother. 2002, 6, 2054–2055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sørum, H.; L’Abée-Lund, T.M.; Solberg, A.; Wold, A. Integron-Containing IncU R Plasmids pRAS1 and pAr-32 from the Fish Pathogen Aeromonas salmonicida. Antimicrob. Agents Chemother. 2003, 4, 1285–1290. [Google Scholar] [CrossRef] [Green Version]
- Lau, S.K.P.; Wong, G.K.M.; Li, M.W.S.; Woo, P.C.Y.; Yuen, K.-Y. Distribution and molecular characterization of tetracycline resistance in Laribacter hongkongensis. J. Antimicrob. Chemother. 2008, 61, 488–497. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Adachi, K.; Akasaka, T.; Ono, N.; Sawai, T. Metal-tetracycline/H+ antiporter of Escherichia coli encoded by a transposon, Tn10. The role of the conserved dipeptide, Ser65-Asp66, in tetracycline transport. J. Biol. Chem. 1990, 265, 15525–15530. [Google Scholar] [CrossRef]
- Bissonnette, L.; Champetier, S.; Buisson, J.P.; Roy, P.H. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: Similarity of the product to transmembrane transport proteins. J. Bacteriol. 1991, 14, 4493–4502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zayko, E.V.; Kuznetsova, O.A.; Bataeva, D.S.; Grudistova, M.A. Identification of antibiotics in meat. Flow cytometry capabilities. All Meat 2020, 5, 56–60. [Google Scholar] [CrossRef]
- Dyall-Smith, M.L.; Liu, Y.; Billman-Jacobe, H. Genome Sequence of an Australian Monophasic Salmonella enterica subsp. enterica Typhimurium Isolate (TW-Stm6) Carrying a Large Plasmid with Multiple Antimicrobial Resistance Genes. Genome Announc. 2017, 5, e00793-17. [Google Scholar] [CrossRef] [Green Version]
- O’Mahony, R.; Quinn, T.; Drudy, D.; Walsh, C.; Whyte, P.; Mattar, S.; Fanning, S. Antimicrobial Resistance in Nontyphoidal Salmonella from Food Sources in Colombia: Evidence for an Unusual Plasmid-Localized Class 1 Integron in Serotypes Typhimurium and Anatum. Microb. Drug Resist. 2006, 12, 269–277. [Google Scholar] [CrossRef] [PubMed]
- NARMS Integrated Report: 2014. Available online: https://www.fda.gov/media/101511/download (accessed on 2 December 2021).
- Li, B.; Yang, X.; Tan, H.; Ke, B.; He, D.; Wang, H.; Chen, Q.; Ke, C.; Zhang, Y. Whole genome sequencing analysis of Salmonella enterica serovar Weltevreden isolated from human stool and contaminated food samples collected from the Southern coastal area of China. Int. J. Food Microbiol. 2018, 266, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Wray, C.; Davies, R.H. Salmonella in Domestic Animals, 2nd ed.; CABI Publishing: Wallingford, UK, 2000; pp. 169–190. [Google Scholar]
- Barilli, E.; Bacci, C.; Villa, Z.S.; Merialdi, G.; D’Incau, M.; Brindani, F.; Vismarra, A. Antimicrobial resistance, biofilm synthesis and virulence genes in Salmonella isolated from pigs bred on intensive farms. Ital. J. Food Saf. 2018, 7, 7223. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A. Importance of integrons in the diffusion of resistance. Veter. Res. 2001, 32, 243–259. [Google Scholar] [CrossRef] [Green Version]
- Carattoli, A.; Villa, L.; Pezzella, C.; Bordi, E.; Visca, P. Expanding Drug Resistance through Integron Acquisition by IncFI Plasmids of Salmonella enterica Typhimurium. Emerg. Infect. Dis. 2001, 7, 444. [Google Scholar] [CrossRef] [PubMed]
- Anonymous. Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella–Part 1: Detection of Salmonella spp; ISO 6579-1; International Organization for Standardization: Geneva, Switzerland, 2017. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Disk Susceptibility Tests, M100S, 29th ed.; CLSI: Wayne, PA, USA, 2019; Volume 39. [Google Scholar]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 6, e1005595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 11, e112963. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The Comprehensive Antibiotic Resistance Database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 12, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In Silico Detection and Typing of Plasmids using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 7, 3895–3903. [Google Scholar] [CrossRef] [Green Version]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, 16–21. [Google Scholar] [CrossRef] [Green Version]

| Sample Type | Number of Samples Analysed | Number of Positive Samples (%) |
|---|---|---|
| Poultry | 112 | 12 (10.7) |
| Pork | 91 | 3 (3.3) |
| Beef | 103 | 7 (6.8) |
| Minced meat | 168 | 10 (6) |
| Total | 474 | 32 (6.8) |
| Antimicrobial Class | Antimicrobial Agent | No. of Strains (%) | ||
|---|---|---|---|---|
| Resistant (R) | Intermediate (I) | Susceptible (S) | ||
| Penicillins | Ampicillin (AMP) | 8 (25.00) | 8 (3.13) | 23 (71.88) |
| Monobactams/carbapenems | Imipenem (IPM) | 6 (18.75) | 2 (6.25) | 24 (75.00) |
| Aminoglycosides | Amikacin (AMK) | 9 (28.13) | 14 (43.75) | 9 (28.13) |
| Streptomycin (STR) | 8 (25.00) | 4 (12.50) | 20 (62.50) | |
| Tobramycin | 5 (15.63) | 1 (3.13) | 26 (81.25) | |
| Cephems | Cefotaxime (CTX) | 6 (18.75) | 6 (18.75) | 20 (62.50) |
| Cefazolin (CFZ) | 15 (46.86) | 8 (25.00) | 9 (28.13) | |
| Folate pathway antagonists | Trimethoprim/ sulfamethoxazole (SXT) | 19 (59.38) | 0 (0) | 13 (40.63) |
| Chloramphenicol (CHL) | 7 (21.88) | 3 (9.38) | 22 (68.75) | |
| Macrolides and azalides | Azithromycin | 12 (37.50) | - | 20 (62.50) |
| β-Lactam/β-lactamase inhibitor combinations | Amoxicillin–clavulanic acid (AMC) | 2 (6.25) | 0 (0) | 30 (93.75) |
| Nitrofuran | Furadonin | 5 (18.75) | 3 (9.38) | 24 (75.00) |
| Tetracyclines | Tetracycline (TET) | 13 (40.63) | 9 (28.13) | 10 (31.25) |
| Antimicrobial Agent | No. of Resistance Isolates by Serotype (Resistance Rate, %) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Salmonella Typhimurium | Salmonella Enteritidis | Salmonella Infantis | Salmonella Kentucky | Salmonella Rissen | Salmonella Reading | Salmonella Give | Salmonella Derby | Salmonella Idikan | Salmonella Indiana | Salmonella Genovar | Total | |
| Ampicillin (AMP) | 25.0 | 16.7 | 33.3 | 0.0 | 50.0 | 0.0 | 100.0 | 0.0 | 0.0 | 100.0 | 0.0 | 25.0 |
| Imipenem (IPM) | 25.0 | 16.7 | 33.3 | 50.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 18.8 |
| Amikacin (AMK) | 0.0 | 16.7 | 33.3 | 0.0 | 50.0 | 66.7 | 0.0 | 50.0 | 0.0 | 100.0 | 0.0 | 28.1 |
| Streptomycin (STR) | 75.0 | 0.0 | 33.3 | 0.0 | 0.0 | 0.0 | 0.0 | 50.0 | 0.0 | 100.0 | 0.0 | 25 |
| Tobramycin (TM) | 25.0 | 0.0 | 22.2 | 0.0 | 0.0 | 0.0 | 0.0 | 50.0 | 0.0 | 100.0 | 0.0 | 15.6 |
| Cefotaxime (CFX) | 25.0 | 33.3 | 11.1 | 0.0 | 0.0 | 33.3 | 0.0 | 50.0 | 0.0 | 0.0 | 0.0 | 18.8 |
| Cefazolin (CFZ) | 50.0 | 50.0 | 44.4 | 50.0 | 50.0 | 66.7 | 100.0 | 50.0 | 0.0 | 0.0 | 0.0 | 50 |
| Trimethoprim/sulfamethoxazole (STX) | 75.0 | 50.0 | 66.7 | 50.0 | 50.0 | 33.3 | 100.0 | 50.0 | 0.0 | 100.0 | 100.0 | 59.34 |
| Chloramphenicol (CHL) | 25.0 | 16.7 | 44.4 | 0.0 | 50.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 21.9 |
| Azithromycin (AZM) | 50.0 | 33.3 | 22.2 | 0.0 | 50.0 | 33.3 | 0.0 | 50.0 | 100.0 | 0.0 | 100.0 | 34.4 |
| Amoxicillin + clavulanic acid (AMK) | 0.0 | 0.0 | 22.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.3 |
| Nitrofurantoin (NIT) | 0.0 | 0.0 | 44.4 | 0.0 | 0.0 | 33.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 15.6 |
| Tetracycline (TET) | 75.0 | 33.3 | 55.6 | 0.0 | 0.0 | 33.3 | 0.0 | 50.0 | 0.0 | 100.0 | 0.0 | 40.6 |
| Serovar | Isolate | Pattern | No. of Antimicrobial Agents | No. of Classes |
|---|---|---|---|---|
| Salmonella Give | S1 | AMP-STX-CFZ | 3 | 3 |
| Salmonella Typhimurium | S3 | STR-AMP-TM-IPM-CHL | 5 | 4 |
| S5 | CTX-STX-AZM-TET-CFZ | 5 | 4 | |
| S14 | STR-STX-AZM-TET-CFZ | 5 | 4 | |
| S25 | STR-STX-TET | 3 | 3 | |
| Salmonella Reading | S23 | CTX-AMK-AZM-CFZ | 4 | 3 |
| S4 | AMK-STX-AZM-TET-NIT | 5 | 5 | |
| S33 | CFZ | 1 | 1 | |
| Salmonella Derby | S6 | STR-AMK-AZM-TET | 4 | 3 |
| S11 | CTX-TM-STX-CFZ | 4 | 3 | |
| Salmonella Idikan | S9 | AZM | 1 | 1 |
| Salmonella Rissen | S10 | CFZ-CHL | 1 | 1 |
| S18 | AMP-AMK-STX-AZM | 4 | 4 | |
| Salmonella Infantis | S26 | STR-IPM-CFZ-NIT | 4 | 4 |
| S15 | AMK-STX-AZM-TET-CFZ | 5 | 5 | |
| SZL 30 | STR-AMP-AMK-TM-STX-IPM-TET-CHL | 8 | 6 | |
| SZL 31 | STR-AMP-AMK-TM-STX-IPM-TET-CHL | 8 | 6 | |
| S34 | AMK-STX-TET-CFZ-NIT | 5 | 5 | |
| S35 | STX | 1 | 1 | |
| S36 | ND | 0 | 0 | |
| S37 | STX-TET-NIT | 3 | 3 | |
| S28 | CTX-AZM-CFZ-CHL-NIT | 5 | 4 | |
| Salmonella Genovar | S16 | STX-AZM | 2 | 2 |
| Salmonella Enteritidis | S7 | AMK-AZM-CFZ | 3 | 3 |
| S8 | CTX-AZM | 2 | 2 | |
| S17 | AMP-STX-TET-CFZ-CHL | 5 | 5 | |
| S20 | STX-IPM-CFZ | 3 | 3 | |
| S27 | CTX-TET | 2 | 2 | |
| S29 | STX | 1 | 1 | |
| Salmonella Kentucky | S32 | ND | 0 | 0 |
| S19 | STX-IMP-CFZ | 3 | 3 | |
| Salmonella Typhimurium | SZL 38 | STR-AMP-AMK-TM-STX-TET | 6 | 4 |
| Strain | Chromosome Size (bp) | Plasmids | |
|---|---|---|---|
| Name | Size (bp) | ||
| SZL 30 | 4,689,375 | pSZL30.1 pSZL30.2 | 276,251 53,986 |
| SZL 31 | 4,689,704 | pSZL31.1 pSZL31.2 | 280,239 53,147 |
| SZL 38 | 5,052,615 | pSZL38.1 | 79,333 |
| Parameter | Strain | ||
|---|---|---|---|
| 30 | 31 | 38 | |
| Predicted genes | 5184 | 5191 | 5320 |
| Protein-coding genes | 5078 | 5085 | 5209 |
| Protein-coding genes with predicted function | 3845 (75.7%) | 3785 (74.4%) | 3851 (73.9%) |
| tRNA genes | 84 | 84 | 89 |
| Resistance Gene | Protein | Antimicrobial Agent | Location | ||
|---|---|---|---|---|---|
| SZL 30 | SZL 31 | SZL 38 | |||
| aadA1 | Aminoglycoside (3″) (9)-adenylyltransferase | STR | pSZL30.2 | - | - |
| aadA2b | - | pSZL31.1 | - | ||
| aph(3″)-Ib | Aminoglycoside 3′-phosphotransferase | - | - | chromosome | |
| aph(6)-Id | Aminoglycoside O-phosphotransferase | - | - | chromosome | |
| blaTEM-1B | Class A β-lactamase | AMP | pSZL30.2 | pSZL31.2 | chromosome |
| aac(6′)-Iaa | Chromosomal encoded aminoglycoside N (6′)-acetyltransferase | AMK, TM | chromosome | chromosome | chromosome |
| sul3 | Dihydropteroate synthase | STX | pSZL30.2 | pSZL31.1 pSZL31.2 | - |
| sul2 | - | - | chromosome | ||
| tet(A) | Tetracycline efflux MSF transporter | TET | pSZL30.1 | pSZL31.1 | - |
| tet(B) | - | - | chromosome | ||
| cmlA1 | Drug efflux MSF transporter Bcr/CflA family | CHL | pSZL30.2 | pSZL31.1 pSZL31.2 | - |
| mefB | MSF efflux transporter | AZM, ERY | pSZL30.2 | chromosome pSZL31.2 | - |
| mdfA/cmr | MSF efflux transporter | RIF, PUR, ERY | chromosome | chromosome | chromosome |
| mdtABC-tolC | MSF efflux transporter | NB, NAL, NOR | chromosome | chromosome | chromosome |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rakitin, A.L.; Yushina, Y.K.; Zaiko, E.V.; Bataeva, D.S.; Kuznetsova, O.A.; Semenova, A.A.; Ermolaeva, S.A.; Beletskiy, A.V.; Kolganova, T.V.; Mardanov, A.V.; et al. Evaluation of Antibiotic Resistance of Salmonella Serotypes and Whole-Genome Sequencing of Multiresistant Strains Isolated from Food Products in Russia. Antibiotics 2022, 11, 1. https://doi.org/10.3390/antibiotics11010001
Rakitin AL, Yushina YK, Zaiko EV, Bataeva DS, Kuznetsova OA, Semenova AA, Ermolaeva SA, Beletskiy AV, Kolganova TV, Mardanov AV, et al. Evaluation of Antibiotic Resistance of Salmonella Serotypes and Whole-Genome Sequencing of Multiresistant Strains Isolated from Food Products in Russia. Antibiotics. 2022; 11(1):1. https://doi.org/10.3390/antibiotics11010001
Chicago/Turabian StyleRakitin, Andrey L., Yulia K. Yushina, Elena V. Zaiko, Dagmara S. Bataeva, Oksana A. Kuznetsova, Anastasia A. Semenova, Svetlana A. Ermolaeva, Aleksey V. Beletskiy, Tat’yana V. Kolganova, Andrey V. Mardanov, and et al. 2022. "Evaluation of Antibiotic Resistance of Salmonella Serotypes and Whole-Genome Sequencing of Multiresistant Strains Isolated from Food Products in Russia" Antibiotics 11, no. 1: 1. https://doi.org/10.3390/antibiotics11010001
APA StyleRakitin, A. L., Yushina, Y. K., Zaiko, E. V., Bataeva, D. S., Kuznetsova, O. A., Semenova, A. A., Ermolaeva, S. A., Beletskiy, A. V., Kolganova, T. V., Mardanov, A. V., Shapovalov, S. O., & Tkachik, T. E. (2022). Evaluation of Antibiotic Resistance of Salmonella Serotypes and Whole-Genome Sequencing of Multiresistant Strains Isolated from Food Products in Russia. Antibiotics, 11(1), 1. https://doi.org/10.3390/antibiotics11010001