Co-Occurrence of blaOXA-23 in the Chromosome and Plasmid: Increased Fitness in Carbapenem-Resistant Acinetobacter baumannii
Abstract
:1. Introduction
2. Results
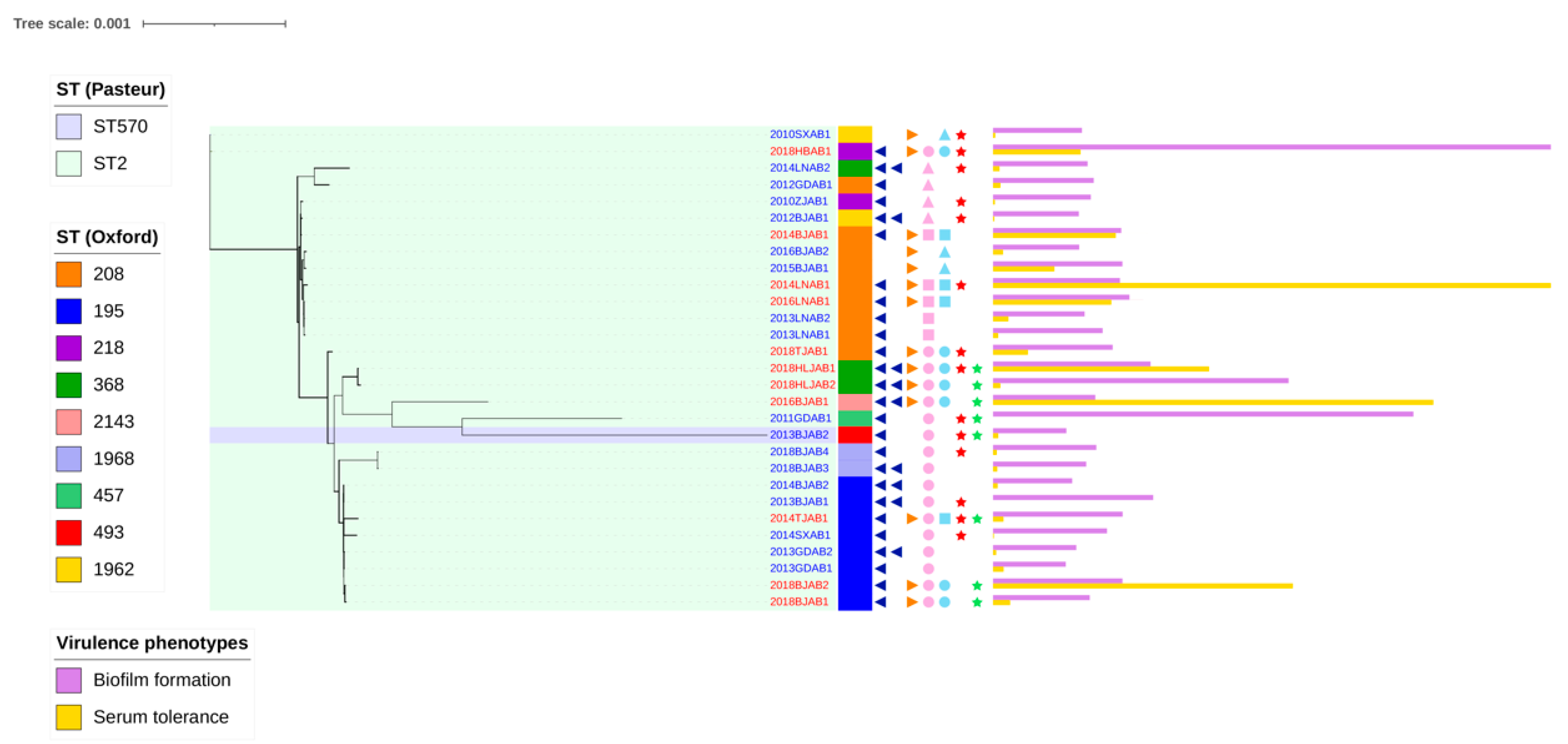
2.1. Prevalence of the Co-Occurrence of blaoxa-23 on Chromosomes and Plasmids
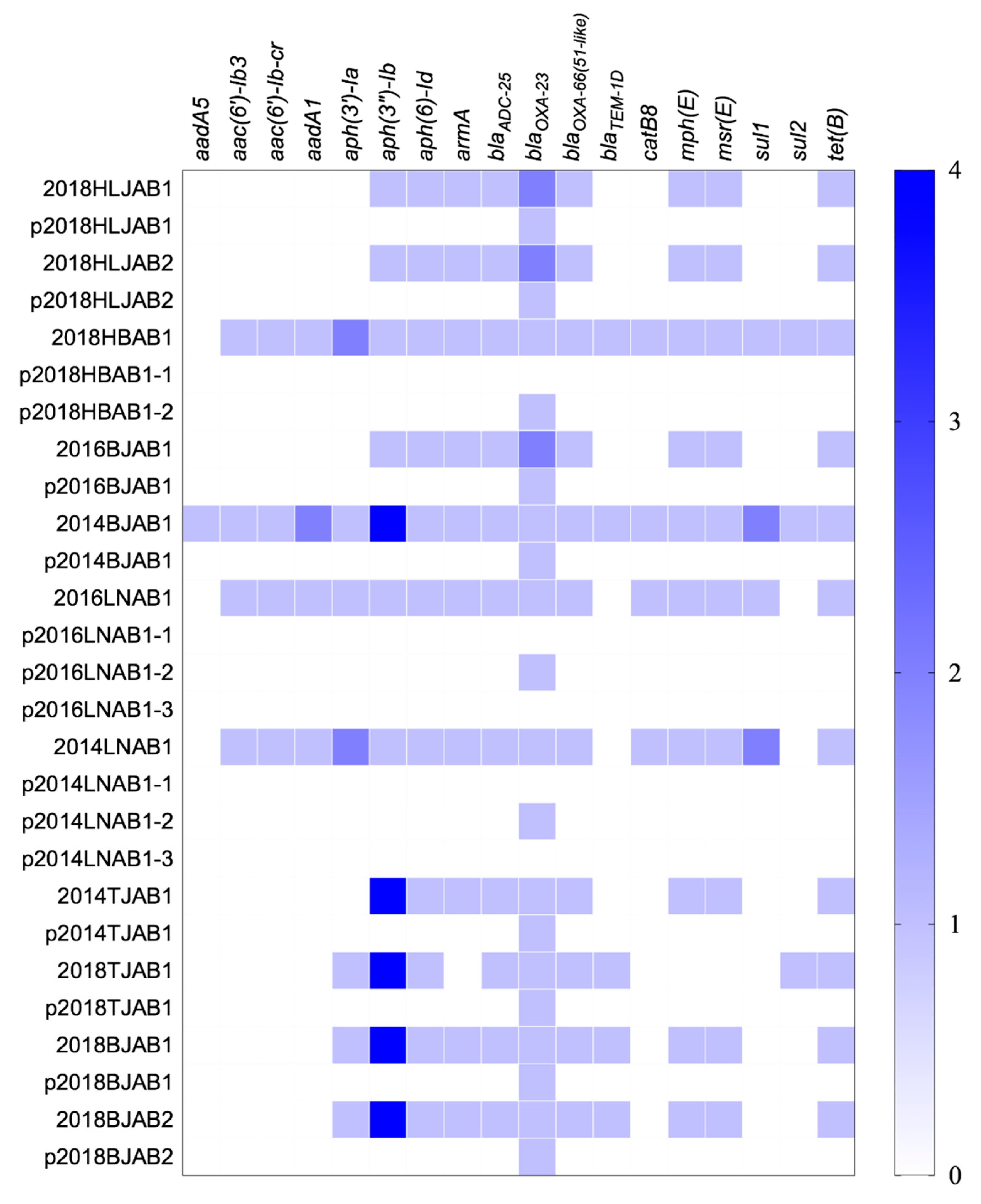
2.2. Resistance Gene Profiles and Correlation between Antibiotic Resistance and blaOXA-23 Location
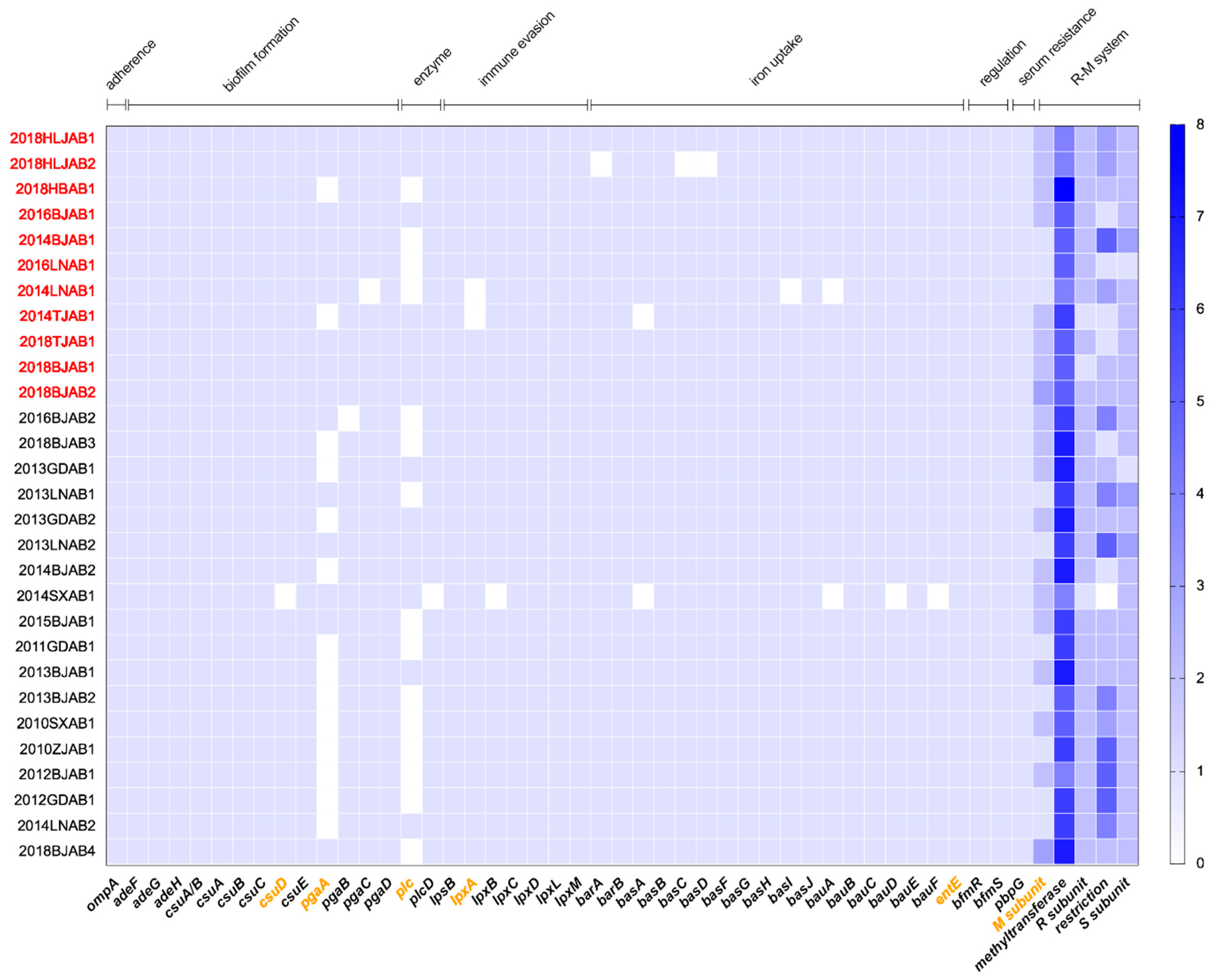
2.3. Comparative Analysis of Virulence-Related Genes and Accessory Genome in CO and SI Isolates
2.4. Mechanism of blaOXA-23 Multiplication
2.5. Plasmid Characteristics of CO Isolates
3. Discussion
4. Materials and Methods
4.1. Bacteria Isolates
4.2. Antimicrobial Susceptibility Testing
4.3. Genome Sequencing and Analyses
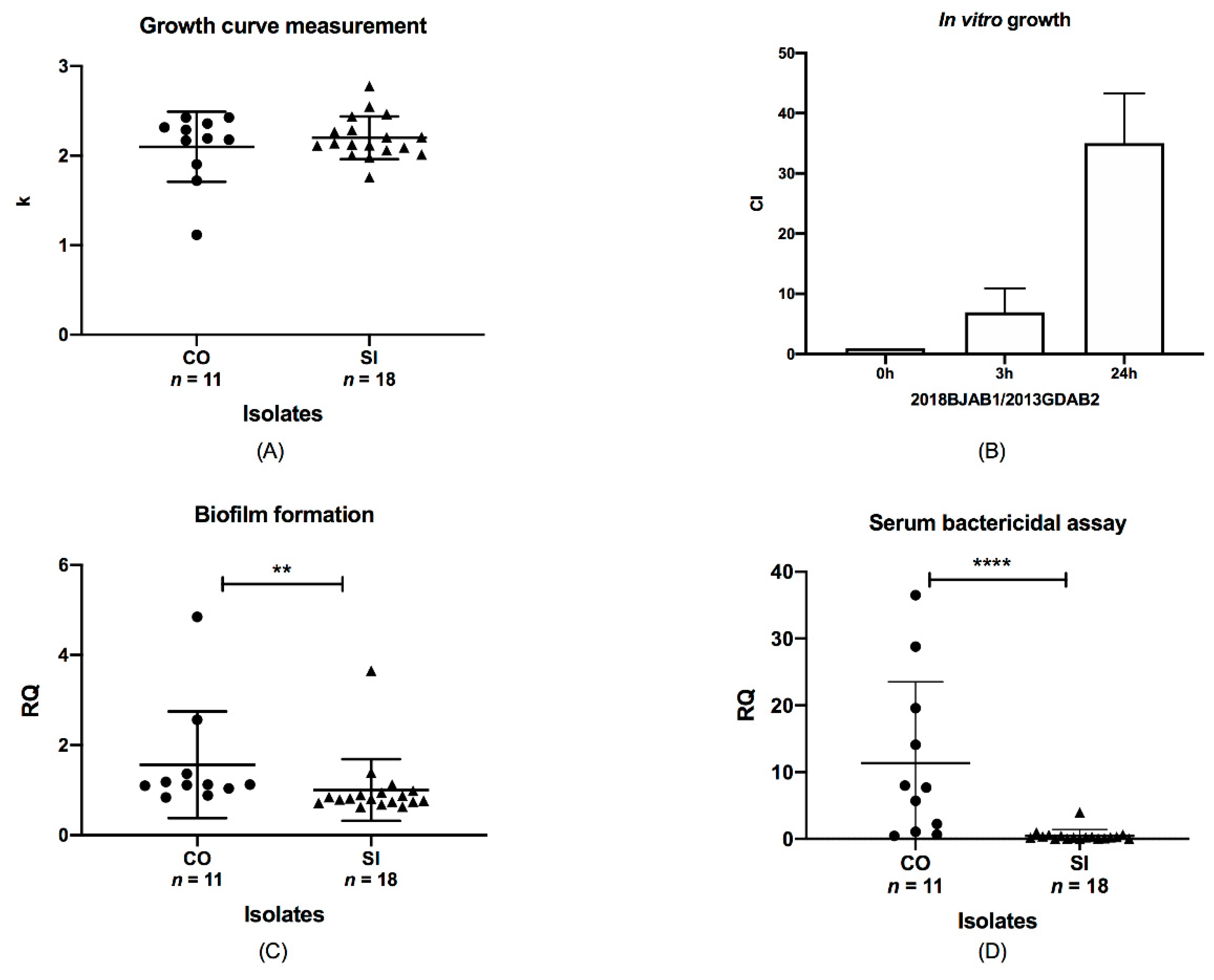
4.4. Growth Curve Assays
4.5. In Vitro Competition Assay
4.6. Biofilm Formation
4.7. Serum Bactericidal Assay
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hu, F.; Guo, Y.; Yang, Y.; Zheng, Y.; Wu, S.; Jiang, X.; Zhu, D.; Wang, F. Resistance reported from China antimicrobial surveillance network (CHINET) in 2018. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 2275–2281. [Google Scholar] [CrossRef] [PubMed]
- Hamidian, M.; Nigro, S.J. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microbiol. Genom. 2019, 5, e000306. [Google Scholar] [CrossRef]
- Codjoe, F.S.; Donkor, E.S. Carbapenem resistance: A review. Med. Sci. 2018, 6, 1. [Google Scholar] [CrossRef] [Green Version]
- Evans, B.A.; Amyes, S.G. OXA beta-lactamases. Clin. Microbiol. Rev. 2014, 27, 241–263. [Google Scholar] [CrossRef] [Green Version]
- Pagano, M.; Martins, A.F.; Barth, A.L. Mobile genetic elements related to carbapenem resistance in Acinetobacter baumannii. Braz. J. Microbiol. 2016, 47, 785–792. [Google Scholar] [CrossRef] [Green Version]
- Nigro, S.J.; Hall, R.M. Structure and context of Acinetobacter transposons carrying the oxa23 carbapenemase gene. J. Antimicrob. Chemother. 2016, 71, 1135–1147. [Google Scholar] [CrossRef] [Green Version]
- Cury, J.; Oliveira, P.H.; de la Cruz, F.; Rocha, E.P.C. Host range and genetic plasticity explain the coexistence of integrative and extrachromosomal mobile genetic elements. Mol. Biol. Evol. 2018, 35, 2230–2239. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Li, X.P.; Fang, L.X.; Sun, R.Y.; He, Y.Z.; Lin, J.; Liao, X.P.; Feng, Y.; Liu, Y.H. Co-occurrence of mcr-1 in the chromosome and on an IncHI2 plasmid: Persistence of colistin resistance in Escherichia coli. Int. J. Antimicrob. Agents 2018, 51, 842–847. [Google Scholar] [CrossRef] [PubMed]
- Ingti, B.; Krishnatreya, D.B.; Maurya, A.P.; Dhar, D.; Chakravarty, A.; Bhattacharjee, A. Role of inducers in detection of blaPDC-mediated oxyimino-cephalosporin resistance in Pseudomonas aeruginosa. Indian J. Med. Res. 2017, 145, 659–664. [Google Scholar]
- Sun, C.; Cui, M.; Zhang, S.; Wang, H.; Song, L.; Zhang, C.; Zhao, Q.; Liu, D.; Wang, Y.; Shen, J.; et al. Plasmid-mediated tigecycline-resistant gene tet(X4) in Escherichia coli from food-producing animals, China, 2008–2018. Emerg. Microbes Infect. 2019, 8, 1524–1527. [Google Scholar] [CrossRef] [Green Version]
- Hua, X.; Shu, J.; Ruan, Z.; Yu, Y.; Feng, Y. Multiplication of blaOXA-23 is common in clinical Acinetobacter baumannii, but does not enhance carbapenem resistance. J. Antimicrob. Chemother. 2016, 71, 3381–3385. [Google Scholar] [CrossRef] [Green Version]
- Bernheim, A.; Sorek, R. The pan-immune system of bacteria: Antiviral defence as a community resource. Nat. Rev. Microbiol. 2020, 18, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Z.; Zhang, H.; Zhao, Y.; Zhang, Z.; Xiao, J. PADS Arsenal: A database of prokaryotic defense systems related genes. Nucleic Acids Res. 2020, 48, D590–D598. [Google Scholar] [CrossRef]
- Zhou, H.; Zhang, T.; Yu, D.; Pi, B.; Yang, Q.; Zhou, J.; Hu, S.; Yu, Y. Genomic analysis of the multidrug-resistant Acinetobacter baumannii strain MDR-ZJ06 widely spread in China. Antimicrob. Agents Chemother. 2011, 55, 4506–4512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.L.; Ji, S.J.; Ruan, Z.; Fu, Y.; Fu, Y.Q.; Wang, Y.F.; Yu, Y.S. Dissemination of blaOXA-23 in Acinetobacter spp. in China: Main roles of conjugative plasmid pAZJ221 and transposon Tn2009. Antimicrob. Agents Chemother. 2015, 59, 1998–2005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef] [Green Version]
- Nasiri, M.J.; Zamani, S.; Fardsanei, F.; Arshadi, M.; Bigverdi, R.; Hajikhani, B.; Goudarzi, H.; Tabarsi, P.; Dabiri, H.; Feizabadi, M.M. Prevalence and mechanisms of carbapenem resistance in acinetobacter baumannii: A comprehensive systematic review of cross-sectional studies from Iran. Microb. Drug Resist. 2020, 26, 270–283. [Google Scholar] [CrossRef]
- Zhen, X.; Lundborg, C.S.; Sun, X.; Gu, S.; Dong, H. Clinical and economic burden of carbapenem-resistant infection or colonization caused by klebsiella pneumoniae, pseudomonas aeruginosa, acinetobacter baumannii: A multicenter study in China. Antibiotics 2020, 9, 514. [Google Scholar] [CrossRef]
- Durao, P.; Balbontin, R.; Gordo, I. Evolutionary mechanisms shaping the maintenance of antibiotic resistance. Trends Microbiol. 2018, 26, 677–691. [Google Scholar] [CrossRef] [Green Version]
- Shang, W.; Hu, Q.; Yuan, W.; Cheng, H.; Yang, J.; Hu, Z.; Yuan, J.; Zhang, X.; Peng, H.; Yang, Y.; et al. Comparative fitness and determinants for the characteristic drug resistance of ST239-MRSA-III-t030 and ST239-MRSA-III-t037 strains isolated in China. Microb. Drug Resist. 2016, 22, 185–192. [Google Scholar] [CrossRef]
- Gama, J.A.; Zilhao, R.; Dionisio, F. Impact of plasmid interactions with the chromosome and other plasmids on the spread of antibiotic resistance. Plasmid 2018, 99, 82–88. [Google Scholar] [CrossRef]
- Choi, A.H.; Slamti, L.; Avci, F.Y.; Pier, G.B.; Maira-Litran, T. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-beta-1-6-N-acetylglucosamine, which is critical for biofilm formation. J. Bacteriol. 2009, 191, 5953–5963. [Google Scholar] [CrossRef] [Green Version]
- Luke, N.R.; Sauberan, S.L.; Russo, T.A.; Beanan, J.M.; Olson, R.; Loehfelm, T.W.; Cox, A.D.; Michael, F.S.; Vinogradov, E.V.; Campagnari, A.A. Identification and characterization of a glycosyltransferase involved in Acinetobacter baumannii lipopolysaccharide core biosynthesis. Infect. Immun. 2010, 78, 2017–2023. [Google Scholar] [CrossRef] [Green Version]
- Ershova, A.S.; Rusinov, I.S.; Spirin, S.A.; Karyagina, A.S.; Alexeevski, A.V. Role of restriction-modification systems in prokaryotic evolution and ecology. Biochemistry 2015, 80, 1373–1386. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Hu, Q.; Wei, R.; Li, R.; Zhao, D.; Ge, M.; Yao, Q.; Yu, X. The XRE family transcriptional regulator SrtR in Streptococcus suis is involved in oxidant tolerance and virulence. Front. Cell. Infect. Microbiol. 2019, 8, 452. [Google Scholar] [CrossRef] [PubMed]
- Lerminiaux, N.A.; Cameron, A.D.S. Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 2019, 65, 34–44. [Google Scholar] [CrossRef]
- Hall, R.M. Integrons and gene cassettes: Hotspots of diversity in bacterial genomes. Ann. N. Y. Acad. Sci. 2012, 1267, 71–78. [Google Scholar] [CrossRef]
- Yoon, E.J.; Kim, J.O.; Yang, J.W.; Kim, H.S.; Lee, K.J.; Jeong, S.H.; Lee, H.; Lee, K. The blaOXA-23-associated transposons in the genome of Acinetobacter spp. represent an epidemiological situation of the species encountering carbapenems. J. Antimicrob. Chemother. 2017, 72, 2708–2714. [Google Scholar] [CrossRef] [Green Version]
- Mugnier, P.D.; Poirel, L.; Nordmann, P. Functional analysis of insertion sequence ISAba1, responsible for genomic plasticity of Acinetobacter baumannii. J. Bacteriol. 2009, 191, 2414–2418. [Google Scholar] [CrossRef] [Green Version]
- Corvec, S.; Poirel, L.; Naas, T.; Drugeon, H.; Nordmann, P. Genetics and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-23 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2007, 51, 1530–1533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, T.F.; Brauer, A.L.; Kirkham, C.; Johnson, A.; Koszelak-Rosenblum, M.; Malkowski, M.G. Role of the zinc uptake ABC transporter of Moraxella catarrhalis in persistence in the respiratory tract. Infect. Immun. 2013, 81, 3406–3413. [Google Scholar] [CrossRef] [Green Version]
- Livneh, Z.; Cohen-Fix, O.; Skaliter, R.; Elizur, T. Replication of damaged DNA and the molecular mechanism of ultraviolet light mutagenesis. Crit. Rev. Biochem. Mol. Biol. 1993, 28, 465–513. [Google Scholar] [CrossRef]
- Kim, D.H.; Jung, S.I.; Kwon, K.T.; Ko, K.S. Occurrence of diverse AbGRI1-type genomic islands in acinetobacter baumannii global clone 2 isolates from South Korea. Antimicrob. Agents Chemother. 2017, 61, e01972-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Octavia, S.; Xu, W.; Ng, O.T.; Marimuthu, K.; Venkatachalam, I.; Cheng, B.; Lin, R.T.P.; Teo, J.W.P. Identification of AbaR4 Acinetobacter baumannii resistance island in clinical isolates of blaOXA-23-positive Proteus mirabilis. J. Antimicrob. Chemother. 2020, 75, 521–525. [Google Scholar] [CrossRef] [PubMed]
- La, M.V.; Jureen, R.; Lin, R.T.; Teo, J.W. Unusual detection of an Acinetobacter class D carbapenemase gene, blaOXA-23, in a clinical Escherichia coli isolate. J. Clin. Microbiol. 2014, 52, 3822–3823. [Google Scholar] [CrossRef] [Green Version]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard, 9th ed.; M07-A9; CLSI: Wayne, PA, USA, 2012. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; 29th informational supplement M100-S29; CLSI: Wayne, PA, USA, 2019. [Google Scholar]
- Roberts, R.J.; Carneiro, M.O.; Schatz, M.C. The advantages of SMRT sequencing. Genome Biol. 2013, 14, 405. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef] [Green Version]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.F.; Petty, N.K.; Zakour, N.L.B.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grant, J.R.; Stothard, P. The CGView server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Wang, Q.; Wang, R.; Zhang, Y.; Wang, X.; Wang, H. Global regulator SoxR is a negative regulator of efflux pump gene expression and affects antibiotic resistance and fitness in Acinetobacter baumannii. Medicine 2017, 96, e7188. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.Y.; Tipton, K.A.; Farokhyfar, M.; Burd, E.M.; Weiss, D.S.; Rather, P.N. A high-frequency phenotypic switch links bacterial virulence and environmental survival in Acinetobacter baumannii. Nat. Microbiol. 2018, 3, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, J.Y.; Lee, H.; Choi, J.Y.; Kim, D.H.; Wi, Y.M.; Peck, K.R.; Ko, K.S. Microbiological features and clinical impact of the type VI secretion system (T6SS) in Acinetobacter baumannii isolates causing bacteremia. Virulence 2017, 8, 1378–1389. [Google Scholar] [CrossRef] [Green Version]

| Isolate ID | Year of Isolation | Location | Source | STs- Pasteur | STs-Oxford | Copy Number of blaOXA-23 in Chromosome | Copy Number of blaOXA-23 in Plasmid * | Plasmid Size (bp) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2018HLJAB1 | 2018 | China: Ha’erbin, Heilongjiang | pus | 2 | 368 | 2 | 1 | 78,023 | |||
| 2018HLJAB2 | 2018 | China: Ha’erbin, Heilongjiang | abdominal fluid | 2 | 368 | 2 | 1 | 78,030 | |||
| 2018HBAB1 | 2018 | China: Wuhan, Hubei | abdominal fluid | 2 | 218 | 1 | 1 | 110,968 | 78,023 (blaOXA-23) | ||
| 2016BJAB1 | 2016 | China: Beijing | sputum | 2 | 2143 | 2 | 1 | 77,943 | |||
| 2014BJAB1 | 2014 | China: Beijing | blood | 2 | 208 | 1 | 1 | 77,530 | |||
| 2016LNAB1 | 2016 | China: Shenyang, Liaoning | cerebrospinal fluid | 2 | 208 | 1 | 1 | 110,967 | 77,530 (blaOXA-23) | 11,205 | |
| 2014LNAB1 | 2014 | China: Shenyang, Liaoning | blood | 2 | 208 | 1 | 1 | 110,998 | 77,537 (blaOXA-23) | 11,194 | |
| 2014TJAB1 | 2014 | China: Tianjin | blood | 2 | 195 | 1 | 1 | 77,531 | |||
| 2018TJAB1 | 2018 | China: Tianjin | blood | 2 | 208 | 1 | 1 | 78,022 | |||
| 2018BJAB1 | 2018 | China: Beijing | sputum | 2 | 195 | 1 | 1 | 78,024 | |||
| 2018BJAB2 | 2018 | China: Beijing | bronchial | 2 | 195 | 1 | 1 | 78,023 | |||
| Isolates | Imipenem | Meropenem | Ceftazidime | Cefepime | TZP | Colistin | Doxycycline | Minocycline | Tigecycline | Amikacin | Ciprofloxacin | Levofloxacin | SXT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CO n = 11 | R% | 100 | 100 | 100 | 100 | 100 | 0 | 100 | 81.8 | 27.3 | 81.8 | 100 | 100 | 45.5 |
| S% | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 18.2 | 72.7 | 18.2 | 0 | 0 | 54.5 | |
| MIC50 | 64 | 64 | >256 | 128 | >256 | 0.5 | 32 | 16 | 4 | >256 | 64 | 16 | 2 | |
| MIC90 | 64 | 64 | >256 | >256 | >256 | 1 | 32 | 32 | 8 | >256 | 128 | 32 | 32 | |
| MIC range | 64 | 16~64 | 64~>256 | 64~>256 | >256 | 0.25~1 | 32~32 | 4~32 | 2~16 | 4~>256 | 64~128 | 8~32 | 0.25~64 | |
| SI n = 18 | R% | 100 | 100 | 94.4 | 100 | 100 | 0 | 94.4 | 50 | 33.3 | 94.4 | 100 | 94.4 | 88.9 |
| S% | 0 | 0 | 5.6 | 0 | 0 | 100 | 5.6 | 50 | 66.7 | 5.6 | 0 | 5.6 | 11.1 | |
| MIC50 | 64 | 32 | 128 | 64 | >256 | 1 | 32 | 8 | 4 | >256 | 64 | 16 | 32 | |
| MIC90 | 64 | 64 | >256 | 256 | >256 | 1 | 32 | 16 | 8 | >256 | 128 | 32 | 128 | |
| MIC range | 16~64 | 16~64 | 4~>256 | 32~>256 | >256 | 0.5~2 | 0.5~64 | 1~16 | 1~16 | 4~>256 | 16~128 | 2~64 | 0.25~128 | |
| Isolate | Tn Type of blaOXA-23 | ST (Oxford Scheme) | Tn Insertion Site in Chromosome | Tn Insertion Site in Plasmid | |||
|---|---|---|---|---|---|---|---|
| Chromosome | Plasmid | Gene Interrupted | Gene Function | Gene Interrupted | Gene Function | ||
| 2018HLJAB1 * | Tn2006/ Tn2006 | Tn2006 | 368 | the promoter region of znuA/ abaR | substrate-binding protein of zinc uptake ABC transporter/AbaR4 | hypothetical protein | unknown |
| 2018HLJAB2 * | Tn2006/ Tn2006 | Tn2006 | 368 | the promoter region of znuA/ abaR | substrate-binding protein of zinc uptake ABC transporter/AbaR4 | hypothetical protein | unknown |
| 2016BJAB1 * | Tn2006/ Tn2006 | Tn2006 | 2143 | the promoter region of znuA/ abaR | substrate-binding protein of zinc uptake ABC transporter/AbaR4 | hypothetical protein | unknown |
| 2018HBAB1 | Tn2006 | Tn2006 | 218 | FMN upstream | reductase | hypothetical protein | unknown |
| 2018TJAB1 | Tn2006 | Tn2006 | 208 | abaR | AbaR4 | hypothetical protein | unknown |
| 2018BJAB1 | Tn2006 | Tn2006 | 195 | abaR | AbaR4 | hypothetical protein | unknown |
| 2018BJAB2 | Tn2006 | Tn2006 | 195 | abaR | AbaR4 | hypothetical protein | unknown |
| 2014TJAB1 | Tn2006 | Tn2009 | 195 | abaR | AbaR25 | hypothetical protein | unknown |
| 2014BJAB1 | Tn2009 | Tn2009 | 208 | Phage related gene | DNA polymerase | hypothetical protein | unknown |
| 2016LNAB1 | Tn2009 | Tn2009 | 208 | Phage related gene | DNA polymerase | hypothetical protein | unknown |
| 2014LNAB1 | Tn2009 | Tn2009 | 208 | Phage related gene | DNA polymerase | hypothetical protein | unknown |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Li, H.; Zhang, J.; Wang, H. Co-Occurrence of blaOXA-23 in the Chromosome and Plasmid: Increased Fitness in Carbapenem-Resistant Acinetobacter baumannii. Antibiotics 2021, 10, 1196. https://doi.org/10.3390/antibiotics10101196
Wang Z, Li H, Zhang J, Wang H. Co-Occurrence of blaOXA-23 in the Chromosome and Plasmid: Increased Fitness in Carbapenem-Resistant Acinetobacter baumannii. Antibiotics. 2021; 10(10):1196. https://doi.org/10.3390/antibiotics10101196
Chicago/Turabian StyleWang, Zhiren, Henan Li, Jiangang Zhang, and Hui Wang. 2021. "Co-Occurrence of blaOXA-23 in the Chromosome and Plasmid: Increased Fitness in Carbapenem-Resistant Acinetobacter baumannii" Antibiotics 10, no. 10: 1196. https://doi.org/10.3390/antibiotics10101196
APA StyleWang, Z., Li, H., Zhang, J., & Wang, H. (2021). Co-Occurrence of blaOXA-23 in the Chromosome and Plasmid: Increased Fitness in Carbapenem-Resistant Acinetobacter baumannii. Antibiotics, 10(10), 1196. https://doi.org/10.3390/antibiotics10101196





