DNA Methyltransferase HsdM Induce Drug Resistance on Mycobacterium tuberculosis via Multiple Effects
Abstract
:1. Introduction
2. Results
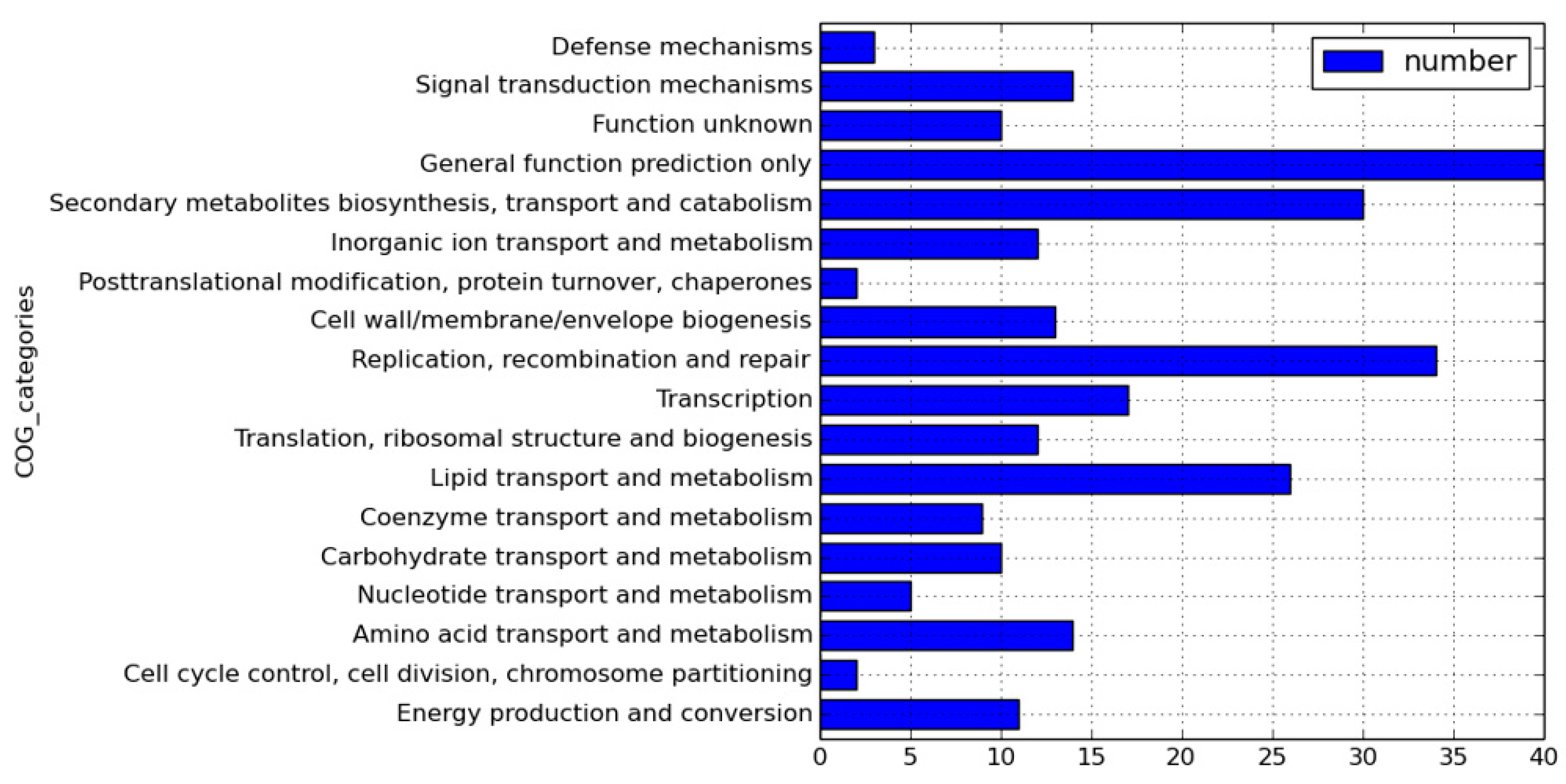
2.1. Genome Sequences of Clinical Strains of M. tuberculosis Using PacBio SMRT Technology
2.2. Genome-Wide Kinetics of Adenine Methylation Did Not Correlate with Antibiotic Treatment
2.3. Genome-Wide Identification of N6-Methyl-Adenine Base Modification by HsdM
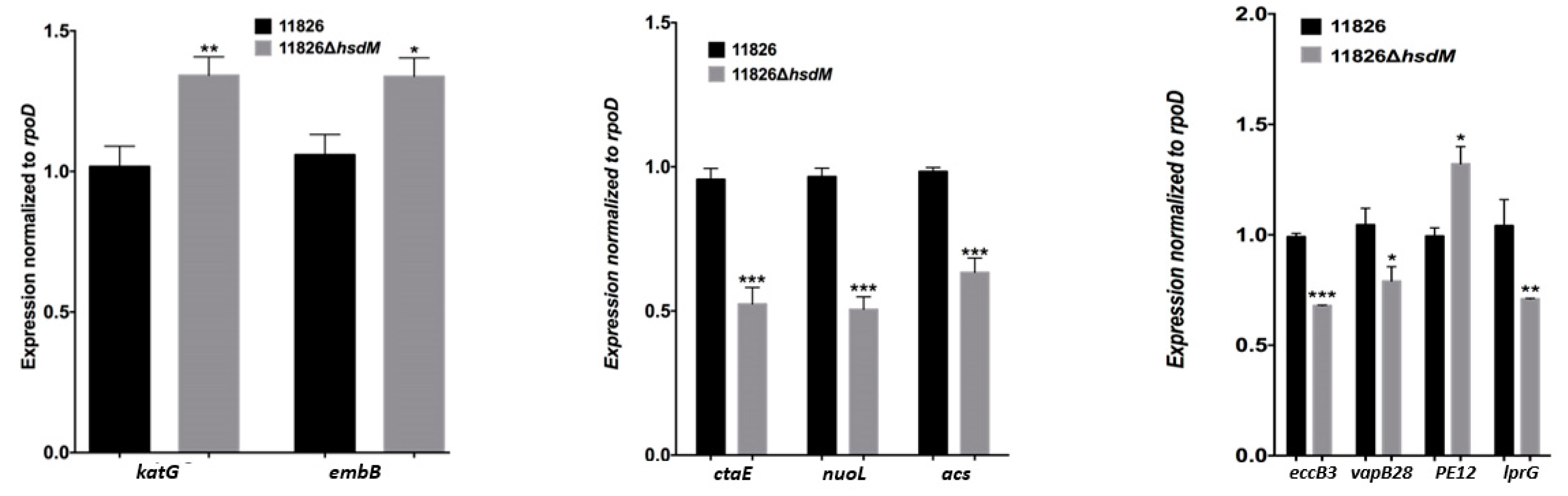
2.4. HsdM Affected M. tuberculosis Growth In Vitro
2.5. HsdM Affected Drug Susceptibility
2.6. Overexpression of HsdM Altered Mutation Rates in M. smegmatis
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Bacterial Strains and Culture Conditions
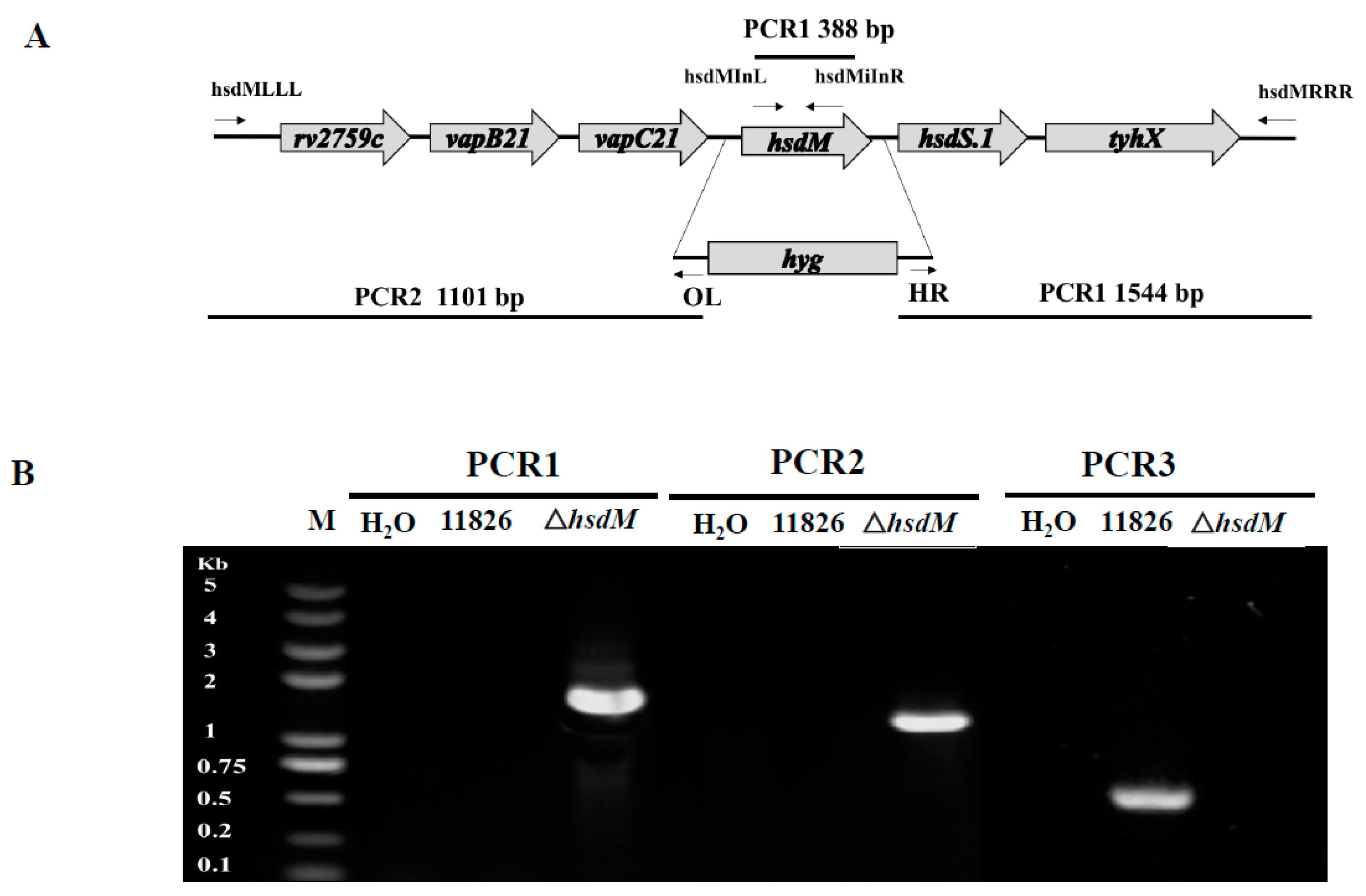
5.2. Generation of the hsdM-Knockout Mutant Strain
5.3. Antibiotic Susceptibility Testing
5.4. SMRT Sequencing
5.5. Bioinformatics Analyses
5.6. De Novo Assembly Details
5.7. RNA Isolation and Quantitative Real-Time PCR
5.8. Mutant Calculation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Global Tuberculosis Report 2020; World Health Organization: Geneva, Switzerland, 2020.
- Gomez-Gonzalez, P.J.; Andreu, N.; Phelan, J.E.; de Sessions, P.F.; Glynn, J.R.; Crampin, A.C.; Campino, S.; Butcher, P.D.; Hibberd, M.L.; Clark, T.G. An integrated whole genome analysis of Mycobacterium tuberculosis reveals insights into relationship between its genome, transcriptome and methylome. Sci. Rep. 2019, 9, 5204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schön, T.; Miotto, P.; Köser, C.U.; Viveiros, M.; Böttger, E.; Cambau, E. Mycobacterium tuberculosis drug-resistance testing: Challenges, recent developments and perspectives. Clin. Microbiol. Infect. 2017, 23, 154–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espinal, M.A.; Kim, S.J.; Suarez, P.G.; Kam, K.M.; Khomenko, A.G.; Migliori, G.B.; Baéz, J.; Kochi, A.; Dye, C.; Raviglione, M.C. Standard short-course chemotherapy for drug-resistant tuberculosis: Treatment outcomes in 6 countries. JAMA 2000, 283, 2537–2545. [Google Scholar] [CrossRef] [Green Version]
- Gunther, G. Multidrug-resistant and extensively drug-resistant tuberculosis: A review of current concepts and future challenges. Clin. Med. 2014, 14, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Casali, N.; Nikolayevskyy, V.; Balabanova, Y.; Harris, S.R.; Ignatyeva, O.; Kontsevaya, I.; Corander, J.; Bryant, J.; Parkhill, J.; Nejentsev, S.; et al. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat. Genet. 2014, 46, 279–286. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Wang, J.; Li, J.; Liu, Y.; Liu, X.; Li, Z.; Kurniyati, K.; Deng, Y.; Wang, G.; Ralph, J.D.; et al. Prevalence of phase variable epigenetic invertons among host-associated bacteria. Nucleic Acids Res. 2020, 48, 11468–11485. [Google Scholar] [CrossRef]
- Vasu, K.; Nagaraja, V. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol. Mol. Biol. Rev. 2013, 77, 53–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shell, S.S.; Prestwich, E.G.; Baek, S.H.; Shah, R.R.; Sassetti, C.M.; Dedon, P.C.; Fortune, S.M. DNA methylation impacts gene expression and ensures hypoxic survival of Mycobacterium tuberculosis. PLoS Pathog. 2013, 9, e1003419. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Zhong, J.; Jia, X.; Liu, G.; Kang, Y.; Dong, M.; Zhang, X.; Li, Q.; Yue, L.; Li, C.; et al. Precision methylome characterization of Mycobacterium tuberculosis complex (MTBC) using PacBio single-molecule real-time (SMRT) technology. Nucleic Acids Res. 2016, 44, 730–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phelan, J.; de Sessions, P.F.; Tientcheu, L.; Perdigao, J.; Machado, D.; Hasan, R.; Hasan, Z.; Bergval, I.L.; Anthony, R.; McNerney, R.; et al. Methylation in Mycobacterium tuberculosis is lineage specific with associated mutations present globally. Sci. Rep. 2018, 8, 160. [Google Scholar] [CrossRef]
- Flusberg, B.A.; Webster, D.R.; Lee, J.H.; Travers, K.J.; Olivares, E.C.; Clark, T.A.; Korlach, J.; Turner, S.W. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 2010, 7, 461–465. [Google Scholar] [CrossRef] [Green Version]
- Cole, S.T.; Brosch, R.; Parkhill, J.; Garnier, T.; Churcher, C.; Harris, D.; Gordon, S.V.; Eiglmeier, K.; Gas, S.; Barry, C.E., III; et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998, 393, 537–544. [Google Scholar] [CrossRef]
- Coscolla, M.; Gagneux, S. Consequences of genomic diversity in Mycobacterium tuberculosis. Semin. Immunol. 2012, 26, 431–444. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Luom, T.; Dong, X.; Sun, G.; Liu, Z.; Gan, M.; Wu, J.; Shen, X.; Gao, Q. Genetic features of Mycobacterium tuberculosis modern Beijing sublineage. Emerg. Microbes Infect. 2016, 5, e14. [Google Scholar] [CrossRef] [Green Version]
- Balbontín, R.; Rowley, G.; Pucciarelli, M.G.; López-Garrido, J.; Wormstone, Y.; Lucchini, S.; García-Del Portillo, F.; Hinton, J.C.; Casadesús, J. DNA adenine methylation regulates virulence gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 2006, 188, 8160–8168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- David, H.L. Probability distribution of drug-resistant mutants in unselected populations of Mycobacterium tuberculosis. Appl. Microbiol. 1970, 20, 810–814. [Google Scholar] [CrossRef] [PubMed]
- Cohen, N.R.; Ross, C.A.; Jain, S.; Shapiro, R.S.; Gutierrez, A.; Belenky, P.; Li, H.; Collins, J.J. A role for the bacterial GATC methylome in antibiotic stress survival. Nat. Genet. 2016, 48, 581–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiner-Oms, Á.; Berney, M.; Boinett, C.; González-Candelas, F.; Young, D.B.; Gagneux, S.; Jacobs, W.R., Jr.; Parkhill, J.; Cortes, T.; Comas, I. Genome-wide mutational biases fuel transcriptional diversity in the Mycobacterium tuberculosis complex. Nat. Commun. 2019, 10, 3994. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Li, H.; Chen, T.; Yu, L.; Guo, H.; Chen, Y.; Chen, M.; Li, Z.; Wu, Z.; Wang, X.; et al. Genome-wide DNA methylation and transcriptome changes in Mycobacterium tuberculosis with rifampicin and isoniazid resistance. Int. J. Clin. Exp. Pathol. 2018, 11, 3036–3045. [Google Scholar]
- Hu, X.; Zhou, X.; Yin, T.; Chen, K.; Hu, Y.; Zhu, B.; Mi, K. The Mycobacterial DNA methyltransferase HsdM decreases intrinsic isoniazid susceptibility. Antibiotics 2021, 10, 1323. [Google Scholar] [CrossRef]
- DebRoy, S.; Shropshire, W.C.; Tran, C.N.; Hao, H.; Gohel, M.; Galloway-Peña, J.; Hanson, B.; Flores, A.R.; Shelburne, S.A. Characterization of the type I restriction modification system broadly conserved among group A streptococci. mSphere 2021, 6, e0079921. [Google Scholar] [CrossRef]
- Finn, M.B.; Ramsey, K.M.; Tolliver, H.J.; Dove, S.L.; Wessels, M.R. Improved transformation efficiency of group A Streptococcus by inactivation of a type I restriction modification system. PLoS ONE 2021, 16, e0248201. [Google Scholar] [CrossRef]
- Bardou, F.; Raynaud, C.; Ramos, C.; Lanéelle, M.A.; Lanŕelle, G. Mechanism of isoniazid uptake in Mycobacterium tuberculosis. Microbiology 1998, 144, 2539–2544. [Google Scholar] [CrossRef] [Green Version]
- Modlin, S.J.; Conkle-Gutierrez, D.; Kim, C.; Mitchell, S.N.; Morrissey, C.; Weinrick, B.C.; Jacobs, W.R.; Ramirez-Busby, S.M.; Hoffner, S.E.; Valafar, F. Drivers and sites of diversity in the DNA adenine methylomes of 93 Mycobacterium tuberculosis complex clinical isolates. eLife 2020, 9, e58542. [Google Scholar] [CrossRef]
- Gu, L.; Chen, Y.; Wang, Q.; Li, X.; Mi, K.; Deng, H. Functional Characterization of Sirtuin-like Protein in Mycobacterium smegmatis. J. Proteome Res. 2015, 14, 4441–4449. [Google Scholar] [CrossRef] [PubMed]
- Vilchèze, C.; Weisbrod, T.R.; Chen, B.; Kremer, L.; Hazbón, M.H.; Wang, F.; Alland, D.; Sacchettini, J.C.; Jacobs, W.R., Jr. Altered NADH/NAD + ratio mediates coresistance to isoniazid and ethionamide in mycobacteria. Antimicrob. Agents Chemother. 2005, 49, 708–720. [Google Scholar] [CrossRef] [Green Version]
- Lobritz, M.A.; Belenky, P.; Porter, C.B.; Gutierrez, A.; Yang, J.H.; Schwarz, E.G.; Dwyer, D.J.; Khalil, A.S.; Collins, J.J. Antibiotic efficacy is linked to bacterial cellular respiration. Proc. Natl. Acad. Sci. USA 2015, 112, 8173–8180. [Google Scholar] [CrossRef] [Green Version]
- Belenky, P.; Ye, J.D.; Porter, C.B.; Cohen, N.R.; Lobritz, M.A.; Ferrante, T.; Jain, S.; Korry, B.J.; Schwarz, E.G.; Walker, G.C.; et al. Bactericidal antibiotics induce toxic metabolic perturbations that lead to cellular damage. Cell Rep. 2015, 13, 968–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bardarov, S.; Bardarov, S.; Pavelka, M.S.; Sambandamurthy, V.; Larsen, M.; Tufariello, J.; Chan, J.; Hatfull, G.; Jacobs, W.R. Specialized transduction: An efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 2002, 148, 3007–3017. [Google Scholar] [CrossRef] [Green Version]
- Franzblau, S.G.; Witzig, R.S.; McLaughlin, J.C.; Torres, P.; Madico, G.; Hernandez, A.; Degnan, M.T.; Cook, M.B.; Quenzer, V.K.; Ferguson, R.M.; et al. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J. Clin. Microbiol. 1998, 36, 362–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chin, C.S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E.; et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hershberg, R.; Lipatov, M.; Small, P.M.; Sheffer, H.; Niemann, S.; Homolka, S.; Roach, J.C.; Kremer, K.; Petrov, D.A.; Feldman, M.W.; et al. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 2008, 6, e311. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Strain Names | Average Read Size (kb) | Sequencing Depth (x) | Genome Size (bp) | Gene Number | SNP | Indel |
|---|---|---|---|---|---|---|
| M. tuberculosis 11495 | 3.9 | 180 | 4,428,395 | 4455 | 1687 | 392 |
| M. tuberculosis 10167 | 2.1 | 240 | 4,418,815 | 4481 | 1677 | 501 |
| M. tuberculosis 11776 | 3.5 | 120 | 4,433,260 | 4501 | 1685 | 488 |
| M. tuberculosis 11826 | 2.5 | 160 | 4,406,742 | 4527 | 1689 | 619 |
| M. tuberculosis 12052 | 2.4 | 150 | 4,421,905 | 4538 | 1655 | 522 |
| M. tuberculosis 12058 | 2.7 | 90 | 4,425,864 | 4490 | 1650 | 459 |
| Strain Name | Methylated Motif | |||||
|---|---|---|---|---|---|---|
| CTCCAG/CTGGAG | CACGCAG | GATN4TTAC | ||||
| No. of Motifs Detected | % Motifs Detected | No. of Motif Detected | % Motifs Detected | No. of Motifs Detected | % Motifs Detected | |
| H37Rv | 99.08 | 1930 | / | / | / | / |
| 11495 | / | / | 99.41 | 839 | 97.84 | 363 |
| 10167 | / | / | 100 | 828 | 77.45 | 285 |
| 11776 | / | / | 99.16 | 825 | 98.4 | 362 |
| 11826 | / | / | 99.27 | 826 | 96.45 | 355 |
| 12052 | / | / | 99.15 | 825 | 94.29 | 347 |
| 12058 | / | / | 95.91 | 798 | 85.59 | 315 |
| Gene Name | Function |
|---|---|
| Respiration-related genes | |
| ctaE | Involved in aerobic respiration, probable cytochrome c oxidase (subunit III) CtaE |
| qcrC | Probable ubiquinol-cytochrome c reductase QcrC (cytochrome c subunit) |
| NuoI | Involved in aerobic/anaerobic respiration |
| cyp126 | Cytochrome P450 126 Cyp126, involved in intermediary metabolism and respiration |
| cyp135B1 | Cytochrome P450 135B1 belongs to a group of heme-thiolate monooxygenases |
| fgd1 | Catalyzes the oxidation of glucose-6-phosphate to 6-phosphogluconolactone using coenzyme F420 (an *-hydroxy-5-deazaflavin derivative) as the electron acceptor |
| frdB | Involved in the interconversion of fumarate and succinate (anaerobic respiration) |
| fdhF | Decomposes formic acid to hydrogen and carbon dioxide under anaerobic conditions in the absence of exogenous electron acceptors |
| Rv1786 | Ferredoxin, an iron-sulfur protein that transfers electrons in a wide variety of metabolic reactions; involved in intermediary metabolism and respiration |
| qor | Rv1454c, a quinone reductase |
| Lipid metabolism-related genes | |
| fadD11 | Rv1550, fatty-acid-CoA ligase |
| fadD12 | Rv1427c, long-chain-fatty-acid—CoA ligase, function unknown, but supposed involvement in lipid degradation |
| fadD16 | Rv0852, possible fatty-acid-CoA ligase FadD16, function unknown, but involved in lipid degradation |
| fadD2 | Rv0270, probable fatty acid-CoA ligase, function unknown, but involved in lipid degradation |
| fadD23 | Rv3826, long-chain-fatty-acid—CoA ligase |
| fadD24 | Rv1529, long-chain-fatty-acid—CoA ligase |
| fadD29 | Rv2950c, long-chain-fatty-acid—CoA ligase |
| fadD35 | Rv2505c, long-chain-fatty-acid—CoA ligase |
| fadE10 | Rv0873, probable acyl-CoA dehydrogenase, function unknown, but involved in lipid degradation |
| lipL | Rv1497, probable esterase, function unknown, but supposed involvement in lipid metabolism |
| Drug resistance-related genes | |
| gyrA | Rv0006, DNA gyrase subunit A, related to fluoroquinolone resistance |
| eis | Rv2416c, enhanced intracellular survival protein, related to kanamycin resistance |
| embB | Rv3795, arabinosyltransferase B, related to ethambutol resistance |
| Rv0194 | Transmembrane multidrug efflux pump, related to multidrug resistance |
| Rv1410c | EmrB/QacA family drug resistance transporter, related to aminoglycosides/tetracycline resistance |
| Rv1877 | EmrB/QacA family drug resistance transporter |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chu, H.; Hu, Y.; Zhang, B.; Sun, Z.; Zhu, B. DNA Methyltransferase HsdM Induce Drug Resistance on Mycobacterium tuberculosis via Multiple Effects. Antibiotics 2021, 10, 1544. https://doi.org/10.3390/antibiotics10121544
Chu H, Hu Y, Zhang B, Sun Z, Zhu B. DNA Methyltransferase HsdM Induce Drug Resistance on Mycobacterium tuberculosis via Multiple Effects. Antibiotics. 2021; 10(12):1544. https://doi.org/10.3390/antibiotics10121544
Chicago/Turabian StyleChu, Hongqian, Yongfei Hu, Bing Zhang, Zhaogang Sun, and Baoli Zhu. 2021. "DNA Methyltransferase HsdM Induce Drug Resistance on Mycobacterium tuberculosis via Multiple Effects" Antibiotics 10, no. 12: 1544. https://doi.org/10.3390/antibiotics10121544
APA StyleChu, H., Hu, Y., Zhang, B., Sun, Z., & Zhu, B. (2021). DNA Methyltransferase HsdM Induce Drug Resistance on Mycobacterium tuberculosis via Multiple Effects. Antibiotics, 10(12), 1544. https://doi.org/10.3390/antibiotics10121544






