In Silico Analysis of PKS and NRPS Gene Clusters in Arisostatin- and Kosinostatin-Producers and Description of Micromonospora okii sp. nov.
Abstract
1. Introduction
2. Results
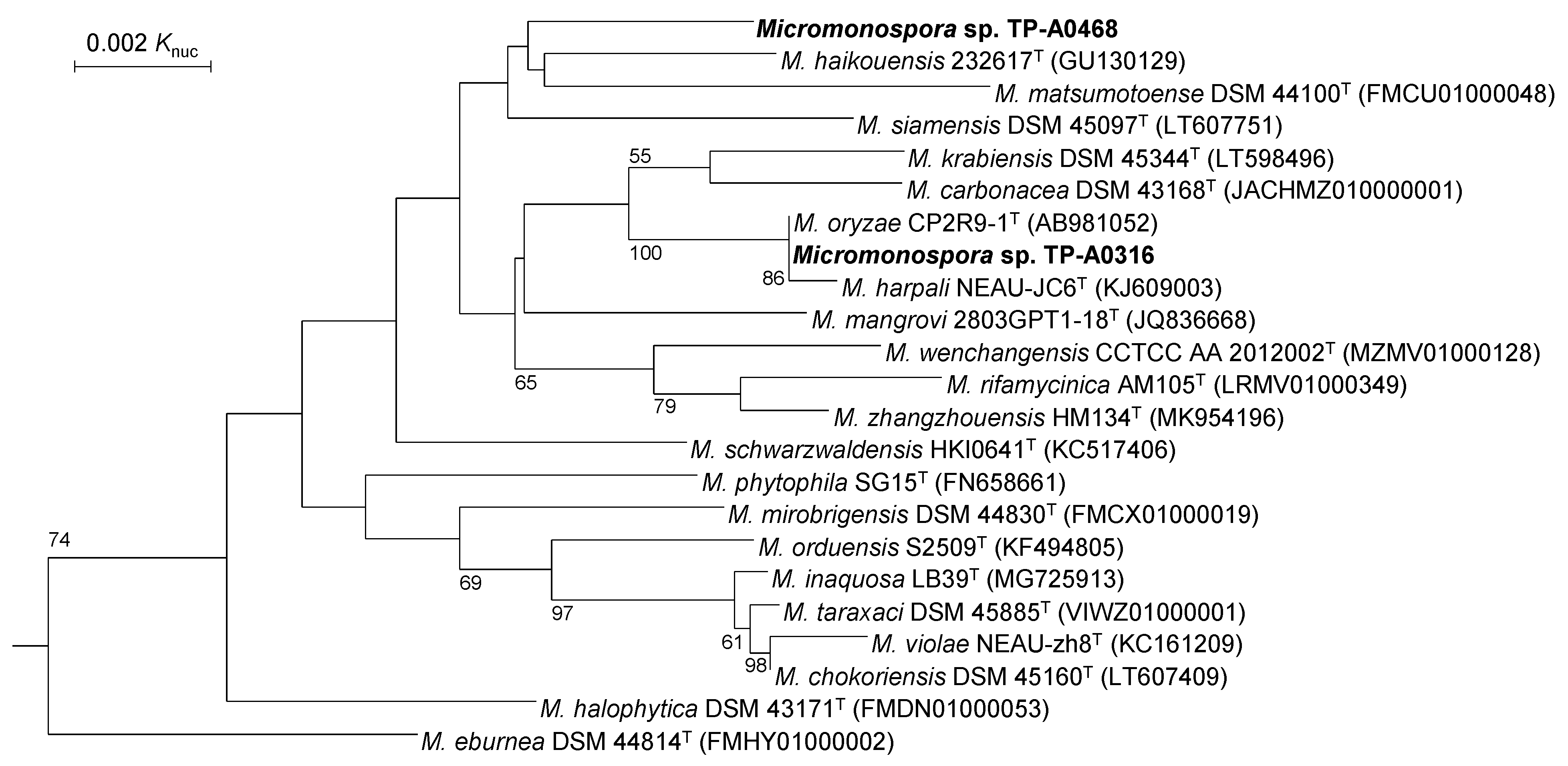
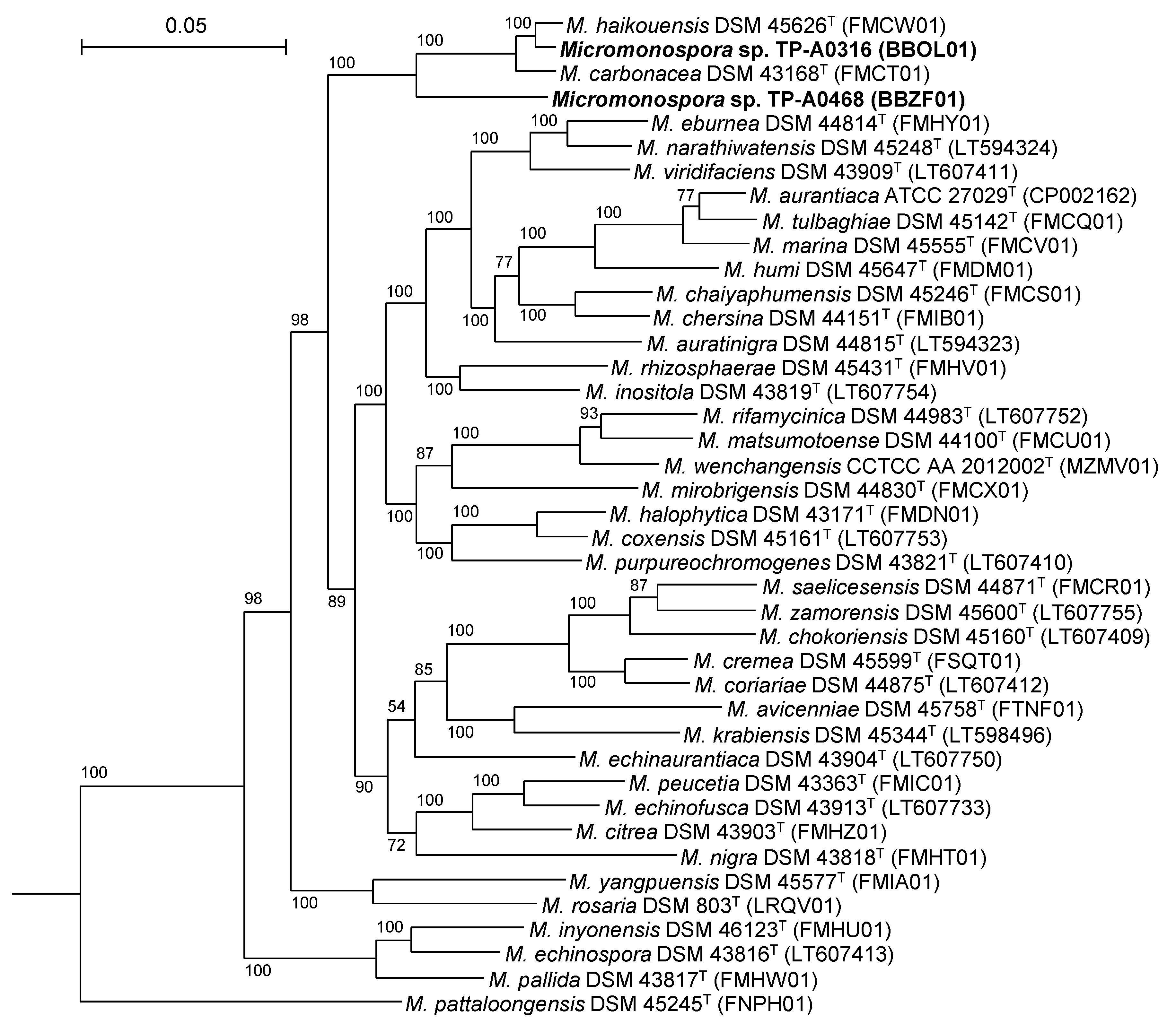
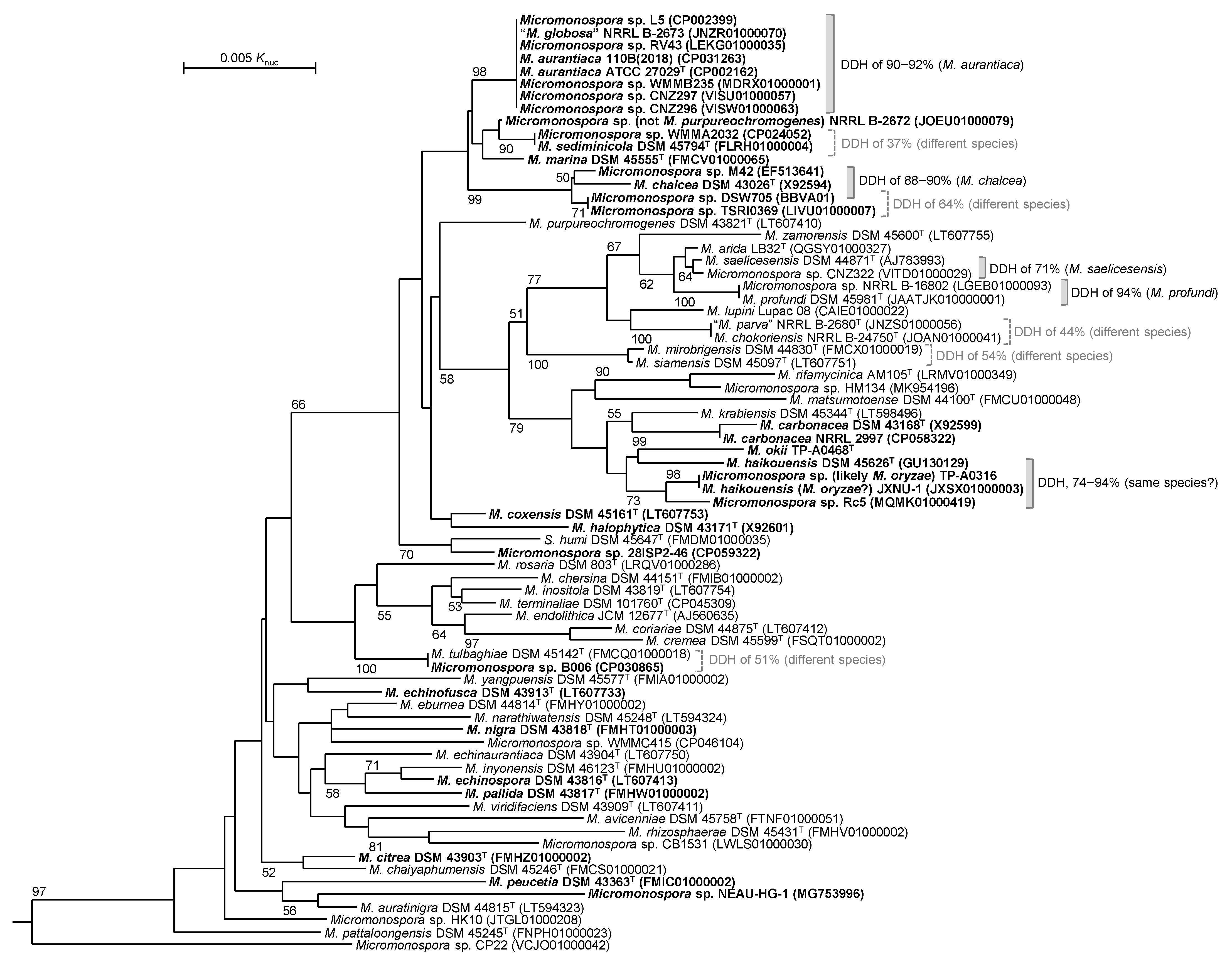
2.1. Classification of Micromonospora sp. TP-A0316 and Micromonospora sp. TP-A0468
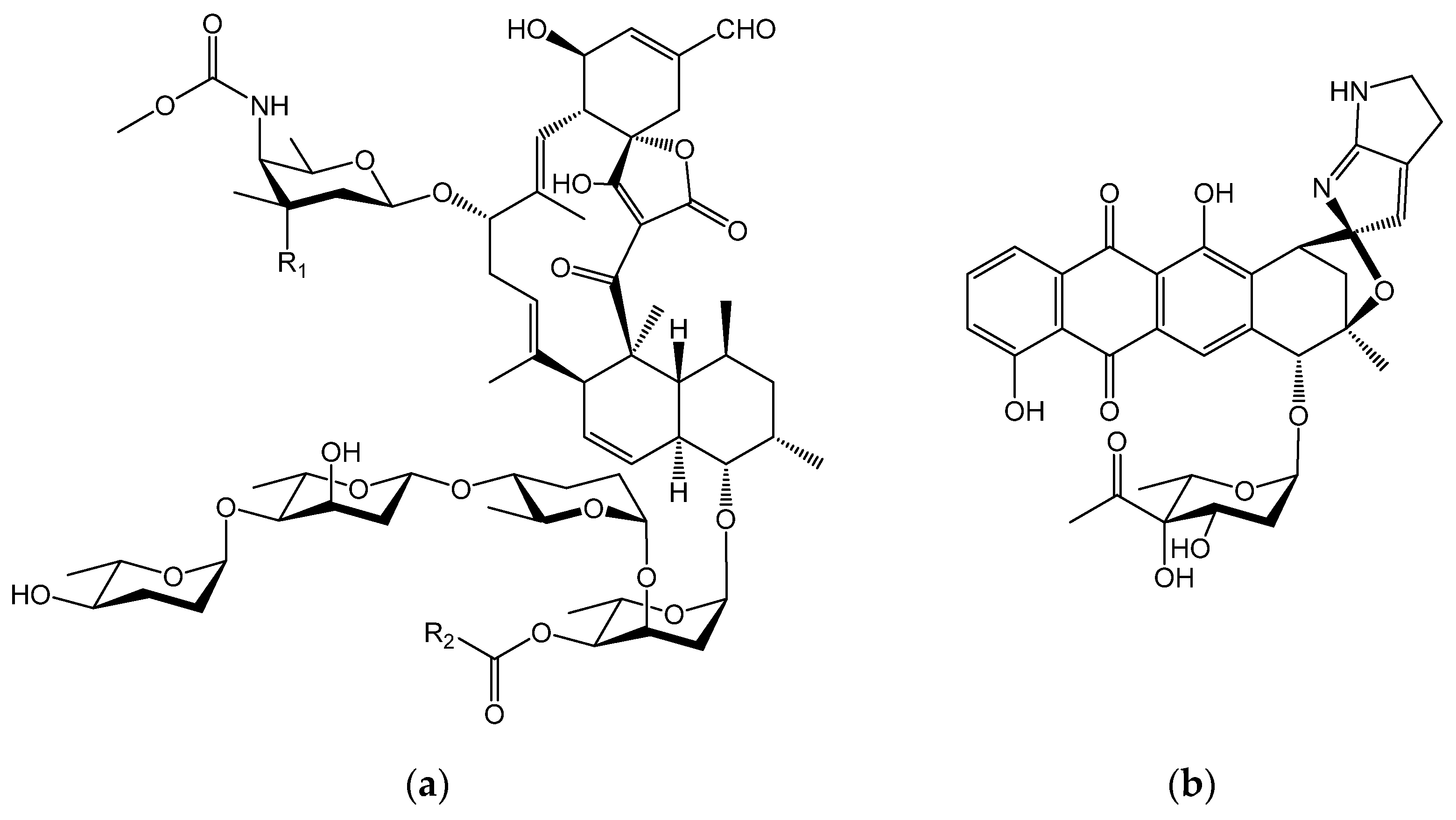
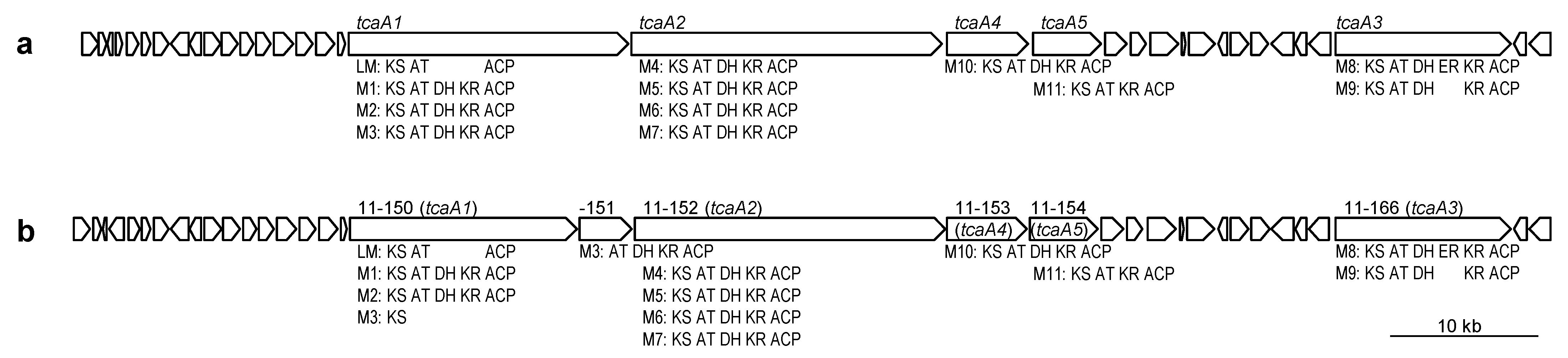
2.2. PKS and NRPS Gene Clusters in Micromonospora sp. TP-A0316 and M. okii TP-A0468T
2.3. Distribution of Quinmuinolidomicin-BGC Orthologs in the Genus Micromonospora
3. Discussion
4. Description of Micromonospora okii sp. nov.
5. Materials and Methods
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef]
- Qi, S.; Gui, M.; Li, H.; Yu, C.; Li, H.; Zeng, Z.; Sun, P. Secondary metabolites from marine Micromonospora: Chemistry and bioactivities. Chem. Biodivers. 2020, 17, e2000024. [Google Scholar] [CrossRef] [PubMed]
- Furumai, T.; Takagi, K.; Igarashi, Y.; Saito, N.; Oki, T. Arisostatins A and B, new members of tetrocarcin class of antibiotics from Micromonospora sp. TP-A0316. I. Taxonomy, fermentation, isolation and biological properties. J. Antibiot. 2000, 53, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Furumai, T.; Igarashi, Y.; Higuchi, H.; Saito, N.; Oki, T. Kosinostatin, a quinocycline antibiotic with antitumor activity from Micromonospora sp. TP-A0468. J. Antibiot. 2002, 55, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Zhang, Y.; Huang, L.; Jia, X.; Zhang, Q.; Zhang, X.; Tang, G.; Liu, W. Cloning and characterization of the tetrocarcin A gene cluster from Micromonospora chalcea NRRL 11289 reveals a highly conserved strategy for tetronate biosynthesis in spirotetronate antibiotics. J. Bacteriol. 2008, 190, 6014–6025. [Google Scholar] [CrossRef]
- Ma, H.; Zhou, Q.; Tang, Y.; Zhang, Z.; Chen, Y.; He, H.; Pan, H.; Tang, M.; Gao, J.; Zhao, S.; et al. Unconventional origin and hybrid system for construction of pyrrolopyrrole moiety in kosinostatin biosynthesis. Chem. Biol. 2013, 20, 796–805. [Google Scholar] [CrossRef]
- Schwarzer, D.; Marahiel, M.A. Multimodular biocatalysts for natural product assembly. Naturwissenschaften 2001, 88, 93–101. [Google Scholar] [CrossRef]
- Meurer, G.; Gerlitz, M.; Wendt-Pienkowski, E.; Vining, L.C.; Rohr, J.; Hutchinson, C.R. Iterative type II polyketide synthases, cyclases and ketoreductases exhibit context-dependent behavior in the biosynthesis of linear and angular decapolyketides. Chem. Biol. 1997, 4, 433–443. [Google Scholar] [CrossRef][Green Version]
- Nett, M.; Ikeda, H.; Moore, B.S. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 2009, 26, 1362–1384. [Google Scholar] [CrossRef]
- Komaki, H.; Oguchi, A.; Tamura, T.; Hamada, M.; Ichikawa, N. Diversity of nonribosomal peptide synthetase and polyketide synthase gene clusters in the genus Acrocarpospora. J. Gen. Appl. Microbiol. 2020, 66, 315–322. [Google Scholar] [CrossRef]
- Komaki, H.; Tamura, T. Polyketide synthase and nonribosomal peptide synthetase gene clusters in type strains of the genus Phytohabitans. Life 2020, 10, 257. [Google Scholar] [CrossRef] [PubMed]
- Komaki, H.; Tamura, T.; Ichikawa, N.; Oguchi, A.; Hamada, M.; Suzuki, K.; Fujita, N. Genome-based analysis of type-I polyketide synthase and nonribosomal peptide synthetase gene clusters in a novel strain taxonomically close to the genus Salinispora. J. Antibiot. 2015, 68, 767–770. [Google Scholar] [CrossRef] [PubMed]
- Kasai, H.; Tamura, T.; Harayama, S. Intrageneric relationships among Micromonospora species deduced from gyrB-based phylogeny and DNA relatedness. Int. J. Syst. Evol. Microbiol. 2000, 50, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Hatano, K.; Nishii, T.; Kasai, H. Taxonomic re-evaluation of whorl-forming Streptomyces (formerly Streptoverticillium) species by using phenotypes, DNA-DNA hybridization and sequences of gyrB, and proposal of Streptomyces luteireticuli (ex Katoh and Arai 1957) corrig., sp. nov., nom. rev. Int. J. Syst. Evol. Microbiol. 2003, 53, 1519–1529. [Google Scholar] [CrossRef] [PubMed]
- Fang, B.; Liu, C.; Guan, X.; Song, J.; Zhao, J.; Liu, H.; Li, C.; Ning, W.; Wang, X.; Xiang, W. Two new species of the genus Micromonospora: Micromonospora palomenae sp. nov. and Micromonospora harpali sp. nov. isolated from the insects. Antonie Van Leeuwenhoek 2015, 108, 141–150. [Google Scholar] [CrossRef]
- Kittiwongwattana, C.; Thanaboripat, D.; Laosinwattana, C.; Koohakan, P.; Parinthawong, N.; Thawai, C. Micromonospora oryzae sp. nov., isolated from roots of upland rice. Int. J. Syst. Evol. Microbiol. 2015, 65, 3818–3823. [Google Scholar] [CrossRef]
- Xie, Q.; Ren, J.; Li, L.; Li, Y.; Deng, Z.; Hong, K. Micromonospora mangrovi sp. nov., isolated from mangrove soil. Antonie Van Leeuwenhoek 2016, 109, 483–491. [Google Scholar] [CrossRef]
- Hashimoto, T.; Hashimoto, J.; Kozone, I.; Amagai, K.; Kawahara, T.; Takahashi, S.; Ikeda, H.; Shin-ya, K. Biosynthesis of quinolidomicin, the largest known macrolide of terrestrial origin: Identification and heterologous expression of a biosynthetic gene cluster over 200 kb. Org. Lett. 2018, 20, 7996–7999. [Google Scholar] [CrossRef]
- McGlinchey, R.P.; Nett, M.; Moore, B.S. Unraveling the biosynthesis of the sporolide cyclohexenone building block. J. Am. Chem. Soc. 2008, 130, 2406–2407. [Google Scholar] [CrossRef]
- Awakawa, T.; Fujita, N.; Hayakawa, M.; Ohnishi, Y.; Horinouchi, S. Characterization of the biosynthesis gene cluster for alkyl-O-dihydrogeranyl-methoxyhydroquinones in Actinoplanes missouriensis. ChemBioChem 2011, 12, 439–448. [Google Scholar] [CrossRef]
- Zhou, Q.; Luo, G.; Zhang, H.; Tang, G. Discovery of 16-demethylrifamycins by removing the predominant polyketide biosynthesis pathway in Micromonospora sp. strain TP-A0468. Appl. Environ. Microbiol. 2019, 85, e02597-18. [Google Scholar] [CrossRef] [PubMed]
- Tao, M.; Wang, L.; Wendt-Pienkowski, E.; George, N.P.; Galm, U.; Zhang, G.; Coughlin, J.M.; Shen, B. The tallysomycin biosynthetic gene cluster from Streptoalloteichus hindustanus E465-94 ATCC 31158 unveiling new insights into the biosynthesis of the bleomycin family of antitumor antibiotics. Mol. Biosyst. 2007, 3, 60–74. [Google Scholar] [CrossRef] [PubMed]
- Braesel, J.; Crnkovic, C.M.; Kunstman, K.J.; Green, S.J.; Maienschein-Cline, M.; Orjala, J.; Murphy, B.T.; Eustaquio, A.S. Complete genome of Micromonospora sp. strain B006 reveals biosynthetic potential of a Lake Michigan actinomycete. J. Nat. Prod. 2018, 81, 2057–2068. [Google Scholar] [CrossRef] [PubMed]
- Komaki, H.; Sakurai, K.; Hosoyama, A.; Kimura, A.; Igarashi, Y.; Tamura, T. Diversity of nonribosomal peptide synthetase and polyketide synthase gene clusters among taxonomically close Streptomyces strains. Sci. Rep. 2018, 8, 6888. [Google Scholar] [CrossRef]
- Wayne, L.G.; Brenner, D.J.; Colwell, R.R.; Grimont, P.A.D.; Kandler, O.; Krichevsky, M.I.; Moore, L.H.; Moore, W.E.C.; Murray, R.G.E.; Stackebrandt, E.; et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 1987, 37, 463–464. [Google Scholar] [CrossRef]
- Weinstein, M.J.; Luedemann, G.M.; Oden, E.M.; Wagman, G.H.; Rosselet, J.P.; Marquez, J.A.; Coniglio, C.T.; Charney, W.; Herzog, H.L.; Black, J. Gentamicin, a new antibiotic complex from Micromonospora. J. Med. Chem. 1963, 6, 463–464. [Google Scholar] [CrossRef]
- Testa, R.T.; Wagman, G.H.; Daniels, P.J.; Weinstein, M.J. Mutamicins; biosynthetically created new sisomicin analogues. J. Antibiot. 1974, 27, 917–921. [Google Scholar] [CrossRef]
- Weinstein, M.J.; Marquez, J.A.; Testa, R.T.; Wagman, G.H.; Oden, E.M.; Waitz, J.A. Antibiotic 6640, a new Micromonospora-produced aminoglycoside antibiotic. J. Antibiot. 1970, 23, 551–554. [Google Scholar] [CrossRef]
- Zhang, F.; Zhao, M.; Braun, D.R.; Ericksen, S.S.; Piotrowski, J.S.; Nelson, J.; Peng, J.; Ananiev, G.E.; Chanana, S.; Barns, K.; et al. A marine microbiome antifungal targets urgent-threat drug-resistant fungi. Science 2020, 370, 974–978. [Google Scholar] [CrossRef]
- Hayakawa, Y.; Matsuoka, M.; Shin-ya, K.; Seto, H. Quinolidomicins A1, A2 and B1, novel 60-membered macrolide antibiotics. I. Taxonomy, fermentation, isolation, physico-chemical properties and biological activity. J. Antibiot. 1993, 46, 1557–1562. [Google Scholar] [CrossRef][Green Version]
- Hayakawa, Y.; Shin-ya, K.; Furihata, K.; Seto, H. Quinolidomicins A1, A2 and B1, novel 60-membered macrolide antibiotics. II. Structure elucidation. J. Antibiot. 1993, 46, 1563–1569. [Google Scholar] [CrossRef][Green Version]
- Komaki, H.; Ichikawa, N.; Oguchi, A.; Hamada, M.; Harunari, E.; Kodani, S.; Fujita, N.; Igarashi, Y. Draft genome sequence of Streptomyces sp. TP-A0867, an alchivemycin producer. Stand. Genom. Sci. 2016, 11, 85. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Ha, S.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Alanjary, M.; Steinke, K.; Ziemert, N. AutoMLST: An automated web server for generating multi-locus species trees highlighting natural product potential. Nucleic Acids Res. 2019, 47, W276–W282. [Google Scholar] [CrossRef] [PubMed]
- Komaki, H.; Ichikawa, N.; Hosoyama, A.; Fujita, N.; Igarashi, Y. Draft genome sequence of marine-derived Streptomyces sp. TP-A0598, a producer of anti-MRSA antibiotic lydicamycins. Stand. Genom. Sci. 2015, 10, 58. [Google Scholar] [CrossRef] [PubMed]
- Komaki, H.; Ishikawa, A.; Ichikawa, N.; Hosoyama, A.; Hamada, M.; Harunari, E.; Nihira, T.; Panbangred, W.; Igarashi, Y. Draft genome sequence of Streptomyces sp. MWW064 for elucidating the rakicidin biosynthetic pathway. Stand. Genom. Sci. 2016, 11, 83. [Google Scholar] [CrossRef] [PubMed]
- Komaki, H.; Sakurai, K.; Hosoyama, A.; Kimura, A.; Trujilo, M.E.; Igarashi, Y.; Tamura, T. Diversity of PKS and NRPS gene clusters between Streptomyces abyssomicinicus sp. nov. and its taxonomic neighbor. J. Antibiot. 2020, 73, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]

| Character | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Melanine formation | + | nd | nd | nd | − |
| Soluble pigment | − | nd | + | nd | − |
| Whole cell sugar | Gal, Xyl, Ara, Glu | Ara, Xyl, Glu | Ara, Glu, Rib, Xyl | Ara, Glu, Xyl | Glu, Xyl, Man |
| Phospholipid | PE, PI | PE, DPG, PIM | DPG, PE, PG, PI, PIMs | DPG, PE, PIM | DPG, PE, PIM |
| Starch hydrolysis | + | + | + | v | + |
| Milk peptonization | + | nd | + | nd | − |
| Cellulose decomposition | − | + | nd | + | − |
| Gelatin liquefaction | + | + | − | nd | − |
| Utilization of carbon source | |||||
| l-Arabinose | + | − | + | − | − |
| d-Fructose | + | v | + | v | − |
| d-Galactose | + | v | + | − | + |
| Inositol | − | nd | − | nd | + |
| Maltose | + | − | nd | + | + |
| d-Mannitol | − | v | − | v | + |
| d-Mannose | + | − | + | + | + |
| d-Raffinose | − | + | w | + | + |
| d-Xylose | − | v | + | v | − |
| Growth temperature (optimum, °C) | 13–41 (25–39) | nd | 20–45 (30) | nd | 15–40 (28) |
| pH for growth (optimum) | 6–10 (7–8) | 5–8.5 | 5–10 (7) | nd | 6–10 (7) |
| NaCl tolerance (%) | <4 | 3 | 4 | 3 | 3.5 |
| Cluster | ORF | Domain Organization | Predicted Product |
|---|---|---|---|
| t1pks-1 | 11-150 (tcaA1) | KS/AT/ACP-KS/AT/DH/KR/ACP-KS/AT/DH/KR/ACP-KS | arisostatins A & B, tetrocarcin A |
| 11-151 (tcaA1) | ATmm/DH/KR/ACP | ||
| 11-152 (tcaA2) | KS/ATm/DH/KR/ACP-KS/AT/DH/KR/ACP-KS/ATm/DH/KR/ACP- | ||
| KS/ATm/DH/KR/ACP | |||
| 11-153 (tcaA4) | KS/ATmm/DH/KR/ACP | ||
| 11-154 (tcaA5) | KS/ATm/KR/ACP | ||
| 11-166 (tcaA3) | KS/AT/DH/ER/KR/ACP- KS/ATmm/DH/KR/ACP | ||
| t1pks-2 * | 1-1073 | CoL/KR/ACP-KS/ATm/DH/KR/ACP-KS/ATmm/DH/ER/KR/ACP | quinolidomicin congener |
| 1-1077 | KS/ATm/DH/KR/ACP | ||
| 1-1078 | KS/ATm/KR/ACP-KS/ATm/KR/ACP-KS/ATm/KR/ACP | ||
| 1-1080 | KS/ATm/DH/ER/KR/ACP-KS/ATmm/DH/ER/KR/ACP-KS/ATm/DH/ER/KR/ACP- | ||
| KS/ATm/KR/ACP | |||
| 1-1081 | KS/ATmm/KR/ACP-KS/ATm/DH/KR/ACP-KS/ATm/DH/KR/ACP- | ||
| KS/ATm/DH/KR/ACP-KS/ATm/KR/ACP-KS/ATm/KR/ACP | |||
| 1-1091 | KS/ATm/KR/ACP-KS/ATmm/DH/KR/ACP | ||
| 1-1092 | KS/ATm/DH/KR/ACP-KS/ATmm/KR/ACP-KS/ATm/KR/ACP | ||
| 1-1093 | KS/ATm/KR/ACP-KS/ATmm/DH/ER/KR/ACP | ||
| 1-1094 | KS/ATm/KR/ACP- KS/ATmm/KR/ACP- KS/ATmm/KR/ACP | ||
| 1-1095 | KS/ATm/DH/KR/ACP-KS/ATmm/DH/KR/ACP | ||
| 1-1096 | KS/ATmm/KR/ACP | ||
| 1-1097 | KS/ATmm/KR/ACP-KS/ATm/KR/ACP | ||
| 1-1098 | KS/ATm/ACP-Te | ||
| t1pks-3 | 4-330 | KS/AT/KR/DH | sporolide |
| t1pks-4P | 14-64 P | KS/AT… | unpredictable |
| 16-1 P | …KR | ||
| 16-2 | KS/AT | ||
| 16-3 | ACP | ||
| t2pks-1 | 4-99 | KSα | aromatic polyketide |
| 4-100 | KSβ (CLF) | ||
| 4-101 | ACP | ||
| t3pks-1 * | 2-674 | KS | alkyl-O-dihydrogeranyl-methoxyhydroquinone |
| nrps-1 | 1-336 | C/A/T-C/Athr/T/E | pentapeptide (x-thr-phe-ser-ile) |
| 1-337 | C/Aphe/T/E-C/Aser/T | ||
| 1-339 | C/Aile/T | ||
| nrps-2 | 4-510 | C-C | tripeptide (x-gly-x) |
| 4-511 | C/A/T-Te | ||
| 4-512 | A | ||
| 4-513 | C/Agly/T | ||
| 4-514 | T | ||
| nrps-3 | 6-252 | C/Acys/T-Te | tetrapeptide (x-x-cys-cys) |
| 6-258 | A-C/Acys/T | ||
| 6-265 | C | ||
| 6-266 | A | ||
| 6-270 | A/T | ||
| nrps-4P | 8-247 | A/T-C | unpredictable |
| 8-248 | A/T | ||
| nrps-5 | 12-31 | C/Aser/T-C/Apro/T-Te | ser-pro |
| pks/nrps-1 | 4-217 4-220 4-223 4-227 4-228 4-229 4-247 | CoL/ACP-KS/ATm/ACP-C/A/T-C Aala/T-C Aglu/T-C/T Athr/T T-C Aser/T-C/T-Te A/T | heptapeptide with polyketide moieties (st-pk-x-ala-glu-y-thr-ser-y) |
| pks/nrps-2 * | 6-50 6-51 6-52 6-53 6-54 6-55 | C/A/T KS ACP C/Aval/T KS/ATm/ACP Aser | tripeptide with polyketide moiety (ser-x-val-pk) |
| pks/nrps-3 * | 6-307 6-310 6-311 6-313 6-314 | A/T-KS/DH A/T-C/T KS/ATm/KR/DH/ACP C/Aasn/T C/Aser/T-Te | pentapeptide with polyketide moiety (x-pk-x-y-pk-asn-ser) |
| pks/nrps-4 | 8-41 8-40 8-39 8-37 | Agly/T-KS/ACP-KS/ATm DH/KR/ACP-KS/ACP-KS/KR/ACP KS/DH/ACP-KS/ATm DH/KR/ACP-AmT | hexaketide with gly |
| M * | Quinolidomicin (qnm) | t1pks-2 | |
|---|---|---|---|
| in M. chalcea AK-AN57 | in Micromonospora sp. TP-A0316 | in M. okii TP-A0468T | |
| (qnmA1) | (1-1073) | (8-118) | |
| L | CoL/ACP | CoL/KR/ACP | CoL/ACP |
| 1 | KS/ATm/DH/KR/ACP | KS/ATm/DH/KR/ACP | KS/ATm/DH/KR/ACP |
| 2 | KS/ATmm/DH/ER/KR/ACP | KS/ATmm/DH/ER/KR/ACP | KS/ATmm/DH/ER/KR/ACP |
| (qnmA2) | (1-1077) | (8-122) | |
| 3 | KS/ATm/DH/KR/ACP | KS/ATm/DH/KR/ACP | KS/ATm/DH/KR/ACP |
| (qnmA3) | (1-1078) | (8-123) | |
| 4 5 6 7 | KS/ATm/DH/KR/ACP KS/ATm/KR/ACP KS/ATm/KR/ACP KS/ATm/KR/ACP | - KS/ATm/KR/ACP KS/ATm/KR/ACP KS/ATm/KR/ACP | - KS/ATm/KR/ACP KS/ATm/KR/ACP KS/ATm/KR/ACP |
| (qnmA4) | (1-1080) | (8-135) | |
| 8 9 10 11 | KS/ATm/DH/ER/KR/ACP KS/AT/DH/ER/KR/ACP KS/ATm/DH/KR/ACP KS/ATm/KR/ACP | KS/ATm/DH/ER/KR/ACP KS/ATmm/DH/ER/KR/ACP KS/ATm/DH/ER/KR/ACP KS/ATm/KR/ACP | KS/ATm/DH/ER/KR/ACP KS/AT/DH/ER/KR/ACP KS/ATm/DH/ER/KR/ACP KS/ATm/KR/ACP |
| (qnmA5) | (1-1081) | (8-136) | |
| 12 | KS/ATm/KR/ACP | KS/ATmm/KR/ACP | KS/ATmm/KR/ACP |
| (8-137) | |||
| 13 | KS/ATm/DH/KR/ACP | KS/ATm/DH/KR/ACP | KS/ATm/DH/KR/ACP |
| 14 | KS/ATm/DH/KR/ACP | KS/ATm/DH/KR/ACP | KS/ATm/DH/KR/ACP |
| 15 | KS/ATm/DH/KR/ACP | KS/ATm/DH/KR/ACP | KS/ATm/DH/KR/ACP |
| 16 | KS/ATm/KR/ACP | KS/ATm/KR/ACP | KS/ATm/KR/ACP |
| 17 | KS/ATm/KR/ACP | KS/ATm/KR/ACP | KS/ATm/KR/ACP |
| (qnmA6) | (1-1091) | (8-146) | |
| 18 19 | KS/ATm/KR/ACP KS/DH/KR/ACP | KS/ATm/KR/ACP KS/ATmm/DH/KR/ACP | KS/ATm/KR/ACP KS/ATmm/DH/KR/ACP |
| (qnmA7) | (1-1092) | (8-147) | |
| 20 21 22 | KS/ATm/DH/KR/ACP KS/ATmm/KR/ACP KS/ATm/KR/ACP | KS/ATm/DH/KR/ACP KS/ATmm/KR/ACP KS/ATm/KR/ACP | KS/ATm/DH/KR/ACP KS/KR/ACP KS/ATm/KR/ACP |
| (qnmA8) | (1-1093) | (8-148) | |
| 23 24 | KS/ATm/KR/ACP KS/ATmm/DH/ER/KR/ACP | KS/ATm/KR/ACP KS/ATmm/DH/ER/KR/ACP | KS/AT/KR/ACP KS/ATmm/DH/ER/KR/ACP |
| (qnmA9) | (1-1094) | (8-149) | |
| 25 | KS/ATm/KR/ACP | KS/ATm/KR/ACP | KS/ATm |
| (8-150) | |||
| 26 27 | KS/ATmm/KR/ACP KS/ATmm/KR/ACP | KS/ATmm/KR/ACP KS/ATmm/KR/ACP | KS/ATmm/KR/ACP KS/ATmm/KR/ACP |
| (qnmA10) | (1-1095) | (8-151) | |
| 28 29 | KS/ATm/DH/KR/ACP KS/ATmm/DH/KR/ACP | KS/ATm/DH/KR/ACP KS/ATmm/DH/KR/ACP | KS/ATm/DH/KR/ACP KS/ATmm/DH/KR/ACP |
| (qnmA11) | (1-1096) | (8-152) | |
| 30 | KS/ATmm/KR/ACP | KS/ATmm/KR/ACP | KS/ATmm/KR/ACP |
| (qnmA12) | (1-1097) | (8-153) | |
| 31 32 | KS/ATmm/KR/ACP KS/ATm/KR/ACP | KS/ATmm/KR/ACP KS/ATm/KR/ACP | KS/ATmm/KR/ACP KS/ATm/KR/ACP |
| (qnmA13) | (1-1098) | (8-154) | |
| 33 | KS/ATm/KR/ACP/Te | KS/ATm/ACP/Te | KS/ATm/ACP/Te |
| Gene Cluster | ORF | Domain Organization | Predicted Product |
|---|---|---|---|
| t1pks-2 * | 8-118 | CoL/ACP-KS/ATm/DH/KR/ACP-KS/ATmm/DH/ER/KR/ACP | quinolidomicin congener |
| 8-122 | KS/ATm/DH/KR/ACP | ||
| 8-123 | KS/ATm/KR/ACP-KS/ATm/KR/ACP-KS/ATm/KR/ACP | ||
| 8-135 | KS/ATm/DH/ER/KR/ACP-KS/AT/DH/ER/KR/ACP- | ||
| KS/ATm/DH/ER/KR/ACP-KS/ATm/KR/ACP | |||
| 8-136 | KS/ATmm/KR/ACP | ||
| 8-137 | KS/ATm/DH/KR/ACP-KS/ATm/DH/KR/ACP-KS/ATm/DH/KR/ACP- | ||
| KS/ATm/KR/ACP-KS/ATm/KR/ACP | |||
| 8-146 8-147 8-148 8-149 8-150 8-151 8-152 8-153 8-154 | KS/ATm/KR/ACP-KS/ATmm/DH/KR/ACP KS/ATm/DH/KR/ACP-KS/KR/ACP-KS/ATm/KR/ACP KS/AT/KR/ACP-KS/ATmm/DH/ER/KR/ACP KS/ATm KS/ATmm/KR/ACP-KS/ATmm/KR/ACP KS/ATm/DH/KR/ACP-KS/ATmm/DH/KR/ACP KS/ATmm/KR/ACP KS/ATmm/KR/ACP-KS/ATm/KR/ACP KS/ATm/ACP-Te | ||
| t1pks-5 | 17-167 | CoL/ACP-KS/ATmm/DH/KR/ACP-KS/ATmm/ACP-KS/ATmm/KR/ACP | 16-demethylrifamycins |
| KS/ATmm/DH/KR/ACP-KS/ATmm/DH/KR/ACP-KS/ATmm/DH/KR/ACP | |||
| 17-166 | KS/ATmm/DH/KR/ACP | ||
| KS/ATmm/DH/KR/ACP | |||
| 17-165 | KS/ATm/DH/KR/ACP-KS/ATm/DH/KR/ACP | ||
| 17-164 | |||
| 17-163 | |||
| t2pks-2 | 8-66 8-67 8-68 | KSα KSβ (CLF) ACP | kosinostatin |
| t2pks-3 | 15-39 15-40 15-41 | KSα KSβ (CLF) ACP | aromatic polyketide |
| t3pks-1 * | 13-182 | KS | alkyl-O-dihydrogeranyl-methoxyhydroquinone |
| t3pks-2 | 9-577 | KS | polyketide with guanidinotide moiety |
| nrps-6P | 8-1 P 8-2 8-14 | …T C/Acys/MT/T Adhb | pyochelin |
| nrps-7 | 9-387 | Aglu/T-TD | glu with β-lactone |
| nrps-8 | 16-60 16-59 16-58 | T Aval C/Apro/T-TD | dipeptide (val-pro) |
| nrps-9 | 19-118 19-110 | A/T C/A/T | dipeptide (x-x) |
| nrps-10 | 20-72 20-83 | A A/T/E | dipeptide (x-x) |
| pks/nrps-2 * | 24-73 24-74 24-75 24-76 24-77 24-78 | C/A/T KS ACP C/Aval/T KS/ATm/ACP Aser | tripeptide with polyketide moiety (ser-x-val-pk) |
| pks/nrps-3 * | 17-59 17-56 17-55 17-52 17-51 | A/T-KS/DH A/T-C/T KS/ATm/KR/DH/ACP C/Aasn/T C/Aser/T-Te | pentapeptide with polyketide moiety (x-pk-x-y-pk-asn-ser) |
| pks/nrps-5 | 21-43 21-44 21-45 21-46 21-47 21-48 21-50 21-51 21-53 21-62 | C/Aasn/T-C/A/T C/Aser/T KS/ATm/MT/KR/ACP C/A/T CoL/T-C/Aser/T-C T-C C/Ab-ala/T-C/Acys/T-C A/T C A/T | methyltallysomycin |
| pks/nrps-6 | 26-56 | A/T-KS/ATm/ACP-C/A/T-C | dipeptide with polyketide moiety (x-pk-x) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Komaki, H.; Ichikawa, N.; Hosoyama, A.; Hamada, M.; Igarashi, Y. In Silico Analysis of PKS and NRPS Gene Clusters in Arisostatin- and Kosinostatin-Producers and Description of Micromonospora okii sp. nov. Antibiotics 2021, 10, 1447. https://doi.org/10.3390/antibiotics10121447
Komaki H, Ichikawa N, Hosoyama A, Hamada M, Igarashi Y. In Silico Analysis of PKS and NRPS Gene Clusters in Arisostatin- and Kosinostatin-Producers and Description of Micromonospora okii sp. nov. Antibiotics. 2021; 10(12):1447. https://doi.org/10.3390/antibiotics10121447
Chicago/Turabian StyleKomaki, Hisayuki, Natsuko Ichikawa, Akira Hosoyama, Moriyuki Hamada, and Yasuhiro Igarashi. 2021. "In Silico Analysis of PKS and NRPS Gene Clusters in Arisostatin- and Kosinostatin-Producers and Description of Micromonospora okii sp. nov." Antibiotics 10, no. 12: 1447. https://doi.org/10.3390/antibiotics10121447
APA StyleKomaki, H., Ichikawa, N., Hosoyama, A., Hamada, M., & Igarashi, Y. (2021). In Silico Analysis of PKS and NRPS Gene Clusters in Arisostatin- and Kosinostatin-Producers and Description of Micromonospora okii sp. nov. Antibiotics, 10(12), 1447. https://doi.org/10.3390/antibiotics10121447





