Abstract
Escherichia coli isolated from meat of different animal species may harbour antimicrobial resistance genes and may thus be a threat to human health. The objectives of this study were to define antimicrobial resistance genes in E. coli isolates from pork, beef, chicken- and turkey meat and analyse whether their resistance genotypes associated with phylogenetic groups or meat species. A total number of 313 E. coli samples were isolated using standard cultural techniques. In 98% of resistant isolates, a dedicated resistance gene could be identified by PCR. Resistance genes detected were tet(A) and tet(B) for tetracycline resistance, strA and aadA1 for streptomycin resistance, sulI and sulII for resistance against sulphonamides, dfr and aphA for kanamycin resistance and blaTEM for ampicillin resistance. One stx1 harbouring E. coli isolated from pork harboured the tet(A) gene and belonged to phylogenetic group B2, whilst another stx1 positive isolate from beef was multi-resistant and tested positive for blaTEM, aphA, strA–B, sulII, and tet(A) and belonged to phylogenetic group A. In conclusion, the distribution of resistance elements was almost identical and statistically indifferent in isolates of different meat species. Phylogenetic groups did not associate with the distribution of resistance genes and a rather low number of diverse resistance genes were detected. Most E. coli populations with different resistance genes against one drug often revealed statistically significant different MIC values.
Keywords:
antibiotic resistance; Escherichia coli; foodborne pathogen; phylogroup; poultry meat; beef; pork 1. Introduction
Antimicrobial resistance (AMR) and its spread is one of the most important health problem nowadays. Indicator microorganisms for AMR such as Escherichia coli (E. coli) are important indicator bacteria for faecal contamination and the occurrence of AMR of animal and human origin. As a commensal and pathogen in warm-blooded animals and humans, E. coli can be isolated from different sources such as faeces, manure, water and food of animal and plant origin.
Isolates from food-producing animals and their food pose a direct link from animals to humans and between locations where the primary antibiotic usage takes place (animal and human medicine). Hence, antibiotic-resistant (ABR) isolates are commonly found and may provide insights into upcoming problems but may also provide foreseeable solutions. The analysis of resistant bacteria by whole genome sequencing is becoming a standard procedure but discrimination is very high and one can often “miss the forest for the trees” while waiting for “cut-off values” for epidemiological applications [1,2].
In E. coli, which has a rather strong phylogenetic structure, the four major phylogroups (A, B1, B2, and D) can provide a framework. The analysis of these phylogenetic groups can be performed by a simple multiplex PCR [3,4,5] but can also be extracted from sequence data. Phylogroups A and B1 are said to be more generalist, which can associate with multidrug resistance and can be a potential transfer vehicle for AMR between animal species, the environment, and humans [6]. Other studies comparing resistant and non-resistant E. coli [7,8] were not able to link specific AMR patterns to certain E. coli phylogroups.
Isolates of groups B1 and A can be found in humans, poultry, ruminants, and pigs [9,10,11,12] whereas groups B2 and D are frequently isolated as extraintestinal pathogens from humans and birds [9,13]. The contamination of food of animal origin by E. coli may happen during slaughter processing steps for instance skinning, defeathering, or evisceration. E. coli isolated from meat may harbour antimicrobial resistance genes but may also be pathogenic to humans for instance STEC and thus be a threat to human health [14,15].
To detect whether the distribution of phylogenetic groups in resistant and non-resistant E. coli differs between most commonly used meat species, we analysed antimicrobial-resistant, -sensitive, and -intermediate E. coli isolated from chicken meat, pork, beef, and turkey meat for antimicrobial resistance genes and defined their phylogenetic group.
2. Results
In this study, we tested a total number of 313 E. coli isolates from different meat species for resistance to most important veterinary drugs in use and defined resistance genes and phylogenetic groups.
2.1. Phenotypical Resistance
Most of the resistant isolates were resistant to tetracycline (n = 230), followed by sulphonamides (n = 142), ampicillin (n = 89), trimethoprim (n = 74), streptomycin (n = 50), and kanamycin (n = 28) by using the disc diffusion assay and the microbroth dilution assay. Fifty-nine isolates exhibited an intermediate phenotype against streptomycin by using the disc diffusion test. Three isolates were intermediate resistant to ampicillin, and one isolate to sulphonamide by using the microbroth dilution assay.
2.2. Tetracycline Resistance Genes
We tested for several genes known to cause resistance against tetracycline in E. coli as are tet(A), tet(B), tet(C), tet(G). Only two of these known resistance genes were detected in our isolates, namely tet(A) and tet(B). All resistant isolates harboured at least one resistance determinant. The tet(A) gene fragment (n = 156) was more frequently detected than the tet(B) gene fragment (n = 76). Two of these isolates (one pork and one beef isolate) harboured both genes. Twenty PCR fragments were sequenced to confirm the partial tetracycline resistance gene sequence. No sequence variations that would have caused an amino acid sequence change were found within these sequences. Comparing the minimum inhibitory tetracycline concentration of the isolates and the detection of resistance genes, a significant difference (p < 0.05) was found within the distribution (Figure 1).
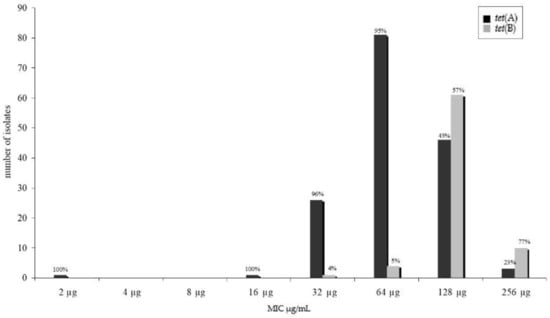
Figure 1.
Distribution of tet(A) and tet(B) gene fragments and resistance phenotype—determined by minimum inhibitory concentration using the microbroth dilution.
Higher minimum inhibitory concentrations were found for isolates harbouring the tet(B) gene fragment. No differences were found between resistance gene distribution and meat species (Figure 2), whereas distribution between tet(A) and tet(B) in minced meat (mixed pork and beef)—with tet(B) as the predominant gene—was significantly different to the meat of the single meat species (p < 0.05).
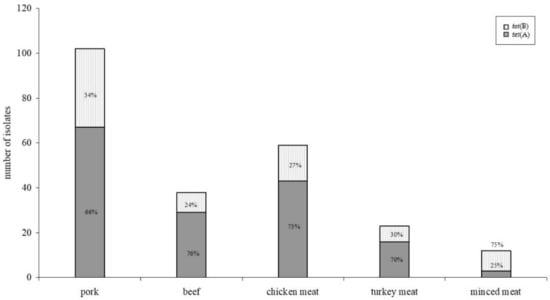
Figure 2.
Distribution of tet(A) and tet(B) gene fragments within different meat species. Despite differences regarding the incidence of resistant isolates in different meat species, no statistically significant (p < 0.05) difference could be detected within the distribution of resistance genes.
2.3. Sulphonamide Resistance Genes
We tested the occurrence of sulI and sulII resistance gene fragments. The sulII gene fragment was found more often (n = 113) than the sulI gene fragment (n = 32) in a total of 142 sulphonamide-resistant E. coli isolates. Three isolates harboured both gene fragments (one pork, one chicken meat, and one minced meat isolate). Sequence analysis was performed in a random sample of sulI (n = 10) and sulII (n = 10) positive isolates and on the only intermediate-resistant isolate (sulII). No differences were detected in the coding sequence of the genes.
2.4. Streptomycin Resistance Genes
Streptomycin resistance in E. coli is recommended to be only tested as a disc diffusion assay [16]. For streptomycin resistance, many isolates were determined as intermediate resistant. Hence, resistance gene detection was performed for all intermediate-resistant isolates as well. In most resistant isolates (n = 50, 94%) the strA–strB gene cluster could be detected by PCR of the strA gene fragment. In three resistant isolates, the aadA1 gene fragment was amplified (6%). In all intermediate isolates, a resistance gene fragment could be detected by PCR (the strA–strB in 40 isolates, the aadA1 in 22 and both genes in three isolates). The strA–strB cluster was associated with higher inhibitory concentrations, as shown in the histogram of the disc diffusion assay (Figure 3).
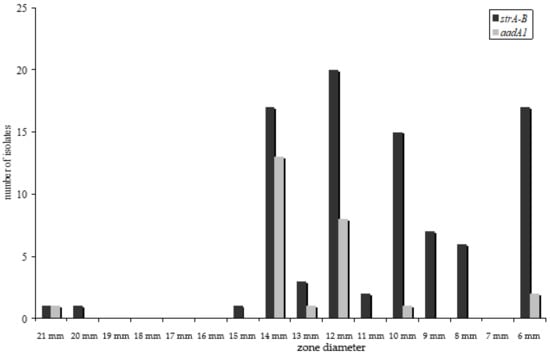
Figure 3.
Distribution of strA–B and aadA1 gene fragments and the resistance phenotype—determined by disc susceptibility testing given by zone diameter. The isolates harbouring the strA–B gene cluster developed higher MIC90.
2.5. Ampicillin Resistance Genes
In this study, we used primers to detect the coding region for different β-lactamases: Temoniera (TEM), cephalomycinase (CMY), Pseudomonas-specific-enzyme group (PSE), active on imipenem (IMP), oxacillinase (OXA). By using distinct primers, we were able to detect the blaTEM gene fragment in all of our resistant isolates. One intermediate isolates harboured the blaTEM gene fragment and another intermediate isolate harboured the blaOXA gene fragment. No resistance gene was found in the third intermediate isolate.
2.6. Trimethoprim Resistance Genes
In 71 isolates, a dfrA, and in 3 isolates (two chicken meat isolates, one minced meat isolate), a dfr7&17 gene fragment could be amplified. In four trimethoprim-resistant isolates (one pork isolate and three chicken meat isolates), none of the tested resistance gene fragments was detected.
2.7. Kanamycin Resistanc Genes
In all 28 resistant isolates, the aphA-1A resistance gene fragment was amplified. Isolates originated from pork (n = 6), beef (n = 5), chicken meat (n = 14), turkey meat (n = 1), and minced meat (n = 2).
2.8. Multi-Resistant Isolates and Gene Association
From all isolates, harbouring resistance elements (phenotypically resistant and intermediate isolates), only 75 carried only one resistance determinant (single resistance) but all the rest of the isolates harboured at least two or more resistance gene elements (n = 190). In more than 82% (n = 61) of the isolates carrying a single resistance gene, this gene was a tetracycline resistance gene—either tet(A) in 46 isolates and tet(B) in 15 isolates. Isolates with multiple resistance gene fragments were found in isolates from all meat species. Resistance genes associated with other resistance genes are stated in Table 1. Most often associated the trimethoprim resistance gene dfrA, the strA–B cluster and the sulII genes. They were associated with each other or with other’s resistance genes. Transformation studies confirmed these results as a combined transfer of pheno- and genotypically resistance could be detected even though this was not the case for every transformant. This has been shown for three plasmids all isolated from three different multidrug-resistant isolates from this study, which were individually transformed into a DH5α-competent strain. Transformants exhibited phenotypical combined resistance to tetracycline, sulphonamides, and streptomycin, combined resistance to tetracycline and streptomycin and single tetracycline resistance. Five other isolated plasmids were not harbouring the expected genes or were simply not transformable into DH5α. In none of the sensitive tested isolates was a resistance gene identified.

Table 1.
Resistance gene association in E. coli isolates from meat.
2.9. Phylogenetic Groups
Phylogenetic group A was the most common group among isolates from all meat sources, varying between 73% in minced meat and 56% in poultry meat. Phylogroup B1 was the second most common group with 28% in beef and poultry, 27% in minced meat, 24% in pork and 17% in turkey meat. Less commonly found were phylogroup D with 0–17% and B2 with only 0–6%. These differences are significant within the meat source (p < 0.05). There were no significant association between phylogenetic groups and the different resistance genes.
3. Discussion
As antimicrobial resistance is a global fear to public health, many industrialised as well as developing countries have established monitoring programs. The main problems in achieving comparable data are, for instance, the different methods used to detect antimicrobial resistance. Genetic methods for the detection of antimicrobial resistance genes appear to be the most discussed approach for overcoming this problem [1,2]. Despite the fact that there are many studies on the dissemination and incidence of resistance genes in diverse microorganisms nowadays, new data are still necessary [9,15,17,18]. In this study, we detected the appropriate resistance gene fragment in 257 out of 261 phenotypically resistant and in 62 of 63 isolates with an intermediate resistance phenotype by PCR—which in both cases is more than 98%. In spite of the large number of known resistance genes, for instance, for tetracycline and ampicillin resistance, in our isolates, only certain genes were commonly found (tet(A) and tet(B); blaTEM). This was rather unexpected as trimmed meat of different animal species may harbour E. coli isolates of animal and of human origin [19,20,21].
Antimicrobial resistance profiles of microorganisms isolated from food of animal origin differ according to the animal species [14]. Therefore, we also expected a diversity in the occurrence of resistance genes in isolates of meat of different animal species. However, the distribution of resistance genes did not differ significantly (Figure 2) for all resistance genes detected. This is even more surprising because it is well known that different resistance genes associate and assign with phenotypically dissimilar minimum inhibitory concentrations of the isolate [21,22,23]. In our study, this difference was statistically significant (p < 0.05) between tet(A) and tet(B) as well as between aadA1 and strA–B harbouring isolates (Figure 1 and Figure 3). No statistically significant divergence could be detected between isolates harbouring either sulI or sulII genes.
Since isolates carrying multiple resistance genes were detected, research has focused on the combined horizontal transfer of resistance genes within or even across bacterial species. Hence, associations between resistance genes are of special interest, as is if and how horizontal transfer appears. In our study, multi-resistant isolates were detected quite often—a single resistance genotype was detected in only 28% of the tested isolates. This single resistance genotype was most often a tet(A) harbouring strain (61%). In all but one intermediate isolates (n = 63) could a responsible resistance gene fragment be detected. The only intermediate resistant isolate in which no resistant gene fragment could be detected was a pork E. coli isolate with intermediate resistance to ampicillin (MIC of 16 µg/mL). As decreased sensitivity to β-lactam antibiotics has been long known to also be due to changes in the membrane structure of E. coli [24], this might be due to this strain as well. These results were confirmed by sequencing a random sample and subsequent alignment. In some of the isolates carrying multiple resistance genes, we could show that isolated plasmids could transfer resistance to the antimicrobial sensitive lab strain DH5α. This was also feasible for combined resistance to tetracycline, sulphonamides, and streptomycin. All the resistant isolates were phylogenetically characterised by phylogenetic grouping. Phylogenetic group A was the most common group among isolates from all meat sources. This varied between 73% in minced meat and 56% in poultry meat. Phylogroup B1 was the second most common group with 28% in beef and poultry, 27% in minced meat, 24% in pork, and 17% in turkey meat. Less commonly found was phylogroup D with 0–17% and B2 with only 0–6%. These differences are significant within the meat source (p < 0.05). There were no significant association between phylogenetic groups and the different resistance genes.
Antimicrobial therapy in farm animals is under discussion as each application inevitably leads to resistance development and the horizontal transfer of resistance genes. Despite the potential involvement of human and animal bacterial pathogens, the transfer of resistance within commensals is important as a reservoir of resistance genes. Even though antimicrobial therapy for animal welfare reasons is required in serious bacterial infections, the prudent use of antimicrobial drugs is necessary.
Food of animal origin may harbour antimicrobial-resistant bacteria and thus the education of consumers in regard to handling, meal preparation and consumption must be a priority in public health.
Our study gives essential information to consider new tools for antimicrobial resistance surveillance using sequence-based methods but the limited number of antimicrobials used to test for resistance in our study comprises a weakness and additional studies are required before solely the surveillance of antimicrobial resistance can be performed using genetic tools. Nevertheless, important antimicrobials used in animals have been tested and we did not consider essential antimicrobials used in humans, even though shigatoxin-producing isolates are clearly pathogens, antimicrobial treatment in humans is not recommended as most antimicrobials trigger toxin production, and thus induce severe disease symptoms—whereas other human infections caused by E. coli are generally not of orally origin.
In conclusion, our results showed a high occurrence of resistant E. coli isolated from poultry meat, pork, and beef; however, a rather low number of diverse resistance genes were detected and they were similarly distributed across all meat species. E. coli populations most often exhibited significantly statistically different MIC values when harbouring different resistance genes.
4. Materials and Methods
Bacterial strains and sampling procedure: 4 intermediate-resistant, 261 resistant, and 48 sensitive E. coli isolates originating from raw meat samples including minced meat were randomly selected and collected throughout Austria’s supermarkets, EU-approved slaughterhouses, butchers, and street markets. E. coli isolates were isolated according to ISO 16649-1. From each plate, between two and three isolates were selected for biochemical analysis and identification and for resistance testing. Only one isolate from each sample was included in the study unless the isolates showed a distinct resistance phenotype. Resistant isolates from pork (102 isolates), beef (41 isolates), chicken meat (82 isolates), turkey meat (23 isolates), 13 from minced meat (pork and beef), 4 intermediate, and 48 sensitive isolates were tested for resistance genes using PCR methods (Table 2).

Table 2.
E. coli isolates from meat and phylogenetic groups.
Biochemical analysis of strains: for the biochemical analysis and identification of E. coli, the API 20E (BioMerieux 20100, bioMérieux Austria GmbH, Vienna, Austria) was used.
Definition of phylogenetic groups: the method described by Clermont et al., 2000 [3], was used to identify four main phylogenetic groups: A, B1, B2, and D. From all isolates, genomic DNA was extracted from bacterial suspensions using standard techniques [25]. For the purpose of multiplex PCR, the KAPA2G Fast Mulitplex Ready Mix (PEQLAB Biotechnologie GMBH, Erlangen, Germany) was used.
Susceptibility testing: isolates that had not been previously phenotypically analysed were tested using the disc diffusion assay and the microbroth dilution assay, as recommended by the Clinical Laboratory Standards Institute with the control strain ATCC 25922 [16]. Resistance was analysed against most important antimicrobial veterinary drug groups as are ampicillin, kanamycin, streptomycin, sulphonamides, tetracycline, and trimethoprim.
PCR of resistance genes was done using primers and conditions stated in Table 3.

Table 3.
Primer and conditions for PCR of tested resistance genes.
The sequencing of PCR products: the sequence was determined using the Big Dye Terminator v. 3.1 cycle sequencing kit and an Applied Biosystems 310 ABI Prism Genetic Analyser.
Transformation studies: on selected isolates with single and multiple detected resistance genes, plasmid isolation was performed using a spin mini preparation tool kit (QIAprep Spin Miniprep Kit, Qiagen, Hilden, Germany). From these plasmids, the PCRs of respective resistance genes were carried out. PCR-positive plasmids were chemically transformed into DH5α using standard techniques [25].
Statistical analysis: statistical analyses were carried out using SPSS (Statistical Package for Social Sciences Version 17.0 SPSS Inc., Somers, NY, USA). To examine differences between isolates of different origins, the occurrence of antimicrobial resistance genes and phenotype, χ2-, and univariable logistic regression tests were used.
Author Contributions
Conceptualisation, F.H., A.S.-P. and A.-B.B.-P.; methodology, F.H., D.S., and A.S.-P.; software, D.K.-J.; validation, F.H., and D.S.; formal analysis, D.K.-J.; investigation, A.S.-P. and F.H.; resources, F.H.; data curation, A.S.-P. and D.S.; writing—original draft preparation, A.S.-P.; writing—review and editing, F.H.; visualisation, A.S.-P.; supervision, F.H. and A.-B.B.-P. All authors have read and agreed to the published version of the manuscript.
Funding
Open Access Funding by the University of Veterinary Medicine Vienna
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. Requests for isolates/plasmids can be send to the corresponding author.
Acknowledgments
Open Access Funding by the University of Veterinary Medicine Vienna.
Conflicts of Interest
The authors declare no conflict of interest.
References
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards); Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Hilbert, F.; et al. Scientific Opinion on the whole genome sequencing and metagenomics for outbreak investigation, source attribution and risk assessment of food-borne microorganisms. EFSA J. 2019, 17, e05898. [Google Scholar] [CrossRef]
- Llarena, A.-K.; Ribeiro-Gonçalves, B.F.; Nuno Silva, D.; Halkilahti, J.; Machado, M.P.; Da Silva, M.S.; Jaakkonen, A.; Isidro, J.; Hämäläinen, C.; Joenperä, J.; et al. INNUENDO: A cross-sectoral platform for the integration of genomics in the surveillance of food-borne pathogens. EFSA Support. Publ. 2018, 15, 142p. [Google Scholar] [CrossRef]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and Simple Determination of the Escherichia coli Phylogenetic Group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef] [PubMed]
- Tartof, S.Y.; Solberg, O.D.; Riley, L.W. Genotypic analyses of uropathogenic Escherichia coli based on fimH single nucleotide polymorphisms (SNPs). J. Med Microbiol. 2007, 56, 1363–1369. [Google Scholar] [CrossRef] [PubMed]
- Weissman, S.J.; Hansen, N.I.; Zaterka-Baxter, K.; Higgins, R.D.; Stoll, B.J. Emergence of Antibiotic Resistance-Associated Clones AmongEscherichia coliRecovered From Newborns with Early-Onset Sepsis and Meningitis in the United States, 2008–2009. J. Pediatr. Infect. Dis. Soc. 2016, 5, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Ratshilingano, M.T.; du Plessis, E.M.; Duvenage, S.; Korsten, L. Characterisation of multidrug resistant Escherichia coli isolated from two commercial lettuce and spinach supply chains. J. Food Prot. 2021. online ahead of print. [Google Scholar] [CrossRef]
- Olesen, B.; Scheutz, F.; Menard, M.; Skov, M.N.; Kolmos, H.J.; Kuskowski, M.A.; Johnson, J.R. Three-Decade Epidemiological Analysis of Escherichia coli O15:K52:H1. J. Clin. Microbiol. 2009, 47, 1857–1862. [Google Scholar] [CrossRef]
- Suzuki, S.; Shibata, N.; Yamane, K.; Wachino, J.-I.; Ito, K.; Arakawa, Y. Change in the prevalence of extended-spectrum- -lactamase-producing Escherichia coli in Japan by clonal spread. J. Antimicrob. Chemother. 2009, 63, 72–79. [Google Scholar] [CrossRef]
- Ciccozzi, M.; Giufre, M.; Accogli, M.; Presti, A.L.; Graziani, C.; Cella, E.; Cerquetti, M. Phylogenetic analysis of multidrug-resistant Escherichia coli clones isolated from humans and poultry. New Microbiol. 2013, 36, 385–394. [Google Scholar]
- Guerra, S.T.; Dalanezi, F.M.; De Paula, C.L.; Hernandes, R.T.; Pantoja, J.C.D.F.; Listoni, F.J.P.; Langoni, H.; Ribeiro, M.G. Putative virulence factors of extra-intestinalEscherichia coliisolated from bovine mastitis with different clinical scores. Lett. Appl. Microbiol. 2019, 68, 403–408. [Google Scholar] [CrossRef]
- Khan, S.B.; Zou, G.; Cheng, Y.-T.; Xiao, R.; Li, L.; Wu, B.; Zhou, R. Phylogenetic grouping and distribution of virulence genes in Escherichia coli along the production and supply chain of pork around Hubei, China. J. Microbiol. Immunol. Infect. 2017, 50, 382–385. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Scheinberg, J.; Dudley, E.G.; Campbell, J.; Roberts, B.; DiMarzio, M.; Debroy, C.; Cutter, C.N. Prevalence and Phylogenetic Characterization of Escherichia coli and Hygiene Indicator Bacteria Isolated from Leafy Green Produce, Beef, and Pork Obtained from Farmers’ Markets in Pennsylvania. J. Food Prot. 2017, 80, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Moulin-Schouleur, M.; Répérant, M.; Laurent, S.; Brée, A.; Mignon-Grasteau, S.; Germon, P.; Rasschaert, D.; Schouler, C. Extraintestinal Pathogenic Escherichia coli Strains of Avian and Human Origin: Link between Phylogenetic Relationships and Common Virulence Patterns. J. Clin. Microbiol. 2007, 45, 3366–3376. [Google Scholar] [CrossRef]
- Mayrhofer, S.; Paulsen, P.; Smulders, F.J.; Hilbert, F. Antimicrobial Resistance in Commensal Escherichia coli Isolated from Muscle Foods as Related to the Veterinary Use of Antimicrobial Agents in Food-Producing Animals in Austria. Microb. Drug Resist. 2006, 12, 278–283. [Google Scholar] [CrossRef]
- Ryu, S.-H.; Lee, J.-H.; Park, S.-H.; Song, M.-O.; Park, S.-H.; Jung, H.-W.; Park, G.-Y.; Choi, S.-M.; Kim, M.-S.; Chae, Y.-Z.; et al. Antimicrobial resistance profiles among Escherichia coli strains isolated from commercial and cooked foods. Int. J. Food Microbiol. 2012, 159, 263–266. [Google Scholar] [CrossRef] [PubMed]
- CLSI 2019. Performance Standards for Antimicrobial Susceptibility Testing M100, 29th ed.; CLSI: Annapolis Junction, MD, USA, 2019. [Google Scholar]
- Furlan, J.P.R.; Stehling, E.G. Multiple sequence types, virulence determinants and antimicrobial resistance genes in multidrug- and colistin-resistant Escherichia coli from agricultural and non-agricultural soils. Environ. Pollut. 2021, 288, 117804. [Google Scholar] [CrossRef]
- Sheikh, A.A.; Checkley, S.; Avery, B.; Chalmers, G.; Bohaychuk, V.; Boerlin, P.; Reid-Smith, R.; Aslam, M. Antimicrobial Resistance and Resistance Genes in Escherichia coli Isolated from Retail Meat Purchased in Alberta, Canada. Foodborne Pathog. Dis. 2012, 9, 625–631. [Google Scholar] [CrossRef]
- Pietsch, M.; Irrgang, A.; Roschanski, N.; Brenner, M.G.; Hamprecht, A.; Rieber, H.; Käsbohrer, A.; Schwarz, S.; Rösler, U.; Kreienbrock, L.; et al. Whole genome analyses of CMY-2-producing Escherichia coli isolates from humans, animals and food in Germany. BMC Genom. 2018, 19, 601. [Google Scholar] [CrossRef]
- Touchon, M.; Perrin, A.; de Sousa, J.A.M.; Vangchhia, B.; Burn, S.; O’Brien, C.L.; Denamur, E.; Gordon, D.; Rocha, E.P. Phylogenetic background and habitat drive the genetic diversification of Escherichia coli. PLoS Genet. 2020, 16, e1008866. [Google Scholar] [CrossRef]
- Wadepohl, K.; Müller, A.; Seinige, D.; Rohn, K.; Blaha, T.; Meemken, D.; Kehrenberg, C. Association of intestinal colonization of ESBL-producing Enterobacteriaceae in poultry slaughterhouse workers with occupational exposure—A German pilot study. PLoS ONE 2020, 15, e0232326. [Google Scholar] [CrossRef] [PubMed]
- Boerlin, P.; Travis, R.; Gyles, C.L.; Reid-Smith, R.; Lim, N.J.H.; Nicholson, V.; McEwen, S.A.; Friendship, R.; Archambault, M. Antimicrobial Resistance and Virulence Genes of Escherichia coli Isolates from Swine in Ontario. Appl. Environ. Microbiol. 2005, 71, 6753–6761. [Google Scholar] [CrossRef]
- Sunde, M.; Norström, M. The genetic background for streptomycin resistance in Escherichia coli influences the distribution of MICs. J. Antimicrob. Chemother. 2005, 56, 87–90. [Google Scholar] [CrossRef]
- Jaffé, A.; A Chabbert, Y.; Derlot, E. Selection and characterization of beta-lactam-resistant Escherichia coli K-12 mutants. Antimicrob. Agents Chemother. 1983, 23, 622–625. [Google Scholar] [CrossRef][Green Version]
- Sambrook, J.F.; Fritsch, E.F.; Maniatis, T. (Eds.) Molecular Cloning: A Laboratory Manual; Cold Spring Habor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Ng, L.-K.; Martin, I.; Alfa, M.; Mulvey, M. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 2001, 15, 209–215. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, S.; White, D.G.; Schroeder, C.M.; Lu, R.; Yang, H.; McDermott, P.F.; Ayers, S.; Meng, J. Characterization of Multiple-Antimicrobial-Resistant Salmonella Serovars Isolated from Retail Meats. Appl. Environ. Microbiol. 2004, 70, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Mazel, D.; Dychinco, B.; Webb, V.A.; Davies, J. Antibiotic Resistance in the ECOR Collection: Integrons and Identification of a Novel aad Gene. Antimicrob. Agents Chemother. 2000, 44, 1568–1574. [Google Scholar] [CrossRef]
- Kerrn, M.B.; Klemmensen, T.; Frimodt-Møller, N.; Espersen, F. Susceptibility of Danish Escherichia coli strains isolated from urinary tract infections and bacteraemia, and distribution of sul genes conferring sulphonamide resistance. J. Antimicrob. Chemother. 2002, 50, 513–516. [Google Scholar] [CrossRef]
- Maidhof, H.; Guerra, B.; Abbas, S.; Elsheikha, H.M.; Whittam, T.S.; Beutin, L. A Multiresistant Clone of Shiga Toxin-Producing Escherichia coli O118:[H16] Is Spread in Cattle and Humans over Different European Countries. Appl. Environ. Microbiol. 2002, 68, 5834–5842. [Google Scholar] [CrossRef]
- Henriques, I.; Moura, A.; Alves, A.; Saavedra, M.J.; Correia, A. Analysing diversity among Î2-lactamase encoding genes in aquatic environments. FEMS Microbiol. Ecol. 2006, 56, 418–429. [Google Scholar] [CrossRef] [PubMed]
- Navia, M.M.; Ruiz, J.; Sanchez-Cespedes, J.; Vila, J. Detection of dihydrofolate reductase genes by PCR and RFLP. Diagn. Microbiol. Infect. Dis. 2003, 46, 295–298. [Google Scholar] [CrossRef]
- Grape, M.; Motakefi, A.; Pavuluri, S.; Kahlmeter, G. Standard and real-time multiplex PCR methods for detection of trimethoprim resistance dfr genes in large collections of bacteria. Clin. Microbiol. Infect. 2007, 13, 1112–1118. [Google Scholar] [CrossRef] [PubMed]
- Sandvang, D.; Aarestrup, F.M. Characterization of Aminoglycoside Resistance Genes and Class 1 Integrons in Porcine and Bovine Gentamicin-ResistantEscherichia coli. Microb. Drug Resist. 2000, 6, 19–27. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).